Mean activities
Top 10 and bottom 10 mean activities.
Mean activities sorted by motif Z-value.
| Motif name | Mean activity | Z-Value | Associated genes | Logo |
|---|---|---|---|---|
| ABF1 | 0.000 | 0.000 |
|
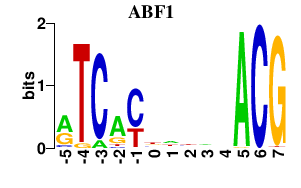
|
| ABF2 | 0.000 | 0.000 |
|
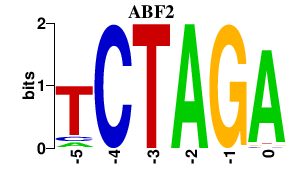
|
| ACE2 | 0.000 | 0.000 |
|
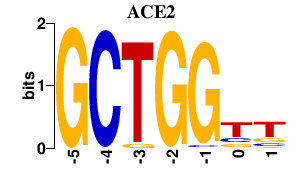
|
| ADR1 | 0.000 | 0.000 |
|
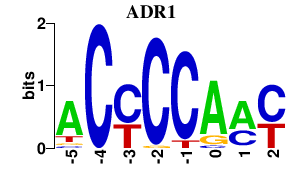
|
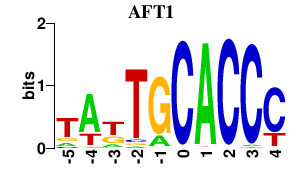
| AFT1 | -0.000 | -0.000 |
|
|
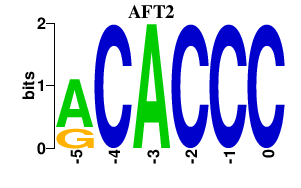
| AFT2 | -0.000 | -0.000 |
|
|
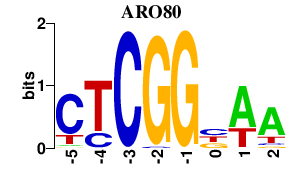
| ARO80 | 0.000 | 0.000 |
|
|
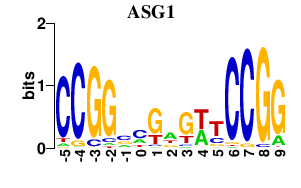
| ASG1 | 0.000 | 0.000 |
|
|
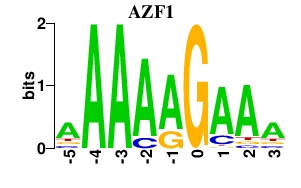
| AZF1 | 0.000 | 0.000 |
|
|
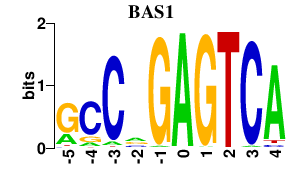
| BAS1 | 0.000 | 0.000 |
|
|
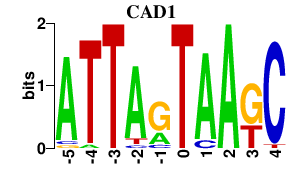
| CAD1 | -0.000 | -0.000 |
|
|
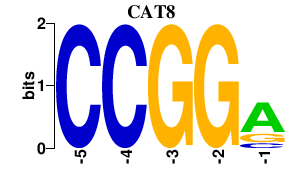
| CAT8 | 0.000 | 0.000 |
|
|
| CBF1 | 0.000 | 0.000 |
|
|
| CEP3 | 0.000 | 0.000 |
|
|
| CHA4 | 0.000 | 0.000 |
|
|
| CIN5 | -0.000 | -0.000 |
|
|
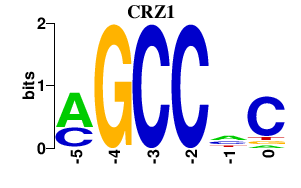
| CRZ1 | 0.000 | 0.000 |
|
|
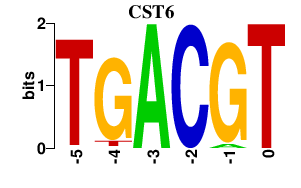
| CST6 | 0.000 | 0.000 |
|
|
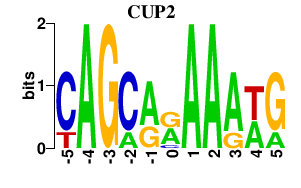
| CUP2 | -0.000 | -0.000 |
|
|
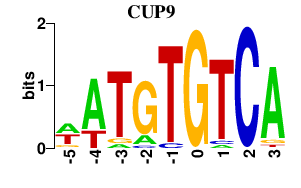
| CUP9 | -0.000 | -0.000 |
|
|
| DAL80 | 0.000 | 0.000 |
|

|
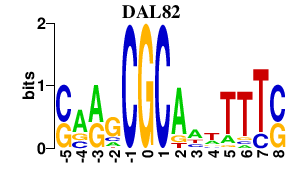
| DAL82 | -0.000 | -0.000 |
|
|
| DOT6 | 0.000 | 0.000 |
|
|
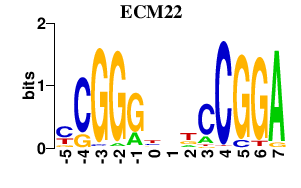
| ECM22 | 0.000 | 0.000 |
|
|
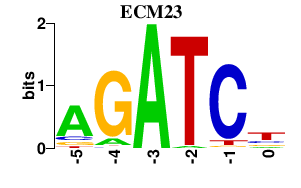
| ECM23 | 0.000 | 0.000 |
|
|
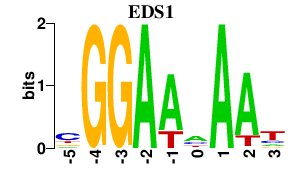
| EDS1 | 0.000 | 0.000 |
|
|
| FHL1 | -0.000 | -0.000 |
|
|
| FKH1 | -0.000 | -0.000 |
|
|
| FKH2 | -0.000 | -0.000 |
|
|
| FZF1 | -0.000 | -0.000 |
|
|
| GAL4 | 0.000 | 0.000 |
|
|
| GAL80 | 0.000 | 0.000 |
|
|
| GAT1 | 0.000 | 0.000 |
|
|
| GAT3 | 0.000 | 0.000 |
|
|
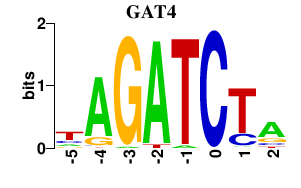
| GAT4 | 0.000 | 0.000 |
|
|
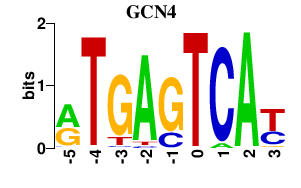
| GCN4 | 0.000 | 0.000 |
|
|
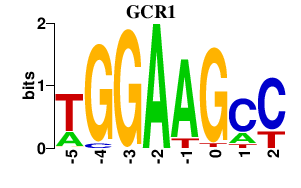
| GCR1 | -0.000 | -0.000 |
|
|
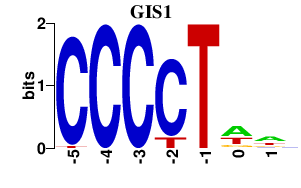
| GIS1 | 0.000 | 0.000 |
|
|
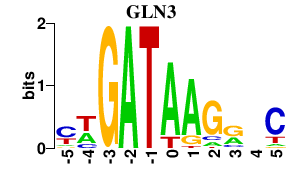
| GLN3 | 0.000 | 0.000 |
|
|
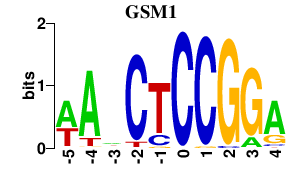
| GSM1 | 0.000 | 0.000 |
|
|
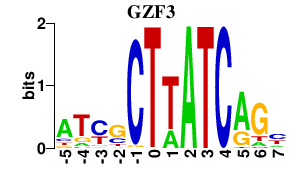
| GZF3 | -0.000 | -0.000 |
|
|
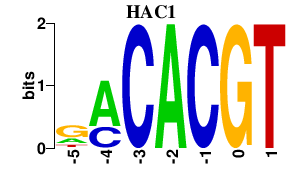
| HAC1 | 0.000 | 0.000 |
|
|
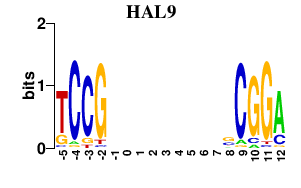
| HAL9 | 0.000 | 0.000 |
|
|
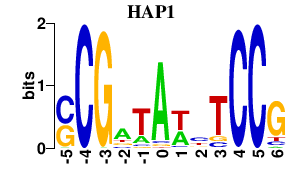
| HAP1 | 0.000 | 0.000 |
|
|
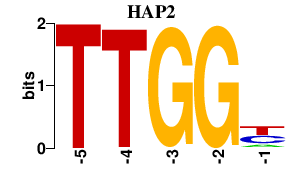
| HAP2 | 0.000 | 0.000 |
|
|
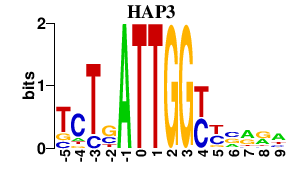
| HAP3 | 0.000 | 0.000 |
|
|
| HAP4 | 0.000 | 0.000 |
|
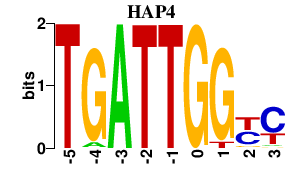
|
| HAP5 | 0.000 | 0.000 |
|
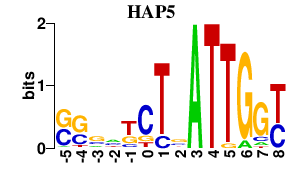
|
| HCM1 | -0.000 | -0.000 |
|
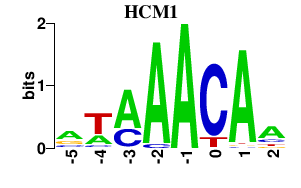
|
| HMRA2 | -0.000 | -0.000 |
|
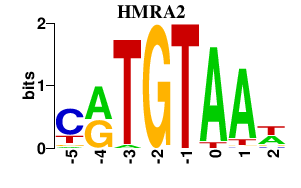
|
| HSF1 | 0.000 | 0.000 |
|
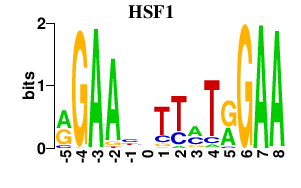
|
| INO2 | -0.000 | -0.000 |
|
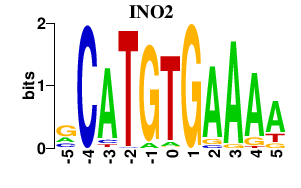
|
| INO4 | -0.000 | -0.000 |
|
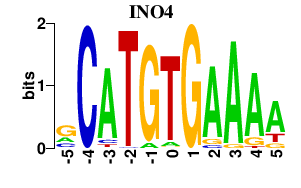
|
| LEU3 | 0.000 | 0.000 |
|
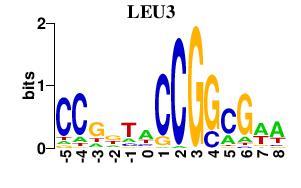
|
| LYS14 | -0.000 | -0.000 |
|
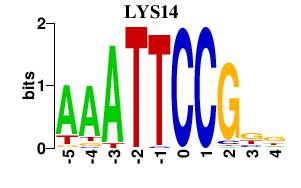
|
| MAC1 | 0.000 | 0.000 |
|
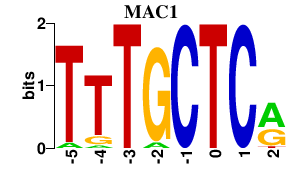
|
| MATALPHA2 | -0.000 | -0.000 |
|
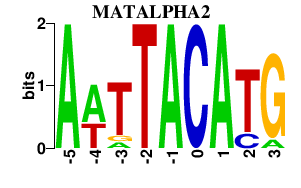
|
| MBP1 | -0.000 | -0.000 |
|
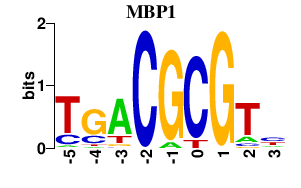
|
| MCM1 | -0.000 | -0.000 |
|
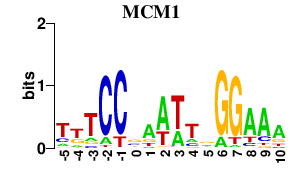
|
| MET31 | -0.000 | -0.000 |
|
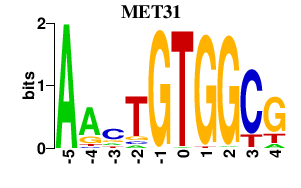
|
| MET32 | -0.000 | -0.000 |
|
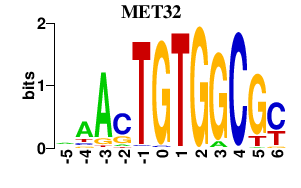
|
| MGA1 | 0.000 | 0.000 |
|
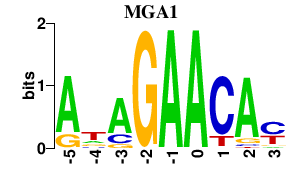
|
| MIG1 | 0.000 | 0.000 |
|
|
| MIG2 | -0.000 | -0.000 |
|
|
| MIG3 | -0.000 | -0.000 |
|
|
| MSN2 | 0.000 | 0.000 |
|
|
| MSN4 | 0.000 | 0.000 |
|
|
| NDT80 | -0.000 | -0.000 |
|
|
| NHP10 | 0.000 | 0.000 |
|
|
| NHP6A | 0.000 | 0.000 |
|
|
| NHP6B | 0.000 | 0.000 |
|
|
| NRG1 | -0.000 | -0.000 |
|
|
| OAF1 | 0.000 | 0.000 |
|
|
| ORC1 | -0.000 | -0.000 |
|
|
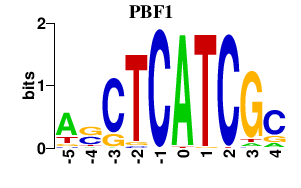
| PBF1 | 0.000 | 0.000 |
|
|
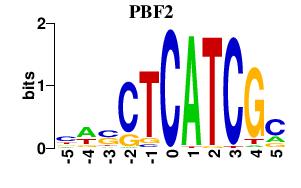
| PBF2 | 0.000 | 0.000 |
|
|
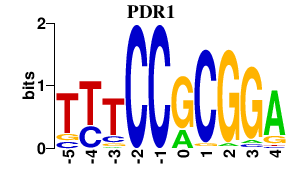
| PDR1 | 0.000 | 0.000 |
|
|
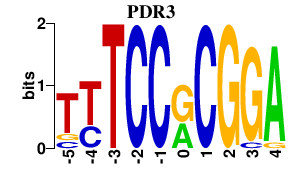
| PDR3 | 0.000 | 0.000 |
|
|
| PDR8 | 0.000 | 0.000 |
|
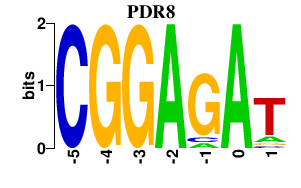
|
| PHD1 | -0.000 | -0.000 |
|
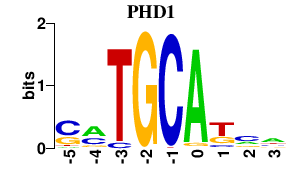
|
| PHO2 | 0.000 | 0.000 |
|
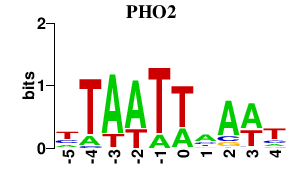
|
| PHO4 | 0.000 | 0.000 |
|
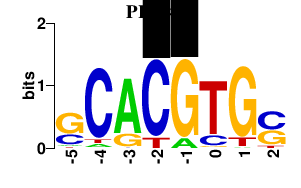
|
| PUT3 | 0.000 | 0.000 |
|
|
| RAP1 | 0.000 | 0.000 |
|
|
| RCS1 | 0.000 | 0.000 |
|
|
| RDR1 | 0.000 | 0.000 |
|
|
| RDS1 | 0.000 | 0.000 |
|
|
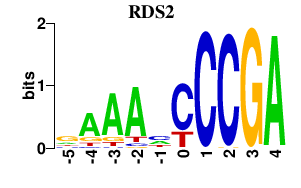
| RDS2 | 0.000 | 0.000 |
|
|
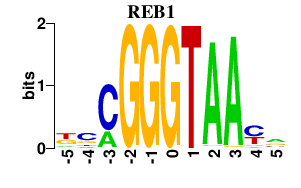
| REB1 | 0.000 | 0.000 |
|
|
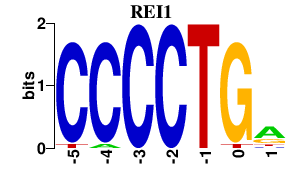
| REI1 | 0.000 | 0.000 |
|
|
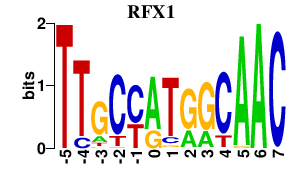
| RFX1 | -0.000 | -0.000 |
|
|
| RGM1 | 0.000 | 0.000 |
|
|
| RGT1 | 0.000 | 0.000 |
|
|
| RIM101 | -0.000 | -0.000 |
|
|
| RLM1 | -0.000 | -0.000 |
|
|
| ROX1 | -0.000 | -0.000 |
|
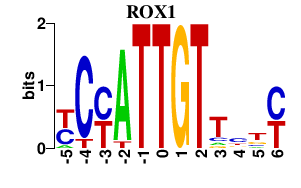
|
| RPH1 | 0.000 | 0.000 |
|
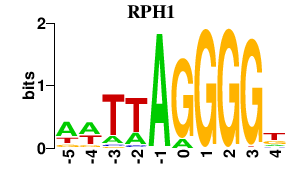
|
| RPN4 | 0.000 | 0.000 |
|
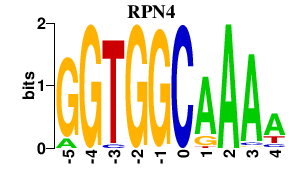
|
| RSC3 | 0.000 | 0.000 |
|
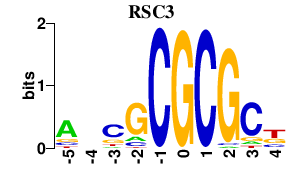
|
| RSC30 | 0.000 | 0.000 |
|
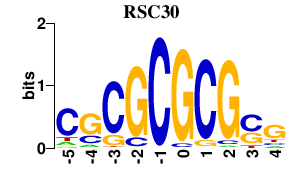
|
| RTG3 | 0.000 | 0.000 |
|
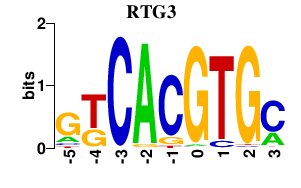
|
| SFL1 | -0.000 | -0.000 |
|
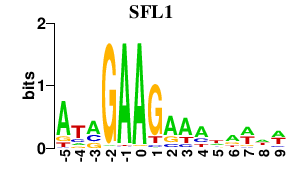
|
| SFP1 | 0.000 | 0.000 |
|
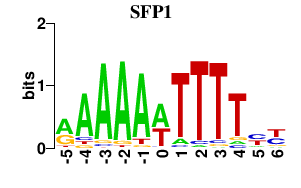
|
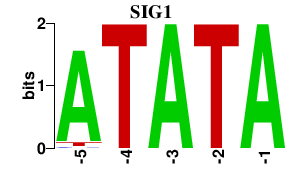
| SIG1 | 0.000 | 0.000 |
|
|
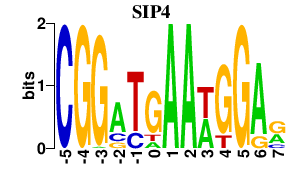
| SIP4 | 0.000 | 0.000 |
|
|
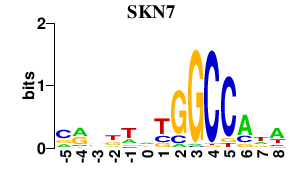
| SKN7 | 0.000 | 0.000 |
|
|
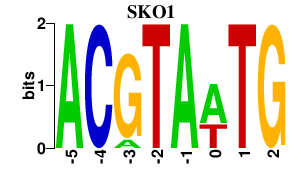
| SKO1 | -0.000 | -0.000 |
|
|
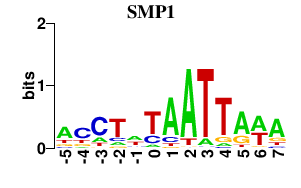
| SMP1 | -0.000 | -0.000 |
|
|
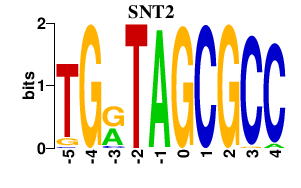
| SNT2 | -0.000 | -0.000 |
|
|
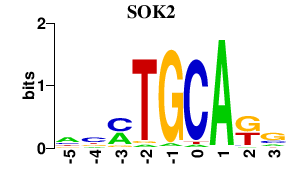
| SOK2 | -0.000 | -0.000 |
|
|
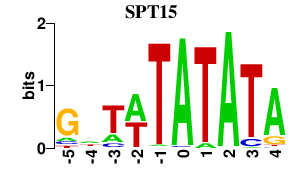
| SPT15 | 0.000 | 0.000 |
|
|
| SRD1 | 0.000 | 0.000 |
|
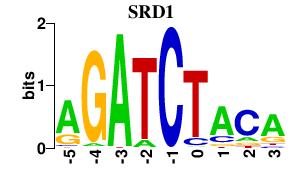
|
| STB1 | -0.000 | -0.000 |
|
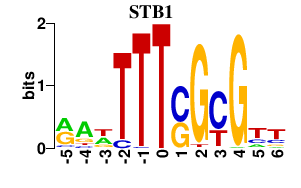
|
| STB3 | 0.000 | 0.000 |
|
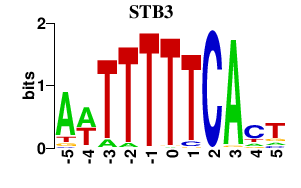
|
| STB4 | 0.000 | 0.000 |
|
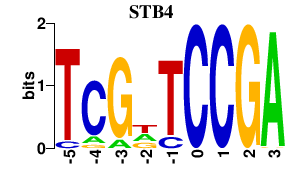
|
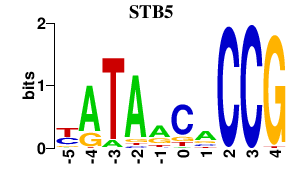
| STB5 | 0.000 | 0.000 |
|
|
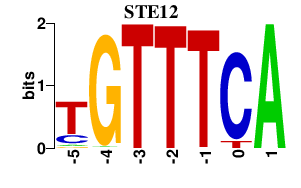
| STE12 | -0.000 | -0.000 |
|
|
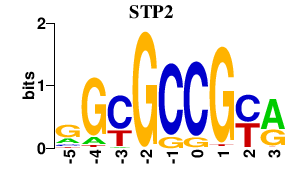
| STP2 | 0.000 | 0.000 |
|
|
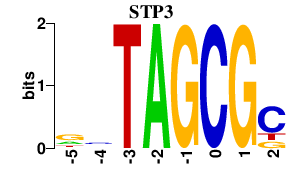
| STP3 | -0.000 | -0.000 |
|
|
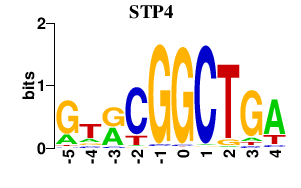
| STP4 | 0.000 | 0.000 |
|
|
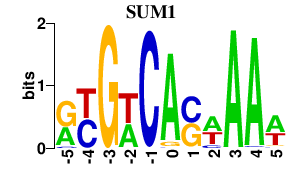
| SUM1 | -0.000 | -0.000 |
|
|
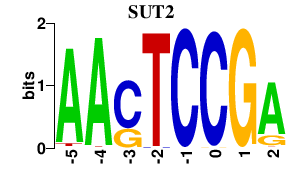
| SUT2 | 0.000 | 0.000 |
|
|
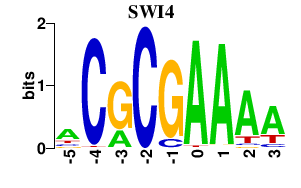
| SWI4 | -0.000 | -0.000 |
|
|
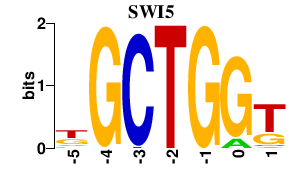
| SWI5 | -0.000 | -0.000 |
|
|
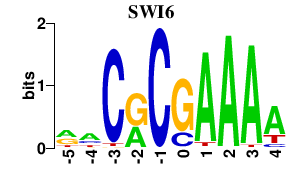
| SWI6 | -0.000 | -0.000 |
|
|
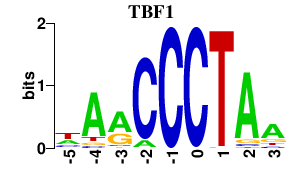
| TBF1 | -0.000 | -0.000 |
|
|
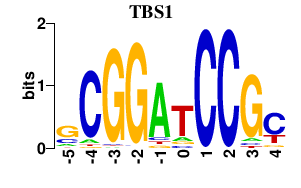
| TBS1 | 0.000 | 0.000 |
|
|
| TEA1 | -0.000 | -0.000 |
|

|
| TEC1 | -0.000 | -0.000 |
|

|
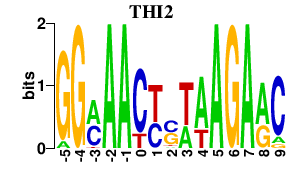
| THI2 | 0.000 | 0.000 |
|
|
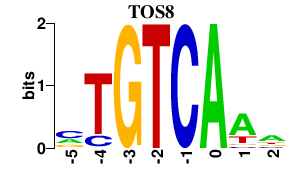
| TOS8 | -0.000 | -0.000 |
|
|
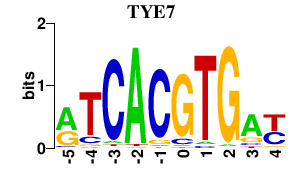
| TYE7 | 0.000 | 0.000 |
|
|
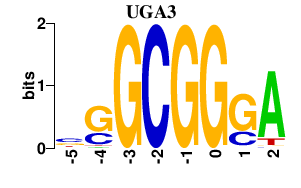
| UGA3 | 0.000 | 0.000 |
|
|
| UME6 | -0.000 | -0.000 |
|
|
| UPC2 | -0.000 | -0.000 |
|
|
| USV1 | 0.000 | 0.000 |
|
|
| XBP1 | 0.000 | 0.000 |
|
|
| YAP1 | -0.000 | -0.000 |
|
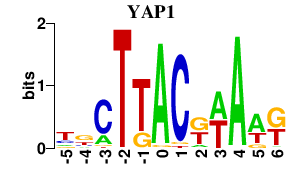
|
| YAP3 | 0.000 | 0.000 |
|
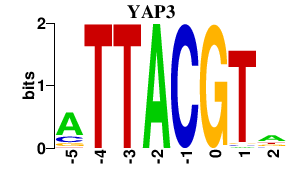
|
| YAP6 | -0.000 | -0.000 |
|
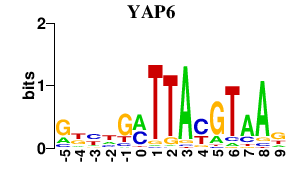
|
| YAP7 | -0.000 | -0.000 |
|
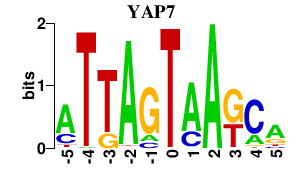
|
| YBL054W | 0.000 | 0.000 |
|
|
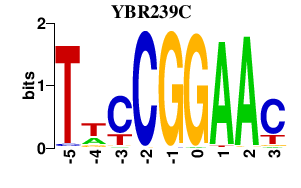
| YBR239C | 0.000 | 0.000 |
|
|
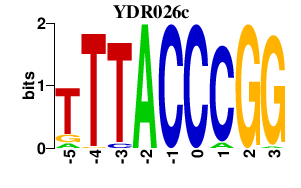
| YDR026c | 0.000 | 0.000 |
|
|
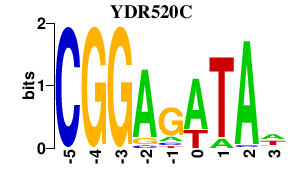
| YDR520C | -0.000 | -0.000 |
|
|
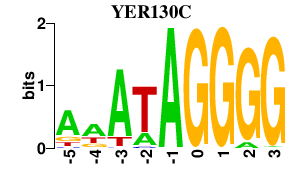
| YER130C | 0.000 | 0.000 |
|
|
| YER184C | 0.000 | 0.000 |
|
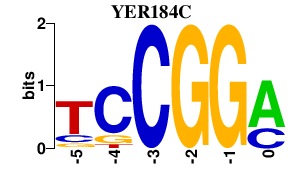
|
| YGR067C | -0.000 | -0.000 |
|
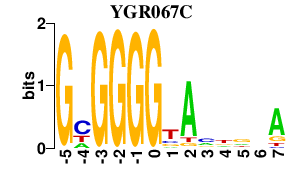
|
| YJL103C | 0.000 | 0.000 |
|
|
| YKL222C | 0.000 | 0.000 |
|
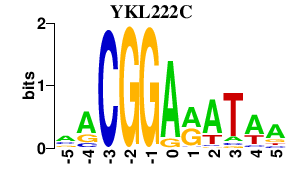
|
| YLL054C | 0.000 | 0.000 |
|
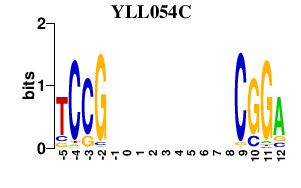
|
| YLR278C | 0.000 | 0.000 |
|
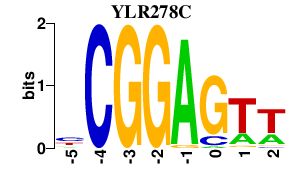
|
| YML081W | 0.000 | 0.000 |
|
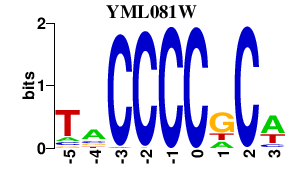
|
| YNR063W | 0.000 | 0.000 |
|
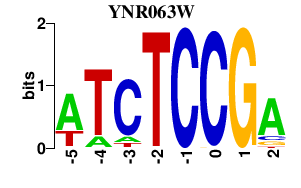
|
| YOX1 | -0.000 | -0.000 |
|
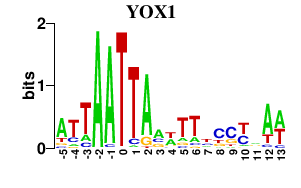
|
| YPL230W | 0.000 | 0.000 |
|
|
| YPR013C | 0.000 | 0.000 |
|
|
| YPR015C | 0.000 | 0.000 |
|
|
| YPR022C | -0.000 | -0.000 |
|
|
| YPR196W | -0.000 | -0.000 |
|
|
| YRM1 | 0.000 | 0.000 |
|
|
| YRR1 | 0.000 | 0.000 |
|
|
| ZMS1 | 0.000 | 0.000 |
|
|