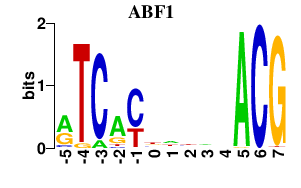
Results for ABF1
Z-value: 3.42
Transcription factors associated with ABF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ABF1
|
S000001595 | DNA binding protein with possible chromatin-reorganizing activity |
Activity-expression correlation:
Activity profile of ABF1 motif
Sorted Z-values of ABF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YPL250W-A | 22.71 |
Identified by fungal homology and RT-PCR |
||
| YKL218C | 17.30 |
SRY1
|
3-hydroxyaspartate dehydratase, deaminates L-threo-3-hydroxyaspartate to form oxaloacetate and ammonia; required for survival in the presence of hydroxyaspartate |
|
| YNL112W | 16.97 |
DBP2
|
Essential ATP-dependent RNA helicase of the DEAD-box protein family, involved in nonsense-mediated mRNA decay and rRNA processing |
|
| YBL039C | 16.85 |
URA7
|
Major CTP synthase isozyme (see also URA8), catalyzes the ATP-dependent transfer of the amide nitrogen from glutamine to UTP, forming CTP, the final step in de novo biosynthesis of pyrimidines; involved in phospholipid biosynthesis |
|
| YIL053W | 16.15 |
RHR2
|
Constitutively expressed isoform of DL-glycerol-3-phosphatase; involved in glycerol biosynthesis, induced in response to both anaerobic and, along with the Hor2p/Gpp2p isoform, osmotic stress |
|
| YKR038C | 15.54 |
KAE1
|
Putative glycoprotease proposed to be in transcription as a component of the EKC protein complex with Bud32p, Cgi121p, Pcc1p, and Gon7p; also identified as a component of the KEOPS protein complex |
|
| YJR145C | 15.04 |
RPS4A
|
Protein component of the small (40S) ribosomal subunit; mutation affects 20S pre-rRNA processing; identical to Rps4Bp and has similarity to rat S4 ribosomal protein |
|
| YLL045C | 14.82 |
RPL8B
|
Ribosomal protein L4 of the large (60S) ribosomal subunit, nearly identical to Rpl8Ap and has similarity to rat L7a ribosomal protein; mutation results in decreased amounts of free 60S subunits |
|
| YPR010C | 14.51 |
RPA135
|
RNA polymerase I subunit A135 |
|
| YHL028W | 13.69 |
WSC4
|
ER membrane protein involved in the translocation of soluble secretory proteins and insertion of membrane proteins into the ER membrane; may also have a role in the stress response but has only partial functional overlap with WSC1-3 |
|
| YOL077C | 13.44 |
BRX1
|
Nucleolar protein, constituent of 66S pre-ribosomal particles; depletion leads to defects in rRNA processing and a block in the assembly of large ribosomal subunits; possesses a sigma(70)-like RNA-binding motif |
|
| YPR119W | 13.29 |
CLB2
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YJR094W-A | 12.89 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YGL123C-A | 11.92 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene RPS2/YGL123W |
||
| YGL123W | 11.84 |
RPS2
|
Protein component of the small (40S) subunit, essential for control of translational accuracy; has similarity to E. coli S5 and rat S2 ribosomal proteins |
|
| YMR217W | 11.80 |
GUA1
|
GMP synthase, an enzyme that catalyzes the second step in the biosynthesis of GMP from inosine 5'-phosphate (IMP); transcription is not subject to regulation by guanine but is negatively regulated by nutrient starvation |
|
| YLR314C | 11.74 |
CDC3
|
Component of the septin ring of the mother-bud neck that is required for cytokinesis; septins recruit proteins to the neck and can act as a barrier to diffusion at the membrane, and they comprise the 10nm filaments seen with EM |
|
| YLR150W | 11.53 |
STM1
|
Protein that binds G4 quadruplex and purine motif triplex nucleic acid; acts with Cdc13p to maintain telomere structure; interacts with ribosomes and subtelomeric Y' DNA; multicopy suppressor of tom1 and pop2 mutations |
|
| YOL101C | 11.45 |
IZH4
|
Membrane protein involved in zinc metabolism, member of the four-protein IZH family, expression induced by fatty acids and altered zinc levels; deletion reduces sensitivity to excess zinc; possible role in sterol metabolism |
|
| YLL044W | 11.42 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; transcription of both YLL044W and the overlapping gene RPL8B is reduced in the gcr1 null mutant |
||
| YKL182W | 11.02 |
FAS1
|
Beta subunit of fatty acid synthetase, which catalyzes the synthesis of long-chain saturated fatty acids; contains acetyltransacylase, dehydratase, enoyl reductase, malonyl transacylase, and palmitoyl transacylase activities |
|
| YGR108W | 10.83 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YGL225W | 10.80 |
VRG4
|
Golgi GDP-mannose transporter; regulates Golgi function and glycosylation in Golgi |
|
| YGR052W | 10.30 |
FMP48
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YMR296C | 10.12 |
LCB1
|
Component of serine palmitoyltransferase, responsible along with Lcb2p for the first committed step in sphingolipid synthesis, which is the condensation of serine with palmitoyl-CoA to form 3-ketosphinganine |
|
| YER036C | 10.07 |
ARB1
|
ATPase of the ATP-binding cassette (ABC) family involved in 40S and 60S ribosome biogenesis, has similarity to Gcn20p; shuttles from nucleus to cytoplasm, physically interacts with Tif6p, Lsg1p |
|
| YNL114C | 9.87 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps the verified ORF RPC19/YNL113W, an RNA polymerase subunit |
||
| YNL118C | 9.78 |
DCP2
|
Catalytic subunit of the Dcp1p-Dcp2p decapping enzyme complex, which removes the 5' cap structure from mRNAs prior to their degradation; member of the Nudix hydrolase family |
|
| YBL063W | 9.78 |
KIP1
|
Kinesin-related motor protein required for mitotic spindle assembly and chromosome segregation; functionally redundant with Cin8p |
|
| YGR185C | 9.73 |
TYS1
|
Cytoplasmic tyrosyl-tRNA synthetase, class I aminoacyl-tRNA synthetase that aminoacylates tRNA(Tyr), required for cytoplasmic protein synthesis, interacts with positions 34 and 35 of the anticodon of tRNATyr |
|
| YLR448W | 9.63 |
RPL6B
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl6Ap and to rat L6 ribosomal protein; binds to 5.8S rRNA |
|
| YOR101W | 9.60 |
RAS1
|
GTPase involved in G-protein signaling in the adenylate cyclase activating pathway, plays a role in cell proliferation; localized to the plasma membrane; homolog of mammalian RAS proto-oncogenes |
|
| YPL062W | 9.53 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YPL062W is not an essential gene; homozygous diploid mutant shows a decrease in glycogen accumulation |
||
| YOR028C | 9.47 |
CIN5
|
Basic leucine zipper transcriptional factor of the yAP-1 family that mediates pleiotropic drug resistance and salt tolerance; localizes constitutively to the nucleus |
|
| YDR002W | 9.47 |
YRB1
|
Ran GTPase binding protein; involved in nuclear protein import and RNA export, ubiquitin-mediated protein degradation during the cell cycle; shuttles between the nucleus and cytoplasm; is essential; homolog of human RanBP1 |
|
| YHR007C | 9.46 |
ERG11
|
Lanosterol 14-alpha-demethylase, catalyzes the C-14 demethylation of lanosterol to form 4,4''-dimethyl cholesta-8,14,24-triene-3-beta-ol in the ergosterol biosynthesis pathway; member of the cytochrome P450 family |
|
| YHR010W | 9.39 |
RPL27A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl27Bp and has similarity to rat L27 ribosomal protein |
|
| YOR029W | 9.27 |
Dubious open reading frame unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YOR025W | 9.13 |
HST3
|
Member of the Sir2 family of NAD(+)-dependent protein deacetylases; involved along with Hst4p in telomeric silencing, cell cycle progression, radiation resistance, genomic stability and short-chain fatty acid metabolism |
|
| YOR342C | 9.06 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and the nucleus |
||
| YLR029C | 8.92 |
RPL15A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl15Bp and has similarity to rat L15 ribosomal protein; binds to 5.8 S rRNA |
|
| YJR063W | 8.85 |
RPA12
|
RNA polymerase I subunit A12.2; contains two zinc binding domains, and the N terminal domain is responsible for anchoring to the RNA pol I complex |
|
| YBR210W | 8.78 |
ERV15
|
Protein involved in export of proteins from the endoplasmic reticulum, has similarity to Erv14p |
|
| YHR174W | 8.78 |
ENO2
|
Enolase II, a phosphopyruvate hydratase that catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate during glycolysis and the reverse reaction during gluconeogenesis; expression is induced in response to glucose |
|
| YHR005C-A | 8.64 |
MRS11
|
Essential protein of the mitochondrial intermembrane space, forms a complex with Tim9p (TIM10 complex) that mediates insertion of hydrophobic proteins at the inner membrane, has homology to Mrs5p, which is also involved in this process |
|
| YDR509W | 8.49 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR508C | 8.46 |
GNP1
|
High-affinity glutamine permease, also transports Leu, Ser, Thr, Cys, Met and Asn; expression is fully dependent on Grr1p and modulated by the Ssy1p-Ptr3p-Ssy5p (SPS) sensor of extracellular amino acids |
|
| YGR050C | 8.37 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR382W | 8.35 |
RPP2B
|
Ribosomal protein P2 beta, a component of the ribosomal stalk, which is involved in the interaction between translational elongation factors and the ribosome; regulates the accumulation of P1 (Rpp1Ap and Rpp1Bp) in the cytoplasm |
|
| YLR257W | 8.30 |
Putative protein of unknown function |
||
| YDR385W | 8.25 |
EFT2
|
Elongation factor 2 (EF-2), also encoded by EFT1; catalyzes ribosomal translocation during protein synthesis; contains diphthamide, the unique posttranslationally modified histidine residue specifically ADP-ribosylated by diphtheria toxin |
|
| YMR106C | 8.16 |
YKU80
|
Subunit of the telomeric Ku complex (Yku70p-Yku80p), involved in telomere length maintenance, structure and telomere position effect; relocates to sites of double-strand cleavage to promote nonhomologous end joining during DSB repair |
|
| YFL022C | 8.11 |
FRS2
|
Alpha subunit of cytoplasmic phenylalanyl-tRNA synthetase, forms a tetramer with Frs1p to form active enzyme; evolutionarily distant from mitochondrial phenylalanyl-tRNA synthetase based on protein sequence, but substrate binding is similar |
|
| YER001W | 8.10 |
MNN1
|
Alpha-1,3-mannosyltransferase, integral membrane glycoprotein of the Golgi complex, required for addition of alpha1,3-mannose linkages to N-linked and O-linked oligosaccharides, one of five S. cerevisiae proteins of the MNN1 family |
|
| YJR123W | 8.04 |
RPS5
|
Protein component of the small (40S) ribosomal subunit, the least basic of the non-acidic ribosomal proteins; phosphorylated in vivo; essential for viability; has similarity to E. coli S7 and rat S5 ribosomal proteins |
|
| YGL036W | 8.01 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YGL036W is not an essential gene |
||
| YJR105W | 7.97 |
ADO1
|
Adenosine kinase, required for the utilization of S-adenosylmethionine (AdoMet); may be involved in recycling adenosine produced through the methyl cycle |
|
| YJL162C | 7.90 |
JJJ2
|
Protein of unknown function, contains a J-domain, which is a region with homology to the E. coli DnaJ protein |
|
| YLR180W | 7.89 |
SAM1
|
S-adenosylmethionine synthetase, catalyzes transfer of the adenosyl group of ATP to the sulfur atom of methionine; one of two differentially regulated isozymes (Sam1p and Sam2p) |
|
| YBR106W | 7.83 |
PHO88
|
Probable membrane protein, involved in phosphate transport; pho88 pho86 double null mutant exhibits enhanced synthesis of repressible acid phosphatase at high inorganic phosphate concentrations |
|
| YOL076W | 7.78 |
MDM20
|
Non-catalytic subunit of the NatB N-terminal acetyltransferase, which catalyzes N-acetylation of proteins with specific N-terminal sequences; involved in mitochondrial inheritance and actin assembly |
|
| YPL037C | 7.74 |
EGD1
|
Subunit beta1 of the nascent polypeptide-associated complex (NAC) involved in protein targeting, associated with cytoplasmic ribosomes; enhances DNA binding of the Gal4p activator; homolog of human BTF3b |
|
| YBR078W | 7.74 |
ECM33
|
GPI-anchored protein of unknown function, has a possible role in apical bud growth; GPI-anchoring on the plasma membrane crucial to function; phosphorylated in mitochondria; similar to Sps2p and Pst1p |
|
| YPR046W | 7.72 |
MCM16
|
Protein involved in kinetochore-microtubule mediated chromosome segregation; binds to centromere DNA |
|
| YKR057W | 7.68 |
RPS21A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps21Bp and has similarity to rat S21 ribosomal protein |
|
| YHL003C | 7.66 |
LAG1
|
Ceramide synthase component, involved in synthesis of ceramide from C26(acyl)-coenzyme A and dihydrosphingosine or phytosphingosine, functionally equivalent to Lac1p |
|
| YKR075C | 7.55 |
Protein of unknown function; similar to YOR062Cp and Reg1p; expression regulated by glucose and Rgt1p; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YGR264C | 7.51 |
MES1
|
Methionyl-tRNA synthetase, forms a complex with glutamyl-tRNA synthetase (Gus1p) and Arc1p, which increases the catalytic efficiency of both tRNA synthetases; also has a role in nuclear export of tRNAs |
|
| YDR094W | 7.51 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified ORF DNF2 |
||
| YJL011C | 7.51 |
RPC17
|
RNA polymerase III subunit C17; physically interacts with C31, C11, and TFIIIB70; may be involved in the recruitment of pol III by the preinitiation complex |
|
| YNL153C | 7.48 |
GIM3
|
Subunit of the heterohexameric cochaperone prefoldin complex which binds specifically to cytosolic chaperonin and transfers target proteins to it |
|
| YGR265W | 7.39 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF MES1/YGR264C, which encodes methionyl-tRNA synthetase |
||
| YAL003W | 7.39 |
EFB1
|
Translation elongation factor 1 beta; stimulates nucleotide exchange to regenerate EF-1 alpha-GTP for the next elongation cycle; part of the EF-1 complex, which facilitates binding of aminoacyl-tRNA to the ribosomal A site |
|
| YGR249W | 7.34 |
MGA1
|
Protein similar to heat shock transcription factor; multicopy suppressor of pseudohyphal growth defects of ammonium permease mutants |
|
| YGL037C | 7.30 |
PNC1
|
Nicotinamidase that converts nicotinamide to nicotinic acid as part of the NAD(+) salvage pathway, required for life span extension by calorie restriction; PNC1 expression responds to all known stimuli that extend replicative life span |
|
| YBR283C | 7.24 |
SSH1
|
Subunit of the Ssh1 translocon complex; Sec61p homolog involved in co-translational pathway of protein translocation; not essential |
|
| YNL113W | 7.16 |
RPC19
|
RNA polymerase subunit, common to RNA polymerases I and III |
|
| YJL014W | 7.11 |
CCT3
|
Subunit of the cytosolic chaperonin Cct ring complex, related to Tcp1p, required for the assembly of actin and tubulins in vivo |
|
| YGL077C | 7.10 |
HNM1
|
Choline/ethanolamine transporter; involved in the uptake of nitrogen mustard and the uptake of glycine betaine during hypersaline stress; co-regulated with phospholipid biosynthetic genes and negatively regulated by choline and myo-inositol |
|
| YLR367W | 7.08 |
RPS22B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps22Ap and has similarity to E. coli S8 and rat S15a ribosomal proteins |
|
| YGR051C | 7.07 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YGR051C is not an essential gene |
||
| YDR144C | 7.05 |
MKC7
|
GPI-anchored aspartyl protease (yapsin) involved in protein processing; shares functions with Yap3p and Kex2p |
|
| YEL001C | 7.02 |
IRC22
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion localizes to the ER; YEL001C is non-essential; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YLR339C | 7.02 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the essential gene RPP0 |
||
| YNL111C | 6.96 |
CYB5
|
Cytochrome b5, involved in the sterol and lipid biosynthesis pathways; acts as an electron donor to support sterol C5-6 desaturation |
|
| YDR095C | 6.93 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNL066W | 6.92 |
SUN4
|
Cell wall protein related to glucanases, possibly involved in cell wall septation; member of the SUN family |
|
| YNL251C | 6.84 |
NRD1
|
RNA-binding protein that interacts with the C-terminal domain of the RNA polymerase II large subunit (Rpo21p), required for transcription termination and 3' end maturation of nonpolyadenylated RNAs |
|
| YLR340W | 6.82 |
RPP0
|
Conserved ribosomal protein P0 similar to rat P0, human P0, and E. coli L10e; shown to be phosphorylated on serine 302 |
|
| YDL014W | 6.73 |
NOP1
|
Nucleolar protein, component of the small subunit processome complex, which is required for processing of pre-18S rRNA; has similarity to mammalian fibrillarin |
|
| YER124C | 6.71 |
DSE1
|
Daughter cell-specific protein, may participate in pathways regulating cell wall metabolism; deletion affects cell separation after division and sensitivity to drugs targeted against the cell wall |
|
| YHR064C | 6.70 |
SSZ1
|
Hsp70 protein that interacts with Zuo1p (a DnaJ homolog) to form a ribosome-associated complex that binds the ribosome via the Zuo1p subunit; also involved in pleiotropic drug resistance via sequential activation of PDR1 and PDR5; binds ATP |
|
| YNL044W | 6.48 |
YIP3
|
Protein localized to COPII vesicles, proposed to be involved in ER to Golgi transport; interacts with members of the Rab GTPase family and Yip1p; also interacts with Rtn1p |
|
| YOL125W | 6.47 |
TRM13
|
2'-O-methyltransferase responsible for modification of tRNA at position 4; exhibits no obvious similarity to other known methyltransferases |
|
| YPL112C | 6.47 |
PEX25
|
Peripheral peroxisomal membrane peroxin required for the regulation of peroxisome size and maintenance, recruits GTPase Rho1p to peroxisomes, induced by oleate, interacts with homologous protein Pex27p |
|
| YMR205C | 6.46 |
PFK2
|
Beta subunit of heterooctameric phosphofructokinase involved in glycolysis, indispensable for anaerobic growth, activated by fructose-2,6-bisphosphate and AMP, mutation inhibits glucose induction of cell cycle-related genes |
|
| YDL081C | 6.46 |
RPP1A
|
Ribosomal stalk protein P1 alpha, involved in the interaction between translational elongation factors and the ribosome; accumulation of P1 in the cytoplasm is regulated by phosphorylation and interaction with the P2 stalk component |
|
| YHR013C | 6.44 |
ARD1
|
Subunit of the N-terminal acetyltransferase NatA (Nat1p, Ard1p, Nat5p); N-terminally acetylates many proteins, which influences multiple processes such as the cell cycle, heat-shock resistance, mating, sporulation, and telomeric silencing |
|
| YOR063W | 6.42 |
RPL3
|
Protein component of the large (60S) ribosomal subunit, has similarity to E. coli L3 and rat L3 ribosomal proteins; involved in the replication and maintenance of killer double stranded RNA virus |
|
| YHR181W | 6.37 |
SVP26
|
Integral membrane protein of the early Golgi apparatus and endoplasmic reticulum, involved in COP II vesicle transport; may also function to promote retention of proteins in the early Golgi compartment |
|
| YPL075W | 6.31 |
GCR1
|
Transcriptional activator of genes involved in glycolysis; DNA-binding protein that interacts and functions with the transcriptional activator Gcr2p |
|
| YLR428C | 6.23 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF CRN1 |
||
| YLR154W-A | 6.23 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR154C | 6.20 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YOR310C | 6.20 |
NOP58
|
Protein involved in pre-rRNA processing, 18S rRNA synthesis, and snoRNA synthesis; component of the small subunit processome complex, which is required for processing of pre-18S rRNA |
|
| YNL189W | 6.18 |
SRP1
|
Karyopherin alpha homolog, forms a dimer with karyopherin beta Kap95p to mediate import of nuclear proteins, binds the nuclear localization signal of the substrate during import; may also play a role in regulation of protein degradation |
|
| YKR074W | 6.10 |
AIM29
|
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm; null mutant displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YLR154W-B | 6.02 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YNL178W | 6.02 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YLR186W | 5.99 |
EMG1
|
Member of the alpha/beta knot fold methyltransferase superfamily; required for maturation of 18S rRNA and for 40S ribosome production; interacts with RNA and with S-adenosylmethionine; associates with spindle/microtubules; forms homodimers |
|
| YLR264W | 5.97 |
RPS28B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps28Ap and has similarity to rat S28 ribosomal protein |
|
| YKL211C | 5.93 |
TRP3
|
Bifunctional enzyme exhibiting both indole-3-glycerol-phosphate synthase and anthranilate synthase activities, forms multifunctional hetero-oligomeric anthranilate synthase:indole-3-glycerol phosphate synthase enzyme complex with Trp2p |
|
| YDR274C | 5.93 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YFR055W | 5.86 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YDL047W | 5.76 |
SIT4
|
Type 2A-related serine-threonine phosphatase that functions in the G1/S transition of the mitotic cycle; cytoplasmic and nuclear protein that modulates functions mediated by Pkc1p including cell wall and actin cytoskeleton organization |
|
| YLR025W | 5.73 |
SNF7
|
One of four subunits of the endosomal sorting complex required for transport III (ESCRT-III); involved in the sorting of transmembrane proteins into the multivesicular body (MVB) pathway; recruited from the cytoplasm to endosomal membranes |
|
| YAL038W | 5.70 |
CDC19
|
Pyruvate kinase, functions as a homotetramer in glycolysis to convert phosphoenolpyruvate to pyruvate, the input for aerobic (TCA cycle) or anaerobic (glucose fermentation) respiration |
|
| YBR162C | 5.68 |
TOS1
|
Covalently-bound cell wall protein of unknown function; identified as a cell cycle regulated SBF target gene; deletion mutants are highly resistant to treatment with beta-1,3-glucanase; has sequence similarity to YJL171C |
|
| YPR187W | 5.61 |
RPO26
|
RNA polymerase subunit ABC23, common to RNA polymerases I, II, and III; part of central core; similar to bacterial omega subunit |
|
| YNL043C | 5.59 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YIP3/YNL044W |
||
| YLR429W | 5.58 |
CRN1
|
Coronin, cortical actin cytoskeletal component that associates with the Arp2p/Arp3p complex to regulate its activity; plays a role in regulation of actin patch assembly |
|
| YGL039W | 5.56 |
Oxidoreductase, catalyzes NADPH-dependent reduction of the bicyclic diketone bicyclo[2.2.2]octane-2,6-dione (BCO2,6D) to the chiral ketoalcohol (1R,4S,6S)-6-hydroxybicyclo[2.2.2]octane-2-one (BCO2one6ol) |
||
| YLR060W | 5.55 |
FRS1
|
Beta subunit of cytoplasmic phenylalanyl-tRNA synthetase, forms a tetramer with Frs2p to generate active enzyme; sequence is evolutionarily distant from mitochondrial phenylalanyl-tRNA synthetase (Msf1p), but substrate binding is similar |
|
| YIL069C | 5.48 |
RPS24B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps24Ap and has similarity to rat S24 ribosomal protein |
|
| YJR006W | 5.48 |
POL31
|
DNA polymerase III (delta) subunit, essential for cell viability; involved in DNA replication and DNA repair |
|
| YDR384C | 5.48 |
ATO3
|
Plasma membrane protein, regulation pattern suggests a possible role in export of ammonia from the cell; phosphorylated in mitochondria; member of the TC 9.B.33 YaaH family of putative transporters |
|
| YGL105W | 5.45 |
ARC1
|
Protein that binds tRNA and methionyl- and glutamyl-tRNA synthetases (Mes1p and Gus1p), delivering tRNA to them, stimulating catalysis, and ensuring their localization to the cytoplasm; also binds quadruplex nucleic acids |
|
| YNL069C | 5.44 |
RPL16B
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, binds to 5.8 S rRNA; has similarity to Rpl16Ap, E. coli L13 and rat L13a ribosomal proteins; transcriptionally regulated by Rap1p |
|
| YML052W | 5.43 |
SUR7
|
Putative integral membrane protein; component of eisosomes; associated with endocytosis, along with Pil1p and Lsp1p; sporulation and plasma membrane sphingolipid content are altered in mutants |
|
| YLR192C | 5.36 |
HCR1
|
Dual function protein involved in translation initiation as a substoichiometric component of eukaryotic translation initiation factor 3 (eIF3) and required for processing of 20S pre-rRNA; binds to eIF3 subunits Rpg1p and Prt1p and 18S rRNA |
|
| YOL041C | 5.36 |
NOP12
|
Nucleolar protein, required for pre-25S rRNA processing; contains an RNA recognition motif (RRM) and has similarity to Nop13p, Nsr1p, and putative orthologs in Drosophila and S. pombe |
|
| YDR087C | 5.36 |
RRP1
|
Essential evolutionarily conserved nucleolar protein necessary for biogenesis of 60S ribosomal subunits and processing of pre-rRNAs to mature rRNAs, associated with several distinct 66S pre-ribosomal particles |
|
| YIL142C-A | 5.35 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL169W | 5.33 |
SUA5
|
Protein required for respiratory growth; null mutation suppresses the Cyc1p translation defect caused by the presence of an aberrant ATG codon upstream of the correct start |
|
| YOR030W | 5.30 |
DFG16
|
Probable multiple transmembrane protein, involved in diploid invasive and pseudohyphal growth upon nitrogen starvation; required for accumulation of processed Rim101p |
|
| YDR074W | 5.30 |
TPS2
|
Phosphatase subunit of the trehalose-6-phosphate synthase/phosphatase complex, which synthesizes the storage carbohydrate trehalose; expression is induced by stress conditions and repressed by the Ras-cAMP pathway |
|
| YOR167C | 5.26 |
RPS28A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps28Bp and has similarity to rat S28 ribosomal protein |
|
| YDL143W | 5.25 |
CCT4
|
Subunit of the cytosolic chaperonin Cct ring complex, related to Tcp1p, required for the assembly of actin and tubulins in vivo |
|
| YER026C | 5.18 |
CHO1
|
Phosphatidylserine synthase, functions in phospholipid biosynthesis; catalyzes the reaction CDP-diaclyglycerol + L-serine = CMP + L-1-phosphatidylserine, transcriptionally repressed by myo-inositol and choline |
|
| YDL208W | 5.15 |
NHP2
|
Nuclear protein related to mammalian high mobility group (HMG) proteins, essential for function of H/ACA-type snoRNPs, which are involved in 18S rRNA processing |
|
| YMR235C | 5.14 |
RNA1
|
GTPase activating protein (GAP) for Gsp1p, involved in nuclear transport |
|
| YPR148C | 5.11 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
||
| YHL029C | 5.10 |
OCA5
|
Cytoplasmic protein required for replication of Brome mosaic virus in S. cerevisiae, which is a model system for studying replication of positive-strand RNA viruses in their natural hosts |
|
| YGR187C | 5.06 |
HGH1
|
Nonessential protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; similar to mammalian BRP16 (Brain protein 16) |
|
| YFR056C | 5.05 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YKL063C | 5.05 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the Golgi |
||
| YEL020C-B | 5.04 |
Dubious open reading frame unlikely to encode a protein; partially overlaps verified gene YEL020W-A; identified by fungal homology and RT-PCR |
||
| YPR170W-B | 5.03 |
Putative protein of unknown function, conserved in fungi; partially overlaps the dubious genes YPR169W-A, YPR170W-A and YRP170C |
||
| YDR099W | 5.01 |
BMH2
|
14-3-3 protein, minor isoform; controls proteome at post-transcriptional level, binds proteins and DNA, involved in regulation of many processes including exocytosis, vesicle transport, Ras/MAPK signaling, and rapamycin-sensitive signaling |
|
| YIL142W | 4.97 |
CCT2
|
Subunit beta of the cytosolic chaperonin Cct ring complex, related to Tcp1p, required for the assembly of actin and tubulins in vivo |
|
| YOR153W | 4.95 |
PDR5
|
Plasma membrane ATP-binding cassette (ABC) transporter, short-lived multidrug transporter actively regulated by Pdr1p; also involved in steroid transport, cation resistance, and cellular detoxification during exponential growth |
|
| YDR044W | 4.92 |
HEM13
|
Coproporphyrinogen III oxidase, an oxygen requiring enzyme that catalyzes the sixth step in the heme biosynthetic pathway; localizes to the mitochondrial inner membrane; transcription is repressed by oxygen and heme (via Rox1p and Hap1p) |
|
| YOR146W | 4.92 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; open reading frame overlaps the verified gene PNO1/YOR145C |
||
| YOR254C | 4.90 |
SEC63
|
Essential subunit of Sec63 complex (Sec63p, Sec62p, Sec66p and Sec72p); with Sec61 complex, Kar2p/BiP and Lhs1p forms a channel competent for SRP-dependent and post-translational SRP-independent protein targeting and import into the ER |
|
| YLR333C | 4.90 |
RPS25B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps25Ap and has similarity to rat S25 ribosomal protein |
|
| YGR037C | 4.87 |
ACB1
|
Acyl-CoA-binding protein, transports newly synthesized acyl-CoA esters from fatty acid synthetase (Fas1p-Fas2p) to acyl-CoA-consuming processes |
|
| YAL053W | 4.84 |
FLC2
|
Putative FAD transporter; required for uptake of FAD into endoplasmic reticulum; involved in cell wall maintenance |
|
| YDR398W | 4.83 |
UTP5
|
Nucleolar protein, component of the small subunit (SSU) processome containing the U3 snoRNA that is involved in processing of pre-18S rRNA |
|
| YMR303C | 4.77 |
ADH2
|
Glucose-repressible alcohol dehydrogenase II, catalyzes the conversion of ethanol to acetaldehyde; involved in the production of certain carboxylate esters; regulated by ADR1 |
|
| YCL063W | 4.77 |
VAC17
|
Protein involved in vacuole inheritance; acts as a vacuole-specific receptor for myosin Myo2p |
|
| YPR163C | 4.76 |
TIF3
|
Translation initiation factor eIF-4B, has RNA annealing activity; contains an RNA recognition motif and binds to single-stranded RNA |
|
| YMR142C | 4.75 |
RPL13B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl13Ap; not essential for viability; has similarity to rat L13 ribosomal protein |
|
| YPR033C | 4.74 |
HTS1
|
Cytoplasmic and mitochondrial histidine tRNA synthetase; encoded by a single nuclear gene that specifies two messages; efficient mitochondrial localization requires both a presequence and an amino-terminal sequence |
|
| YGL097W | 4.73 |
SRM1
|
Nucleotide exchange factor for Gsp1p, localizes to the nucleus, required for nucleocytoplasmic trafficking of macromolecules; suppressor of the pheromone response pathway; potentially phosphorylated by Cdc28p |
|
| YDL084W | 4.73 |
SUB2
|
Component of the TREX complex required for nuclear mRNA export; member of the DEAD-box RNA helicase superfamily and is involved in early and late steps of spliceosome assembly; homolog of the human splicing factor hUAP56 |
|
| YJL183W | 4.70 |
MNN11
|
Subunit of a Golgi mannosyltransferase complex that also contains Anp1p, Mnn9p, Mnn10p, and Hoc1p, and mediates elongation of the polysaccharide mannan backbone; has homology to Mnn10p |
|
| YOR357C | 4.69 |
SNX3
|
Sorting nexin required to maintain late-Golgi resident enzymes in their proper location by recycling molecules from the prevacuolar compartment; contains a PX domain and sequence similarity to human Snx3p |
|
| YNL119W | 4.69 |
NCS2
|
Protein with a role in urmylation and in invasive and pseudohyphal growth; inhibits replication of Brome mosaic virus in S. cerevisiae, which is a model system for studying replication of positive-strand RNA viruses in their natural hosts |
|
| YDR120C | 4.68 |
TRM1
|
tRNA methyltransferase, localizes to both the nucleus and mitochondrion to produce the modified base N2,N2-dimethylguanosine in tRNAs in both compartments |
|
| YPL160W | 4.67 |
CDC60
|
Cytosolic leucyl tRNA synthetase, ligates leucine to the appropriate tRNA |
|
| YLR292C | 4.66 |
SEC72
|
Non-essential subunit of Sec63 complex (Sec63p, Sec62p, Sec66p and Sec72p); with Sec61 complex, Kar2p/BiP and Lhs1p forms a channel competent for SRP-dependent and post-translational SRP-independent protein targeting and import into the ER |
|
| YHR196W | 4.66 |
UTP9
|
Nucleolar protein, component of the small subunit (SSU) processome containing the U3 snoRNA that is involved in processing of pre-18S rRNA |
|
| YBL032W | 4.65 |
HEK2
|
RNA binding protein with similarity to hnRNP-K that localizes to the cytoplasm and to subtelomeric DNA; required for the proper localization of ASH1 mRNA; involved in the regulation of telomere position effect and telomere length |
|
| YER012W | 4.62 |
PRE1
|
Beta 4 subunit of the 20S proteasome; localizes to the nucleus throughout the cell cycle |
|
| YEL027W | 4.59 |
CUP5
|
Proteolipid subunit of the vacuolar H(+)-ATPase V0 sector (subunit c; dicyclohexylcarbodiimide binding subunit); required for vacuolar acidification and important for copper and iron metal ion homeostasis |
|
| YMR143W | 4.58 |
RPS16A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps16Bp and has similarity to E. coli S9 and rat S16 ribosomal proteins |
|
| YGR103W | 4.58 |
NOP7
|
Nucleolar protein involved in rRNA processing and 60S ribosomal subunit biogenesis; constituent of several different pre-ribosomal particles; required for exit from G0 and the initiation of cell proliferation |
|
| YBR031W | 4.55 |
RPL4A
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, nearly identical to Rpl4Bp and has similarity to E. coli L4 and rat L4 ribosomal proteins |
|
| YLR175W | 4.55 |
CBF5
|
Pseudouridine synthase catalytic subunit of box H/ACA small nucleolar ribonucleoprotein particles (snoRNPs), acts on both large and small rRNAs and on snRNA U2 |
|
| YIL021W | 4.54 |
RPB3
|
RNA polymerase II third largest subunit B44, part of central core; similar to prokaryotic alpha subunit |
|
| YKL030W | 4.54 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; partially overlaps the verified gene MAE1 |
||
| YPL086C | 4.54 |
ELP3
|
Subunit of Elongator complex, which is required for modification of wobble nucleosides in tRNA; exhibits histone acetyltransferase activity that is directed to histones H3 and H4; disruption confers resistance to K. lactis zymotoxin |
|
| YPR165W | 4.53 |
RHO1
|
GTP-binding protein of the rho subfamily of Ras-like proteins, involved in establishment of cell polarity; regulates protein kinase C (Pkc1p) and the cell wall synthesizing enzyme 1,3-beta-glucan synthase (Fks1p and Gsc2p) |
|
| YLL011W | 4.52 |
SOF1
|
Essential protein required for biogenesis of 40S (small) ribosomal subunit; has similarity to the beta subunit of trimeric G-proteins and the splicing factor Prp4p |
|
| YER003C | 4.52 |
PMI40
|
Mannose-6-phosphate isomerase, catalyzes the interconversion of fructose-6-P and mannose-6-P; required for early steps in protein mannosylation |
|
| YLR453C | 4.51 |
RIF2
|
Protein that binds to the Rap1p C-terminus and acts synergistically with Rif1p to help control telomere length and establish telomeric silencing; deletion results in telomere elongation |
|
| YPL210C | 4.50 |
SRP72
|
Core component of the signal recognition particle (SRP) ribonucleoprotein (RNP) complex that functions in targeting nascent secretory proteins to the endoplasmic reticulum (ER) membrane |
|
| YDR454C | 4.48 |
GUK1
|
Guanylate kinase, converts GMP to GDP; required for growth and mannose outer chain elongation of cell wall N-linked glycoproteins |
|
| YKL099C | 4.47 |
UTP11
|
Nucleolar protein, component of the small subunit (SSU) processome containing the U3 snoRNA that is involved in processing of pre-18S rRNA |
|
| YLR249W | 4.46 |
YEF3
|
Translational elongation factor 3, stimulates the binding of aminoacyl-tRNA (AA-tRNA) to ribosomes by releasing EF-1 alpha from the ribosomal complex; contains two ABC cassettes; binds and hydrolyses ATP |
|
| YJL115W | 4.46 |
ASF1
|
Nucleosome assembly factor, involved in chromatin assembly and disassembly, anti-silencing protein that causes derepression of silent loci when overexpressed; plays a role in regulating Ty1 transposition |
|
| YOR087W | 4.46 |
YVC1
|
Vacuolar cation channel, mediates release of Ca(2+) from the vacuole in response to hyperosmotic shock |
|
| YOR092W | 4.43 |
ECM3
|
Non-essential protein of unknown function; involved in signal transduction and the genotoxic response; induced rapidly in response to treatment with 8-methoxypsoralen and UVA irradiation |
|
| YNR016C | 4.42 |
ACC1
|
Acetyl-CoA carboxylase, biotin containing enzyme that catalyzes the carboxylation of acetyl-CoA to form malonyl-CoA; required for de novo biosynthesis of long-chain fatty acids |
|
| YIR033W | 4.39 |
MGA2
|
ER membrane protein involved in regulation of OLE1 transcription, acts with homolog Spt23p; inactive ER form dimerizes and one subunit is then activated by ubiquitin/proteasome-dependent processing followed by nuclear targeting |
|
| YNL002C | 4.39 |
RLP7
|
Nucleolar protein with similarity to large ribosomal subunit L7 proteins; constituent of 66S pre-ribosomal particles; plays an essential role in processing of precursors to the large ribosomal subunit RNAs |
|
| YDL229W | 4.37 |
SSB1
|
Cytoplasmic ATPase that is a ribosome-associated molecular chaperone, functions with J-protein partner Zuo1p; may be involved in folding of newly-made polypeptide chains; member of the HSP70 family; interacts with phosphatase subunit Reg1p |
|
| YBR011C | 4.36 |
IPP1
|
Cytoplasmic inorganic pyrophosphatase (PPase), catalyzes the rapid exchange of oxygens from Pi with water, highly expressed and essential for viability, active-site residues show identity to those from E. coli PPase |
|
| YLR276C | 4.34 |
DBP9
|
ATP-dependent RNA helicase of the DEAD-box family involved in biogenesis of the 60S ribosomal subunit |
|
| YPL012W | 4.34 |
RRP12
|
Protein required for export of the ribosomal subunits; associates with the RNA components of the pre-ribosomes; contains HEAT-repeats |