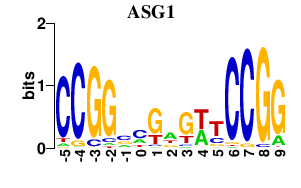
Results for ASG1
Z-value: 0.54
Transcription factors associated with ASG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ASG1
|
S000001392 | Zinc cluster protein proposed to be a transcriptional regulator |
Activity-expression correlation:
Activity profile of ASG1 motif
Sorted Z-values of ASG1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YLR154W-A | 3.71 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR154W-B | 3.06 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR154W-C | 1.89 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YAR035W | 1.05 |
YAT1
|
Outer mitochondrial carnitine acetyltransferase, minor ethanol-inducible enzyme involved in transport of activated acyl groups from the cytoplasm into the mitochondrial matrix; phosphorylated |
|
| YBR056W-A | 0.74 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR056C-B |
||
| YKR097W | 0.66 |
PCK1
|
Phosphoenolpyruvate carboxykinase, key enzyme in gluconeogenesis, catalyzes early reaction in carbohydrate biosynthesis, glucose represses transcription and accelerates mRNA degradation, regulated by Mcm1p and Cat8p, located in the cytosol |
|
| YFR016C | 0.52 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and bud; interacts with Spa2p; YFL016C is not an essential gene |
||
| YNL065W | 0.48 |
AQR1
|
Plasma membrane multidrug transporter of the major facilitator superfamily, confers resistance to short-chain monocarboxylic acids and quinidine; involved in the excretion of excess amino acids |
|
| YPL250W-A | 0.45 |
Identified by fungal homology and RT-PCR |
||
| YDL067C | 0.43 |
COX9
|
Subunit VIIa of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain |
|
| YOR348C | 0.42 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YNR017W | 0.38 |
TIM23
|
Essential protein of the mitochondrial inner membrane, component of the mitochondrial import system |
|
| YGR280C | 0.36 |
PXR1
|
Essential protein involved in rRNA and snoRNA maturation; competes with TLC1 RNA for binding to Est2p, suggesting a role in negative regulation of telomerase; human homolog inhibits telomerase; contains a G-patch RNA interacting domain |
|
| YDL034W | 0.33 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps with verified gene GPR1/YDL035C; YDL034W is not an essential gene |
||
| YEL012W | 0.33 |
UBC8
|
Ubiquitin-conjugating enzyme that negatively regulates gluconeogenesis by mediating the glucose-induced ubiquitination of fructose-1,6-bisphosphatase (FBPase); cytoplasmic enzyme that catalyzes the ubiquitination of histones in vitro |
|
| YLR055C | 0.32 |
SPT8
|
Subunit of the SAGA transcriptional regulatory complex but not present in SAGA-like complex SLIK/SALSA, required for SAGA-mediated inhibition at some promoters |
|
| YMR135W-A | 0.32 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOL085W-A | 0.31 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YOL085C |
||
| YJL045W | 0.30 |
Minor succinate dehydrogenase isozyme; homologous to Sdh1p, the major isozyme reponsible for the oxidation of succinate and transfer of electrons to ubiquinone; induced during the diauxic shift in a Cat8p-dependent manner |
||
| YJR078W | 0.30 |
BNA2
|
Putative tryptophan 2,3-dioxygenase or indoleamine 2,3-dioxygenase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p and Aft2p |
|
| YIL057C | 0.29 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YPR126C | 0.29 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR016C | 0.29 |
SOK2
|
Nuclear protein that plays a regulatory role in the cyclic AMP (cAMP)-dependent protein kinase (PKA) signal transduction pathway; negatively regulates pseudohyphal differentiation; homologous to several transcription factors |
|
| YLR044C | 0.29 |
PDC1
|
Major of three pyruvate decarboxylase isozymes, key enzyme in alcoholic fermentation, decarboxylates pyruvate to acetaldehyde; subject to glucose-, ethanol-, and autoregulation; involved in amino acid catabolism |
|
| YCR027C | 0.28 |
RHB1
|
Putative Rheb-related GTPase involved in regulating canavanine resistance and arginine uptake; member of the Ras superfamily of G-proteins |
|
| YFL014W | 0.28 |
HSP12
|
Plasma membrane localized protein that protects membranes from desiccation; induced by heat shock, oxidative stress, osmostress, stationary phase entry, glucose depletion, oleate and alcohol; regulated by the HOG and Ras-Pka pathways |
|
| YKR041W | 0.27 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YOL040C | 0.27 |
RPS15
|
Protein component of the small (40S) ribosomal subunit; has similarity to E. coli S19 and rat S15 ribosomal proteins |
|
| YJR147W | 0.27 |
HMS2
|
Protein with similarity to heat shock transcription factors; overexpression suppresses the pseudohyphal filamentation defect of a diploid mep1 mep2 homozygous null mutant |
|
| YDR441C | 0.27 |
APT2
|
Apparent pseudogene, not transcribed or translated under normal conditions; encodes a protein with similarity to adenine phosphoribosyltransferase, but artificially expressed protein exhibits no enzymatic activity |
|
| YPR008W | 0.26 |
HAA1
|
Transcriptional activator involved in the transcription of TPO2, HSP30 and other genes encoding membrane stress proteins; involved in adaptation to weak acid stress |
|
| YKL029C | 0.26 |
MAE1
|
Mitochondrial malic enzyme, catalyzes the oxidative decarboxylation of malate to pyruvate, which is a key intermediate in sugar metabolism and a precursor for synthesis of several amino acids |
|
| YOR050C | 0.26 |
Hypothetical protein |
||
| YFR034C | 0.25 |
PHO4
|
Basic helix-loop-helix (bHLH) transcription factor of the myc-family; binds cooperatively with Pho2p to the PHO5 promoter; function is regulated by phosphorylation at multiple sites and by phosphate availability |
|
| YMR303C | 0.24 |
ADH2
|
Glucose-repressible alcohol dehydrogenase II, catalyzes the conversion of ethanol to acetaldehyde; involved in the production of certain carboxylate esters; regulated by ADR1 |
|
| YEL049W | 0.24 |
PAU2
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YJR146W | 0.24 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene HMS2 |
||
| YOL039W | 0.22 |
RPP2A
|
Ribosomal protein P2 alpha, a component of the ribosomal stalk, which is involved in the interaction between translational elongation factors and the ribosome; regulates the accumulation of P1 (Rpp1Ap and Rpp1Bp) in the cytoplasm |
|
| YOL006C | 0.21 |
TOP1
|
Topoisomerase I, nuclear enzyme that relieves torsional strain in DNA by cleaving and re-sealing the phosphodiester backbone; relaxes both positively and negatively supercoiled DNA; functions in replication, transcription, and recombination |
|
| YDR259C | 0.21 |
YAP6
|
Putative basic leucine zipper (bZIP) transcription factor; overexpression increases sodium and lithium tolerance |
|
| YLR223C | 0.21 |
IFH1
|
Essential protein with a highly acidic N-terminal domain; IFH1 exhibits genetic interactions with FHL1, overexpression interferes with silencing at telomeres and HM loci; potential Cdc28p substrate |
|
| YKL103C | 0.20 |
LAP4
|
Vacuolar aminopeptidase, often used as a marker protein in studies of autophagy and cytosol to vacuole targeting (CVT) pathway |
|
| YOL086C | 0.20 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YJR077C | 0.20 |
MIR1
|
Mitochondrial phosphate carrier, imports inorganic phosphate into mitochondria; functionally redundant with Pic2p but more abundant than Pic2p under normal conditions; phosphorylated |
|
| YKL050C | 0.19 |
Protein of unknown function; the YKL050W protein is a target of the SCFCdc4 ubiquitin ligase complex and YKL050W transcription is regulated by Azf1p |
||
| YCR016W | 0.19 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus and nucleus; YCR016W is not an essential gene |
||
| YIL100W | 0.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps the dubious ORF YIL100C-A |
||
| YKL165C-A | 0.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YML089C | 0.18 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression induced by calcium shortage |
||
| YGL056C | 0.18 |
SDS23
|
One of two S. cerevisiae homologs (Sds23p and Sds24p) of the Schizosaccharomyces pombe Sds23 protein, which genetic studies have implicated in APC/cyclosome regulation |
|
| YKL101W | 0.18 |
HSL1
|
Nim1p-related protein kinase that regulates the morphogenesis and septin checkpoints; associates with the assembled septin filament; required along with Hsl7p for bud neck recruitment, phosphorylation, and degradation of Swe1p |
|
| YAR070C | 0.18 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR002W | 0.18 |
PDH1
|
Mitochondrial protein that participates in respiration, induced by diauxic shift; homologous to E. coli PrpD, may take part in the conversion of 2-methylcitrate to 2-methylisocitrate |
|
| YHR096C | 0.17 |
HXT5
|
Hexose transporter with moderate affinity for glucose, induced in the presence of non-fermentable carbon sources, induced by a decrease in growth rate, contains an extended N-terminal domain relative to other HXTs |
|
| YIL099W | 0.17 |
SGA1
|
Intracellular sporulation-specific glucoamylase involved in glycogen degradation; induced during starvation of a/a diploids late in sporulation, but dispensable for sporulation |
|
| YER028C | 0.16 |
MIG3
|
Probable transcriptional repressor involved in response to toxic agents such as hydroxyurea that inhibit ribonucleotide reductase; phosphorylation by Snf1p or the Mec1p pathway inactivates Mig3p, allowing induction of damage response genes |
|
| YKR042W | 0.16 |
UTH1
|
Mitochondrial outer membrane and cell wall localized SUN family member required for mitochondrial autophagy; involved in the oxidative stress response, life span during starvation, mitochondrial biogenesis, and cell death |
|
| YPL182C | 0.16 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene CTI6/YPL181W |
||
| YMR009W | 0.16 |
ADI1
|
Acireductone dioxygenease involved in the methionine salvage pathway; ortholog of human MTCBP-1; transcribed with YMR010W and regulated post-transcriptionally by RNase III (Rnt1p) cleavage; ADI1 mRNA is induced in heat shock conditions |
|
| YAL039C | 0.16 |
CYC3
|
Cytochrome c heme lyase (holocytochrome c synthase), attaches heme to apo-cytochrome c (Cyc1p or Cyc7p) in the mitochondrial intermembrane space; human ortholog may have a role in microphthalmia with linear skin defects (MLS) |
|
| YIL149C | 0.15 |
MLP2
|
Myosin-like protein associated with the nuclear envelope, connects the nuclear pore complex with the nuclear interior; involved in the Tel1p pathway that controls telomere length |
|
| YER011W | 0.15 |
TIR1
|
Cell wall mannoprotein of the Srp1p/Tip1p family of serine-alanine-rich proteins; expression is downregulated at acidic pH and induced by cold shock and anaerobiosis; abundance is increased in cells cultured without shaking |
|
| YPL128C | 0.15 |
TBF1
|
Telobox-containing general regulatory factor; binds to TTAGGG repeats within subtelomeric anti-silencing regions (STARs) and possibly throughout the genome and mediates their insulating capacity by blocking silent chromatin propagation |
|
| YML025C | 0.15 |
YML6
|
Mitochondrial ribosomal protein of the large subunit, has similarity to E. coli L4 ribosomal protein and human mitoribosomal MRP-L4 protein; essential for viability, unlike most other mitoribosomal proteins |
|
| YFR034W-A | 0.14 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YFR035C; identified by gene-trapping, microarray expression analysis, and genome-wide homology searching |
||
| YNL208W | 0.14 |
Protein of unknown function; may interact with ribosomes, based on co-purification experiments; authentic, non-tagged protein is detected in purified mitochondria in high-throughput studies; potential orthologs found in other fungi |
||
| YOL012C | 0.14 |
HTZ1
|
Histone variant H2AZ, exchanged for histone H2A in nucleosomes by the SWR1 complex; involved in transcriptional regulation through prevention of the spread of silent heterochromatin |
|
| YOR226C | 0.14 |
ISU2
|
Conserved protein of the mitochondrial matrix, required for synthesis of mitochondrial and cytosolic iron-sulfur proteins, performs a scaffolding function in mitochondria during Fe/S cluster assembly; isu1 isu2 double mutant is inviable |
|
| YDR508C | 0.14 |
GNP1
|
High-affinity glutamine permease, also transports Leu, Ser, Thr, Cys, Met and Asn; expression is fully dependent on Grr1p and modulated by the Ssy1p-Ptr3p-Ssy5p (SPS) sensor of extracellular amino acids |
|
| YMR043W | 0.14 |
MCM1
|
Transcription factor involved in cell-type-specific transcription and pheromone response; plays a central role in the formation of both repressor and activator complexes |
|
| YDL182W | 0.14 |
LYS20
|
Homocitrate synthase isozyme, catalyzes the condensation of acetyl-CoA and alpha-ketoglutarate to form homocitrate, which is the first step in the lysine biosynthesis pathway; highly similar to the other isozyme, Lys21p |
|
| YKL097C | 0.13 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species |
||
| YCL023C | 0.13 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified ORF KCC4 |
||
| YHR021W-A | 0.13 |
ECM12
|
Non-essential protein of unknown function |
|
| YBR114W | 0.13 |
RAD16
|
Protein that recognizes and binds damaged DNA in an ATP-dependent manner (with Rad7p) during nucleotide excision repair; subunit of Nucleotide Excision Repair Factor 4 (NEF4) and the Elongin-Cullin-Socs (ECS) ligase complex |
|
| YML090W | 0.13 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YML089C; exhibits growth defect on a non-fermentable (respiratory) carbon source |
||
| YLL028W | 0.12 |
TPO1
|
Polyamine transporter that recognizes spermine, putrescine, and spermidine; catalyzes uptake of polyamines at alkaline pH and excretion at acidic pH; phosphorylation enhances activity and sorting to the plasma membrane |
|
| YOR230W | 0.12 |
WTM1
|
Transcriptional modulator involved in regulation of meiosis, silencing, and expression of RNR genes; required for nuclear localization of the ribonucleotide reductase small subunit Rnr2p and Rnr4p; contains WD repeats |
|
| YBR002C | 0.12 |
RER2
|
Cis-prenyltransferase involved in dolichol synthesis; participates in endoplasmic reticulum (ER) protein sorting |
|
| YAR050W | 0.12 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YJL041W | 0.12 |
NSP1
|
Essential component of the nuclear pore complex, which mediates nuclear import and export |
|
| YIL101C | 0.12 |
XBP1
|
Transcriptional repressor that binds to promoter sequences of the cyclin genes, CYS3, and SMF2; expression is induced by stress or starvation during mitosis, and late in meiosis; member of the Swi4p/Mbp1p family; potential Cdc28p substrate |
|
| YKR067W | 0.12 |
GPT2
|
Glycerol-3-phosphate/dihydroxyacetone phosphate dual substrate-specific sn-1 acyltransferase located in lipid particles and the ER; involved in the stepwise acylation of glycerol-3-phosphate and dihydroxyacetone in lipid biosynthesis |
|
| YLR205C | 0.12 |
HMX1
|
ER localized, heme-binding peroxidase involved in the degradation of heme; does not exhibit heme oxygenase activity despite similarity to heme oxygenases; expression regulated by AFT1 |
|
| YKL166C | 0.11 |
TPK3
|
cAMP-dependent protein kinase catalytic subunit; promotes vegetative growth in response to nutrients via the Ras-cAMP signaling pathway; inhibited by regulatory subunit Bcy1p in the absence of cAMP; partially redundant with Tpk1p and Tpk2p |
|
| YER177W | 0.11 |
BMH1
|
14-3-3 protein, major isoform; controls proteome at post-transcriptional level, binds proteins and DNA, involved in regulation of many processes including exocytosis, vesicle transport, Ras/MAPK signaling, and rapamycin-sensitive signaling |
|
| YJL136C | 0.11 |
RPS21B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps21Ap and has similarity to rat S21 ribosomal protein |
|
| YPL181W | 0.11 |
CTI6
|
Protein that relieves transcriptional repression by binding to the Cyc8p-Tup1p corepressor and recruiting the SAGA complex to the repressed promoter; contains a PHD finger domain |
|
| YOR071C | 0.11 |
NRT1
|
High-affinity nicotinamide riboside transporter; also transports thiamine with low affinity; shares sequence similarity with Thi7p and Thi72p; proposed to be involved in 5-fluorocytosine sensitivity |
|
| YOL084W | 0.11 |
PHM7
|
Protein of unknown function, expression is regulated by phosphate levels; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery and vacuole |
|
| YMR011W | 0.10 |
HXT2
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YOR200W | 0.10 |
Hypothetical protein |
||
| YOL054W | 0.10 |
PSH1
|
Nuclear protein, putative RNA polymerase II elongation factor; isolated as Pob3p/Spt16p-binding protein |
|
| YGL117W | 0.10 |
Putative protein of unknown function |
||
| YHR033W | 0.10 |
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm |
||
| YKL063C | 0.10 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the Golgi |
||
| YDR380W | 0.10 |
ARO10
|
Phenylpyruvate decarboxylase, catalyzes decarboxylation of phenylpyruvate to phenylacetaldehyde, which is the first specific step in the Ehrlich pathway |
|
| YIL100C-A | 0.10 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR271C | 0.10 |
FSF1
|
Putative protein, predicted to be an alpha-isopropylmalate carrier; belongs to the sideroblastic-associated protein family; non-tagged protein is detected in purified mitochondria; likely to play a role in iron homeostasis |
|
| YKL028W | 0.10 |
TFA1
|
TFIIE large subunit, involved in recruitment of RNA polymerase II to the promoter, activation of TFIIH, and promoter opening |
|
| YLL060C | 0.10 |
GTT2
|
Glutathione S-transferase capable of homodimerization; functional overlap with Gtt2p, Grx1p, and Grx2p |
|
| YOR072W | 0.10 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOR072W-A; diploid deletion strains are methotrexate, paraquat and wortmannin sensitive |
||
| YNL103W | 0.10 |
MET4
|
Leucine-zipper transcriptional activator, responsible for the regulation of the sulfur amino acid pathway, requires different combinations of the auxiliary factors Cbf1p, Met28p, Met31p and Met32p |
|
| YER088C | 0.10 |
DOT6
|
Protein of unknown function, involved in telomeric gene silencing and filamentation |
|
| YHR200W | 0.10 |
RPN10
|
Non-ATPase base subunit of the 19S regulatory particle (RP) of the 26S proteasome; N-terminus plays a role in maintaining the structural integrity of the RP; binds selectively to polyubiquitin chains; homolog of the mammalian S5a protein |
|
| YJL115W | 0.09 |
ASF1
|
Nucleosome assembly factor, involved in chromatin assembly and disassembly, anti-silencing protein that causes derepression of silent loci when overexpressed; plays a role in regulating Ty1 transposition |
|
| YOR199W | 0.09 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR259C | 0.09 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF TNA1/YGR260W |
||
| YML091C | 0.09 |
RPM2
|
Protein subunit of mitochondrial RNase P, has roles in nuclear transcription, cytoplasmic and mitochondrial RNA processing, and mitochondrial translation; distributed to mitochondria, cytoplasmic processing bodies, and the nucleus |
|
| YGL035C | 0.09 |
MIG1
|
Transcription factor involved in glucose repression; sequence specific DNA binding protein containing two Cys2His2 zinc finger motifs; regulated by the SNF1 kinase and the GLC7 phosphatase |
|
| YMR276W | 0.09 |
DSK2
|
Nuclear-enriched ubiquitin-like polyubiquitin-binding protein, required for spindle pole body (SPB) duplication and for transit through the G2/M phase of the cell cycle, involved in proteolysis, interacts with the proteasome |
|
| YNR002C | 0.09 |
ATO2
|
Putative transmembrane protein involved in export of ammonia, a starvation signal that promotes cell death in aging colonies; phosphorylated in mitochondria; member of the TC 9.B.33 YaaH family; homolog of Ady2p and Y. lipolytica Gpr1p |
|
| YFL016C | 0.09 |
MDJ1
|
Co-chaperone that stimulates the ATPase activity of the HSP70 protein Ssc1p; involved in protein folding/refolding in the mitochodrial matrix; required for proteolysis of misfolded proteins; member of the HSP40 (DnaJ) family of chaperones |
|
| YDL210W | 0.09 |
UGA4
|
Permease that serves as a gamma-aminobutyrate (GABA) transport protein involved in the utilization of GABA as a nitrogen source; catalyzes the transport of putrescine and delta-aminolevulinic acid (ALA); localized to the vacuolar membrane |
|
| YCL024W | 0.09 |
KCC4
|
Protein kinase of the bud neck involved in the septin checkpoint, associates with septin proteins, negatively regulates Swe1p by phosphorylation, shows structural homology to bud neck kinases Gin4p and Hsl1p |
|
| YGL149W | 0.08 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF INO80/YGL150C. |
||
| YCR008W | 0.08 |
SAT4
|
Ser/Thr protein kinase involved in salt tolerance; funtions in regulation of Trk1p-Trk2p potassium transporter; partially redundant with Hal5p; has similarity to Npr1p |
|
| YIL123W | 0.08 |
SIM1
|
Protein of the SUN family (Sim1p, Uth1p, Nca3p, Sun4p) that may participate in DNA replication, promoter contains SCB regulation box at -300 bp indicating that expression may be cell cycle-regulated |
|
| YCR010C | 0.08 |
ADY2
|
Acetate transporter required for normal sporulation; phosphorylated in mitochondria |
|
| YBL103C | 0.08 |
RTG3
|
Basic helix-loop-helix-leucine zipper (bHLH/Zip) transcription factor that forms a complex with another bHLH/Zip protein, Rtg1p, to activate the retrograde (RTG) and TOR pathways |
|
| YDR275W | 0.08 |
BSC2
|
Protein of unknown function, ORF exhibits genomic organization compatible with a translational readthrough-dependent mode of expression |
|
| YOR157C | 0.08 |
PUP1
|
Endopeptidase with trypsin-like activity that cleaves after basic residues; beta 2 subunit of 20S proteasome, synthesized as a proprotein before being proteolytically processed for assembly into 20S particle; human homolog is subunit Z |
|
| YBR083W | 0.08 |
TEC1
|
Transcription factor required for full Ty1 epxression, Ty1-mediated gene activation, and haploid invasive and diploid pseudohyphal growth; TEA/ATTS DNA-binding domain family member |
|
| YFL021W | 0.08 |
GAT1
|
Transcriptional activator of genes involved in nitrogen catabolite repression; contains a GATA-1-type zinc finger DNA-binding motif; activity and localization regulated by nitrogen limitation and Ure2p |
|
| YKL096W-A | 0.08 |
CWP2
|
Covalently linked cell wall mannoprotein, major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored |
|
| YFR028C | 0.08 |
CDC14
|
Protein phosphatase required for mitotic exit; located in the nucleolus until liberated by the FEAR and Mitotic Exit Network in anaphase, enabling it to act on key substrates to effect a decrease in CDK/B-cyclin activity and mitotic exit |
|
| YER010C | 0.08 |
Protein of unknown function, forms a ring-shaped homotrimer; has similarity to members of the prokaryotic rraA family; possibly involved in a phosphotransfer reaction |
||
| YJL134W | 0.08 |
LCB3
|
Long-chain base-1-phosphate phosphatase with specificity for dihydrosphingosine-1-phosphate, regulates ceramide and long-chain base phosphates levels, involved in incorporation of exogenous long chain bases in sphingolipids |
|
| YGL228W | 0.08 |
SHE10
|
Putative glycosylphosphatidylinositol (GPI)-anchored protein of unknown function; overexpression causes growth arrest |
|
| YHR032W-A | 0.08 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YHR032C-A |
||
| YGR251W | 0.07 |
Putative protein of unknown function; deletion mutant has defects in pre-rRNA processing; green fluorescent protein (GFP)-fusion protein localizes to both the nucleus and the nucleolus; YGR251W is an essential gene |
||
| YBL022C | 0.07 |
PIM1
|
ATP-dependent Lon protease, involved in degradation of misfolded proteins in mitochondria; required for biogenesis and maintenance of mitochondria |
|
| YDR156W | 0.07 |
RPA14
|
RNA polymerase I subunit A14 |
|
| YGL088W | 0.07 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps snR10, a snoRNA required for preRNA processing |
||
| YJL215C | 0.07 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR119W-A | 0.07 |
Putative protein of unknown function |
||
| YPL230W | 0.07 |
USV1
|
Putative transcription factor containing a C2H2 zinc finger; mutation affects transcriptional regulation of genes involved in protein folding, ATP binding, and cell wall biosynthesis |
|
| YNR070W | 0.07 |
Putative transporter of the ATP-binding cassette (ABC) family, implicated in pleiotropic drug resistance; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YER167W | 0.07 |
BCK2
|
Protein rich in serine and threonine residues involved in protein kinase C signaling pathway, which controls cell integrity; overproduction suppresses pkc1 mutations |
|
| YNL290W | 0.07 |
RFC3
|
Subunit of heteropentameric Replication factor C (RF-C), which is a DNA binding protein and ATPase that acts as a clamp loader of the proliferating cell nuclear antigen (PCNA) processivity factor for DNA polymerases delta and epsilon |
|
| YGL089C | 0.07 |
MF(ALPHA)2
|
Mating pheromone alpha-factor, made by alpha cells; interacts with mating type a cells to induce cell cycle arrest and other responses leading to mating; also encoded by MF(ALPHA)1, which is more highly expressed than MF(ALPHA)2 |
|
| YBR113W | 0.07 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene CYC8 |
||
| YER116C | 0.07 |
SLX8
|
Subunit of the Slx5-Slx8 substrate-specific ubiquitin ligase complex; stimulated by prior attachment of SUMO to the substrate |
|
| YGR260W | 0.07 |
TNA1
|
High affinity nicotinic acid plasma membrane permease, responsible for uptake of low levels of nicotinic acid; expression of the gene increases in the absence of extracellular nicotinic acid or para-aminobenzoate (PABA) |
|
| YBL015W | 0.07 |
ACH1
|
Acetyl-coA hydrolase, primarily localized to mitochondria; phosphorylated; required for acetate utilization and for diploid pseudohyphal growth |
|
| YER117W | 0.06 |
RPL23B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl23Ap and has similarity to E. coli L14 and rat L23 ribosomal proteins |
|
| YOR072W-A | 0.06 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YOR072W; originally identified by fungal homology and RT-PCR |
||
| YBR007C | 0.06 |
DSF2
|
Deletion suppressor of mpt5 mutation |
|
| YDR094W | 0.06 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified ORF DNF2 |
||
| YNL104C | 0.06 |
LEU4
|
Alpha-isopropylmalate synthase (2-isopropylmalate synthase); the main isozyme responsible for the first step in the leucine biosynthesis pathway |
|
| YNL042W | 0.06 |
BOP3
|
Protein of unknown function, potential Cdc28p substrate; overproduction suppresses a pam1 slv3 double null mutation and confers resistance to methylmercury |
|
| YGL045W | 0.06 |
RIM8
|
Protein of unknown function, involved in the proteolytic activation of Rim101p in response to alkaline pH; has similarity to A. nidulans PalF |
|
| YPR052C | 0.06 |
NHP6A
|
High-mobility group non-histone chromatin protein, functionally redundant with Nhp6Bp; homologous to mammalian high mobility group proteins 1 and 2; acts to recruit transcription factor Rcs1p to certain promoters |
|
| YML031C-A | 0.06 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF NDC1/YML031W; questionable ORF from MIPS |
||
| YMR275C | 0.06 |
BUL1
|
Ubiquitin-binding component of the Rsp5p E3-ubiquitin ligase complex, functional homolog of Bul2p, disruption causes temperature-sensitive growth, overexpression causes missorting of amino acid permeases |
|
| YDL211C | 0.06 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YBR112C | 0.06 |
CYC8
|
General transcriptional co-repressor, acts together with Tup1p; also acts as part of a transcriptional co-activator complex that recruits the SWI/SNF and SAGA complexes to promoters |
|
| YDR348C | 0.06 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery and bud neck; potential Cdc28p substrate |
||
| YDL235C | 0.06 |
YPD1
|
Phosphorelay intermediate protein, phosphorylated by the plasma membrane sensor Sln1p in response to osmotic stress and then in turn phosphorylates the response regulators Ssk1p in the cytosol and Skn7p in the nucleus |
|
| YPR006C | 0.06 |
ICL2
|
2-methylisocitrate lyase of the mitochondrial matrix, functions in the methylcitrate cycle to catalyze the conversion of 2-methylisocitrate to succinate and pyruvate; ICL2 transcription is repressed by glucose and induced by ethanol |
|
| YPR192W | 0.06 |
AQY1
|
Spore-specific water channel that mediates the transport of water across cell membranes, developmentally controlled; may play a role in spore maturation, probably by allowing water outflow, may be involved in freeze tolerance |
|
| YJR052W | 0.05 |
RAD7
|
Protein that recognizes and binds damaged DNA in an ATP-dependent manner (with Rad16p) during nucleotide excision repair; subunit of Nucleotide Excision Repair Factor 4 (NEF4) and the Elongin-Cullin-Socs (ECS) ligase complex |
|
| YMR070W | 0.05 |
MOT3
|
Nuclear transcription factor with two Cys2-His2 zinc fingers; involved in repression of a subset of hypoxic genes by Rox1p, repression of several DAN/TIR genes during aerobic growth, and repression of ergosterol biosynthetic genes |
|
| YCR025C | 0.05 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YCR025C is not an essential gene |
||
| YFR055W | 0.05 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YKL082C | 0.05 |
RRP14
|
Essential protein, constituent of 66S pre-ribosomal particles; interacts with proteins involved in ribosomal biogenesis and cell polarity; member of the SURF-6 family |
|
| YOR375C | 0.05 |
GDH1
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh3p; expression regulated by nitrogen and carbon sources |
|
| YHR208W | 0.05 |
BAT1
|
Mitochondrial branched-chain amino acid aminotransferase, homolog of murine ECA39; highly expressed during logarithmic phase and repressed during stationary phase |
|
| YNL014W | 0.05 |
HEF3
|
Translational elongation factor EF-3; paralog of YEF3 and member of the ABC superfamily; stimulates EF-1 alpha-dependent binding of aminoacyl-tRNA by the ribosome; normally expressed in zinc deficient cells |
|
| YOR369C | 0.05 |
RPS12
|
Protein component of the small (40S) ribosomal subunit; has similarity to rat ribosomal protein S12 |
|
| YMR052W | 0.05 |
FAR3
|
Protein involved in G1 cell cycle arrest in response to pheromone, in a pathway different from the Far1p-dependent pathway; interacts with Far7p, Far8p, Far9p, Far10p, and Far11p |
|
| YDL246C | 0.05 |
SOR2
|
Protein of unknown function, computational analysis of large-scale protein-protein interaction data suggests a possible role in fructose or mannose metabolism |
|
| YKL187C | 0.05 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in a phosphorylated state in highly purified mitochondria in high-throughput studies |
||
| YDR256C | 0.05 |
CTA1
|
Catalase A, breaks down hydrogen peroxide in the peroxisomal matrix formed by acyl-CoA oxidase (Pox1p) during fatty acid beta-oxidation |
|
| YGR086C | 0.05 |
PIL1
|
Primary component of eisosomes, which are large immobile cell cortex structures associated with endocytosis; null mutants show activation of Pkc1p/Ypk1p stress resistance pathways; detected in phosphorylated state in mitochondria |
|
| YKL109W | 0.05 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YMR135C | 0.05 |
GID8
|
Protein of unknown function, involved in proteasome-dependent catabolite inactivation of fructose-1,6-bisphosphatase; contains LisH and CTLH domains, like Vid30p; dosage-dependent regulator of START |
|
| YJL116C | 0.05 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YML054C-A | 0.05 |
Putative protein of unknown function |
||
| YDL086W | 0.05 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YDL086W is not an essential gene |
||
| YNL068C | 0.05 |
FKH2
|
Forkhead family transcription factor with a major role in the expression of G2/M phase genes; positively regulates transcriptional elongation; negative role in chromatin silencing at HML and HMR; substrate of the Cdc28p/Clb5p kinase |
|
| YLR254C | 0.05 |
NDL1
|
Homolog of nuclear distribution factor NudE, NUDEL; interacts with Pac1p and regulates dynein targeting to microtubule plus ends |
|
| YMR224C | 0.05 |
MRE11
|
Subunit of a complex with Rad50p and Xrs2p (MRX complex) that functions in repair of DNA double-strand breaks and in telomere stability, exhibits nuclease activity that appears to be required for MRX function; widely conserved |
|
| YDR155C | 0.05 |
CPR1
|
Cytoplasmic peptidyl-prolyl cis-trans isomerase (cyclophilin), catalyzes the cis-trans isomerization of peptide bonds N-terminal to proline residues; binds the drug cyclosporin A |
|
| YOR013W | 0.05 |
IRC11
|
Dubious opening reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YOR012C; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YMR049C | 0.05 |
ERB1
|
Protein required for maturation of the 25S and 5.8S ribosomal RNAs; constituent of 66S pre-ribosomal particles; homologous to mammalian Bop1 |
|
| YML007W | 0.05 |
YAP1
|
Basic leucine zipper (bZIP) transcription factor required for oxidative stress tolerance; activated by H2O2 through the multistep formation of disulfide bonds and transit from the cytoplasm to the nucleus; mediates resistance to cadmium |
|
| YHR048W | 0.04 |
YHK8
|
Presumed antiporter of the DHA1 family of multidrug resistance transporters; contains 12 predicted transmembrane spans; expression of gene is up-regulated in cells exhibiting reduced susceptibility to azoles |
|
| YPR054W | 0.04 |
SMK1
|
Middle sporulation-specific mitogen-activated protein kinase (MAPK) required for production of the outer spore wall layers; negatively regulates activity of the glucan synthase subunit Gsc2p |
|
| YDL066W | 0.04 |
IDP1
|
Mitochondrial NADP-specific isocitrate dehydrogenase, catalyzes the oxidation of isocitrate to alpha-ketoglutarate; not required for mitochondrial respiration and may function to divert alpha-ketoglutarate to biosynthetic processes |
|
| YOR198C | 0.04 |
BFR1
|
Component of mRNP complexes associated with polyribosomes; implicated in secretion and nuclear segregation; multicopy suppressor of BFA (Brefeldin A) sensitivity |
|
| YJL198W | 0.04 |
PHO90
|
Low-affinity phosphate transporter; deletion of pho84, pho87, pho89, pho90, and pho91 causes synthetic lethality; transcription independent of Pi and Pho4p activity; overexpression results in vigorous growth |
|
| YJR148W | 0.04 |
BAT2
|
Cytosolic branched-chain amino acid aminotransferase, homolog of murine ECA39; highly expressed during stationary phase and repressed during logarithmic phase |
|
| YDL139C | 0.04 |
SCM3
|
Nonhistone component of centromeric chromatin that binds stoichiometrically to CenH3-H4 histones, required for kinetochore assembly; contains nuclear export signal (NES); required for G2/M progression and localization of Cse4p |
|
| YLR312W-A | 0.04 |
MRPL15
|
Mitochondrial ribosomal protein of the large subunit |
|
| YFR029W | 0.04 |
PTR3
|
Component of the SPS plasma membrane amino acid sensor system (Ssy1p-Ptr3p-Ssy5p), which senses external amino acid concentration and transmits intracellular signals that result in regulation of expression of amino acid permease genes |
|
| YDR274C | 0.04 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOL060C | 0.04 |
MAM3
|
Protein required for normal mitochondrial morphology, has similarity to hemolysins |
|
| YLL043W | 0.04 |
FPS1
|
Plasma membrane channel, member of major intrinsic protein (MIP) family; involved in efflux of glycerol and in uptake of acetic acid and the trivalent metalloids arsenite and antimonite; phosphorylated by Hog1p MAPK under acetate stress |
|
| YHR199C-A | 0.04 |
Putative protein of unknown function; transcribed sequence appears to contain an intron |
Network of associatons between targets according to the STRING database.
First level regulatory network of ASG1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.3 | 1.9 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.1 | 0.5 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.1 | 0.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.4 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.1 | 0.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.5 | GO:0032973 | amino acid export(GO:0032973) |
| 0.1 | 0.2 | GO:0019543 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 0.1 | 0.4 | GO:0000947 | amino acid catabolic process to alcohol via Ehrlich pathway(GO:0000947) |
| 0.1 | 0.3 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.3 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 0.1 | 0.2 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) |
| 0.1 | 0.1 | GO:0016094 | polyprenol metabolic process(GO:0016093) polyprenol biosynthetic process(GO:0016094) dolichol metabolic process(GO:0019348) dolichol biosynthetic process(GO:0019408) |
| 0.1 | 0.3 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.1 | 0.2 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.1 | 0.2 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.2 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.2 | GO:0090282 | positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.0 | 0.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0009411 | response to UV(GO:0009411) cellular response to radiation(GO:0071478) cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.1 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.1 | GO:0072417 | cellular response to biotic stimulus(GO:0071216) response to cell cycle checkpoint signaling(GO:0072396) response to mitotic cell cycle checkpoint signaling(GO:0072414) response to spindle checkpoint signaling(GO:0072417) response to mitotic spindle checkpoint signaling(GO:0072476) response to mitotic cell cycle spindle assembly checkpoint signaling(GO:0072479) response to spindle assembly checkpoint signaling(GO:0072485) negative regulation of protein import into nucleus during spindle assembly checkpoint(GO:1901925) |
| 0.0 | 0.2 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.2 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.0 | 0.1 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) aggregation of unicellular organisms(GO:0098630) cell aggregation(GO:0098743) |
| 0.0 | 0.2 | GO:0001308 | negative regulation of chromatin silencing involved in replicative cell aging(GO:0001308) |
| 0.0 | 0.1 | GO:0051457 | negative regulation of transcription from RNA polymerase II promoter involved in meiotic cell cycle(GO:0010674) maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0036170 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0100070 | regulation of fatty acid biosynthetic process by transcription from RNA polymerase II promoter(GO:0100070) regulation of fatty acid biosynthetic process by regulation of transcription from RNA polymerase II promoter(GO:2000531) |
| 0.0 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000461) |
| 0.0 | 0.3 | GO:0022610 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0051597 | response to methylmercury(GO:0051597) cellular response to methylmercury(GO:0071406) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.5 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.1 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:1902931 | negative regulation of alcohol biosynthetic process(GO:1902931) |
| 0.0 | 0.3 | GO:0006407 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.0 | GO:0001315 | age-dependent response to reactive oxygen species(GO:0001315) |
| 0.0 | 0.2 | GO:0015758 | glucose transport(GO:0015758) |
| 0.0 | 0.1 | GO:0061186 | negative regulation of chromatin silencing at silent mating-type cassette(GO:0061186) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.0 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.0 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 0.0 | GO:0000304 | response to singlet oxygen(GO:0000304) |
| 0.0 | 0.1 | GO:0001666 | response to hypoxia(GO:0001666) chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.0 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.0 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 0.1 | GO:0090294 | nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) nitrogen catabolite activation of transcription(GO:0090294) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.1 | GO:0007234 | osmosensory signaling via phosphorelay pathway(GO:0007234) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.0 | GO:0023021 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.1 | GO:0071400 | response to lipid(GO:0033993) response to oleic acid(GO:0034201) response to fatty acid(GO:0070542) cellular response to lipid(GO:0071396) cellular response to fatty acid(GO:0071398) cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0000321 | re-entry into mitotic cell cycle after pheromone arrest(GO:0000321) |
| 0.0 | 0.1 | GO:0019740 | nitrogen utilization(GO:0019740) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.0 | GO:0043200 | response to amino acid(GO:0043200) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.0 | 0.2 | GO:0032545 | CURI complex(GO:0032545) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0030869 | RENT complex(GO:0030869) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.3 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0033698 | Rpd3L complex(GO:0033698) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) Rad17 RFC-like complex(GO:0031389) Elg1 RFC-like complex(GO:0031391) |
| 0.0 | 0.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.0 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0016406 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.1 | 0.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.4 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 0.1 | 0.4 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.2 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.0 | 0.1 | GO:0015606 | spermidine transmembrane transporter activity(GO:0015606) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0015489 | putrescine transmembrane transporter activity(GO:0015489) |
| 0.0 | 0.4 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.0 | 0.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.0 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.2 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.0 | 0.1 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.1 | GO:0005186 | mating pheromone activity(GO:0000772) pheromone activity(GO:0005186) |
| 0.0 | 0.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.0 | GO:0051880 | double-stranded telomeric DNA binding(GO:0003691) G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.0 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.0 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.0 | 0.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0033592 | RNA strand annealing activity(GO:0033592) annealing activity(GO:0097617) |
| 0.0 | 0.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.0 | 0.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |