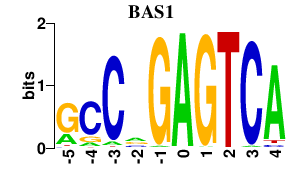
Results for BAS1
Z-value: 0.83
Transcription factors associated with BAS1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BAS1
|
S000001807 | Myb-related transcription factor |
Activity-expression correlation:
Activity profile of BAS1 motif
Sorted Z-values of BAS1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YMR096W | 6.93 |
SNZ1
|
Protein involved in vitamin B6 biosynthesis; member of a stationary phase-induced gene family; coregulated with SNO1; interacts with Sno1p and with Yhr198p, perhaps as a multiprotein complex containing other Snz and Sno proteins |
|
| YKL217W | 6.20 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YIL057C | 5.01 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YMR095C | 4.39 |
SNO1
|
Protein of unconfirmed function, involved in pyridoxine metabolism; expression is induced during stationary phase; forms a putative glutamine amidotransferase complex with Snz1p, with Sno1p serving as the glutaminase |
|
| YBR296C | 3.65 |
PHO89
|
Na+/Pi cotransporter, active in early growth phase; similar to phosphate transporters of Neurospora crassa; transcription regulated by inorganic phosphate concentrations and Pho4p |
|
| YJR078W | 3.60 |
BNA2
|
Putative tryptophan 2,3-dioxygenase or indoleamine 2,3-dioxygenase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p and Aft2p |
|
| YLR356W | 3.25 |
Putative protein of unknown function with similarity to SCM4; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR356W is not an essential gene |
||
| YLR174W | 2.95 |
IDP2
|
Cytosolic NADP-specific isocitrate dehydrogenase, catalyzes oxidation of isocitrate to alpha-ketoglutarate; levels are elevated during growth on non-fermentable carbon sources and reduced during growth on glucose |
|
| YMR206W | 2.92 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YDL222C | 2.80 |
FMP45
|
Integral membrane protein localized to mitochondria (untagged protein) and eisosomes, immobile patches at the cortex associated with endocytosis; sporulation and sphingolipid content are altered in mutants; has homologs SUR7 and YNL194C |
|
| YAR035W | 2.77 |
YAT1
|
Outer mitochondrial carnitine acetyltransferase, minor ethanol-inducible enzyme involved in transport of activated acyl groups from the cytoplasm into the mitochondrial matrix; phosphorylated |
|
| YMR189W | 2.15 |
GCV2
|
P subunit of the mitochondrial glycine decarboxylase complex, required for the catabolism of glycine to 5,10-methylene-THF; expression is regulated by levels of 5,10-methylene-THF in the cytoplasm |
|
| YGR065C | 2.06 |
VHT1
|
High-affinity plasma membrane H+-biotin (vitamin H) symporter; mutation results in fatty acid auxotrophy; 12 transmembrane domain containing major facilitator subfamily member; mRNA levels negatively regulated by iron deprivation and biotin |
|
| YMR081C | 2.02 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YGR067C | 1.94 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YNR073C | 1.91 |
Putative mannitol dehydrogenase |
||
| YHL036W | 1.85 |
MUP3
|
Low affinity methionine permease, similar to Mup1p |
|
| YJR077C | 1.75 |
MIR1
|
Mitochondrial phosphate carrier, imports inorganic phosphate into mitochondria; functionally redundant with Pic2p but more abundant than Pic2p under normal conditions; phosphorylated |
|
| YHR096C | 1.75 |
HXT5
|
Hexose transporter with moderate affinity for glucose, induced in the presence of non-fermentable carbon sources, induced by a decrease in growth rate, contains an extended N-terminal domain relative to other HXTs |
|
| YER091C-A | 1.71 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YIL099W | 1.70 |
SGA1
|
Intracellular sporulation-specific glucoamylase involved in glycogen degradation; induced during starvation of a/a diploids late in sporulation, but dispensable for sporulation |
|
| YER069W | 1.69 |
ARG5,6
|
Protein that is processed in the mitochondrion to yield acetylglutamate kinase and N-acetyl-gamma-glutamyl-phosphate reductase, which catalyze the 2nd and 3rd steps in arginine biosynthesis; enzymes form a complex with Arg2p |
|
| YDR019C | 1.68 |
GCV1
|
T subunit of the mitochondrial glycine decarboxylase complex, required for the catabolism of glycine to 5,10-methylene-THF; expression is regulated by levels of levels of 5,10-methylene-THF in the cytoplasm |
|
| YER068C-A | 1.61 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YKL163W | 1.61 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YFR053C | 1.60 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YCL030C | 1.58 |
HIS4
|
Multifunctional enzyme containing phosphoribosyl-ATP pyrophosphatase, phosphoribosyl-AMP cyclohydrolase, and histidinol dehydrogenase activities; catalyzes the second, third, ninth and tenth steps in histidine biosynthesis |
|
| YGL062W | 1.52 |
PYC1
|
Pyruvate carboxylase isoform, cytoplasmic enzyme that converts pyruvate to oxaloacetate; highly similar to isoform Pyc2p but differentially regulated; mutations in the human homolog are associated with lactic acidosis |
|
| YPL262W | 1.47 |
FUM1
|
Fumarase, converts fumaric acid to L-malic acid in the TCA cycle; cytosolic and mitochondrial localization determined by the N-terminal mitochondrial targeting sequence and protein conformation; phosphorylated in mitochondria |
|
| YOR152C | 1.42 |
Putative protein of unknown function; has no similarity to any known protein; YOR152C is not an essential gene |
||
| YMR120C | 1.39 |
ADE17
|
Enzyme of 'de novo' purine biosynthesis containing both 5-aminoimidazole-4-carboxamide ribonucleotide transformylase and inosine monophosphate cyclohydrolase activities, isozyme of Ade16p; ade16 ade17 mutants require adenine and histidine |
|
| YHR053C | 1.37 |
CUP1-1
|
Metallothionein, binds copper and mediates resistance to high concentrations of copper and cadmium; locus is variably amplified in different strains, with two copies, CUP1-1 and CUP1-2, in the genomic sequence reference strain S288C |
|
| YER084W | 1.37 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YFL030W | 1.32 |
AGX1
|
Alanine:glyoxylate aminotransferase (AGT), catalyzes the synthesis of glycine from glyoxylate, which is one of three pathways for glycine biosynthesis in yeast; has similarity to mammalian and plant alanine:glyoxylate aminotransferases |
|
| YOL084W | 1.32 |
PHM7
|
Protein of unknown function, expression is regulated by phosphate levels; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery and vacuole |
|
| YGL184C | 1.28 |
STR3
|
Cystathionine beta-lyase, converts cystathionine into homocysteine |
|
| YIL101C | 1.22 |
XBP1
|
Transcriptional repressor that binds to promoter sequences of the cyclin genes, CYS3, and SMF2; expression is induced by stress or starvation during mitosis, and late in meiosis; member of the Swi4p/Mbp1p family; potential Cdc28p substrate |
|
| YER162C | 1.18 |
RAD4
|
Protein that recognizes and binds damaged DNA (with Rad23p) during nucleotide excision repair; subunit of Nuclear Excision Repair Factor 2 (NEF2); homolog of human XPC protein |
|
| YBR230C | 1.14 |
OM14
|
Integral mitochondrial outer membrane protein; abundance is decreased in cells grown in glucose relative to other carbon sources; appears to contain 3 alpha-helical transmembrane segments; ORF encodes a 97-basepair intron |
|
| YLR164W | 1.12 |
Mitochondrial inner membrane of unknown function; similar to Tim18p and Sdh4p; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
||
| YMR056C | 1.12 |
AAC1
|
Mitochondrial inner membrane ADP/ATP translocator, exchanges cytosolic ADP for mitochondrially synthesized ATP; phosphorylated; Aac1p is a minor isoform while Pet9p is the major ADP/ATP translocator |
|
| YGR144W | 1.10 |
THI4
|
Thiazole synthase, catalyzes formation of the thiazole moiety of thiamin pyrophosphate; required for thiamine biosynthesis and for mitochondrial genome stability |
|
| YJL130C | 1.09 |
URA2
|
Bifunctional carbamoylphosphate synthetase (CPSase)-aspartate transcarbamylase (ATCase), catalyzes the first two enzymatic steps in the de novo biosynthesis of pyrimidines; both activities are subject to feedback inhibition by UTP |
|
| YPL278C | 1.09 |
Putative protein of unknown function; gene expression regulated by copper levels |
||
| YJR150C | 1.08 |
DAN1
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YER039C-A | 1.05 |
Putative protein of unknown function; YER039C-A is not an essential gene |
||
| YER076C | 1.05 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; analysis of HA-tagged protein suggests a membrane localization |
||
| YJR046W | 1.03 |
TAH11
|
DNA replication licensing factor, required for pre-replication complex assembly |
|
| YLL019C | 1.02 |
KNS1
|
Nonessential putative protein kinase of unknown cellular role; member of the LAMMER family of protein kinases, which are serine/threonine kinases also capable of phosphorylating tyrosine residues |
|
| YMR118C | 1.02 |
Protein of unknown function with similarity to succinate dehydrogenase cytochrome b subunit; YMR118C is not an essential gene |
||
| YKL016C | 1.02 |
ATP7
|
Subunit d of the stator stalk of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YMR107W | 1.01 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YMR188C | 0.98 |
MRPS17
|
Mitochondrial ribosomal protein of the small subunit |
|
| YLR058C | 0.97 |
SHM2
|
Cytosolic serine hydroxymethyltransferase, converts serine to glycine plus 5,10 methylenetetrahydrofolate; major isoform involved in generating precursors for purine, pyrimidine, amino acid, and lipid biosynthesis |
|
| YOR128C | 0.97 |
ADE2
|
Phosphoribosylaminoimidazole carboxylase, catalyzes a step in the 'de novo' purine nucleotide biosynthetic pathway; red pigment accumulates in mutant cells deprived of adenine |
|
| YNR050C | 0.95 |
LYS9
|
Saccharopine dehydrogenase (NADP+, L-glutamate-forming); catalyzes the formation of saccharopine from alpha-aminoadipate 6-semialdehyde, which is the seventh step in lysine biosynthesis pathway |
|
| YPL250C | 0.94 |
ICY2
|
Protein of unknown function; mobilized into polysomes upon a shift from a fermentable to nonfermentable carbon source; potential Cdc28p substrate |
|
| YLR122C | 0.93 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR123C |
||
| YGR043C | 0.93 |
NQM1
|
Protein of unknown function; transcription is repressed by Mot1p and induced by alpha-factor and during diauxic shift; null mutant non-quiescent cells exhibit reduced reproductive capacity |
|
| YDL170W | 0.93 |
UGA3
|
Transcriptional activator necessary for gamma-aminobutyrate (GABA)-dependent induction of GABA genes (such as UGA1, UGA2, UGA4); zinc-finger transcription factor of the Zn(2)-Cys(6) binuclear cluster domain type; localized to the nucleus |
|
| YPR026W | 0.92 |
ATH1
|
Acid trehalase required for utilization of extracellular trehalose |
|
| YBR056W-A | 0.92 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR056C-B |
||
| YER076W-A | 0.89 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YER076C |
||
| YDR263C | 0.88 |
DIN7
|
Mitochondrial nuclease functioning in DNA repair and replication, modulates the stability of the mitochondrial genome, induced by exposure to mutagens, also induced during meiosis at a time nearly coincident with commitment to recombination |
|
| YPL135W | 0.84 |
ISU1
|
Conserved protein of the mitochondrial matrix, performs a scaffolding function during assembly of iron-sulfur clusters, interacts physically and functionally with yeast frataxin (Yfh1p); isu1 isu2 double mutant is inviable |
|
| YER061C | 0.83 |
CEM1
|
Mitochondrial beta-keto-acyl synthase with possible role in fatty acid synthesis; required for mitochondrial respiration |
|
| YLR123C | 0.82 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR122C; contains characteristic aminoacyl-tRNA motif |
||
| YDR186C | 0.81 |
Putative protein of unknown function; may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YGR087C | 0.79 |
PDC6
|
Minor isoform of pyruvate decarboxylase, decarboxylates pyruvate to acetaldehyde, involved in amino acid catabolism; transcription is glucose- and ethanol-dependent, and is strongly induced during sulfur limitation |
|
| YPR027C | 0.78 |
Putative protein of unknown function |
||
| YER101C | 0.77 |
AST2
|
Protein that may have a role in targeting of plasma membrane [H+]ATPase (Pma1p) to the plasma membrane, as suggested by analysis of genetic interactions |
|
| YOR343C | 0.74 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YJR038C | 0.74 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJR154W | 0.73 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YHR092C | 0.72 |
HXT4
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YOR227W | 0.72 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YNL144C | 0.72 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YNL144C is not an essential gene |
||
| YOR381W | 0.71 |
FRE3
|
Ferric reductase, reduces siderophore-bound iron prior to uptake by transporters; expression induced by low iron levels |
|
| YJR146W | 0.71 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene HMS2 |
||
| YOL085W-A | 0.71 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YOL085C |
||
| YOR221C | 0.70 |
MCT1
|
Predicted malonyl-CoA:ACP transferase, putative component of a type-II mitochondrial fatty acid synthase that produces intermediates for phospholipid remodeling |
|
| YJR045C | 0.70 |
SSC1
|
Mitochondrial matrix ATPase, subunit of the presequence translocase-associated protein import motor (PAM) and of SceI endonuclease; involved in protein folding and translocation into the matrix; phosphorylated; member of HSP70 family |
|
| YDL130W-A | 0.69 |
STF1
|
Protein involved in regulation of the mitochondrial F1F0-ATP synthase; Stf1p and Stf2p may act as stabilizing factors that enhance inhibitory action of the Inh1p protein |
|
| YMR135C | 0.68 |
GID8
|
Protein of unknown function, involved in proteasome-dependent catabolite inactivation of fructose-1,6-bisphosphatase; contains LisH and CTLH domains, like Vid30p; dosage-dependent regulator of START |
|
| YDR453C | 0.65 |
TSA2
|
Stress inducible cytoplasmic thioredoxin peroxidase; cooperates with Tsa1p in the removal of reactive oxygen, nitrogen and sulfur species using thioredoxin as hydrogen donor; deletion enhances the mutator phenotype of tsa1 mutants |
|
| YDR463W | 0.65 |
STP1
|
Transcription factor, activated by proteolytic processing in response to signals from the SPS sensor system for external amino acids; activates transcription of amino acid permease genes and may have a role in tRNA processing |
|
| YDL234C | 0.65 |
GYP7
|
GTPase-activating protein for yeast Rab family members including: Ypt7p (most effective), Ypt1p, Ypt31p, and Ypt32p (in vitro); involved in vesicle mediated protein trafficking |
|
| YKR080W | 0.64 |
MTD1
|
NAD-dependent 5,10-methylenetetrahydrafolate dehydrogenase, plays a catalytic role in oxidation of cytoplasmic one-carbon units; expression is regulated by Bas1p and Bas2p, repressed by adenine, and may be induced by inositol and choline |
|
| YHR139C | 0.64 |
SPS100
|
Protein required for spore wall maturation; expressed during sporulation; may be a component of the spore wall |
|
| YMR182C | 0.63 |
RGM1
|
Putative transcriptional repressor with proline-rich zinc fingers; overproduction impairs cell growth |
|
| YGL236C | 0.63 |
MTO1
|
Mitochondrial protein, forms a heterodimer complex with Mss1p that performs the 5-carboxymethylaminomethyl modification of the wobble uridine base in mitochondrial tRNAs; required for respiration in paromomycin-resistant 15S rRNA mutants |
|
| YMR194C-B | 0.63 |
Putative protein of unknown function |
||
| YOR011W-A | 0.62 |
Putative protein of unknown function |
||
| YOR072W | 0.62 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOR072W-A; diploid deletion strains are methotrexate, paraquat and wortmannin sensitive |
||
| YHR150W | 0.59 |
PEX28
|
Peroxisomal integral membrane peroxin, involved in the regulation of peroxisomal size, number and distribution; genetic interactions suggest that Pex28p and Pex29p act at steps upstream of those mediated by Pex30p, Pex31p, and Pex32p |
|
| YDR298C | 0.59 |
ATP5
|
Subunit 5 of the stator stalk of mitochondrial F1F0 ATP synthase, which is an evolutionarily conserved enzyme complex required for ATP synthesis; homologous to bovine subunit OSCP (oligomycin sensitivity-conferring protein); phosphorylated |
|
| YJR160C | 0.59 |
MPH3
|
Alpha-glucoside permease, transports maltose, maltotriose, alpha-methylglucoside, and turanose; identical to Mph2p; encoded in a subtelomeric position in a region likely to have undergone duplication |
|
| YJR095W | 0.58 |
SFC1
|
Mitochondrial succinate-fumarate transporter, transports succinate into and fumarate out of the mitochondrion; required for ethanol and acetate utilization |
|
| YLR125W | 0.58 |
Putative protein of unknown function; mutant has decreased Ty3 transposition; YLR125W is not an essential gene |
||
| YOL154W | 0.58 |
ZPS1
|
Putative GPI-anchored protein; transcription is induced under low-zinc conditions, as mediated by the Zap1p transcription factor, and at alkaline pH |
|
| YDL244W | 0.57 |
THI13
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YNL333W | 0.57 |
SNZ2
|
Member of a stationary phase-induced gene family; transcription of SNZ2 is induced prior to diauxic shift, and also in the absence of thiamin in a Thi2p-dependent manner; forms a coregulated gene pair with SNO2; interacts with Thi11p |
|
| YKR009C | 0.57 |
FOX2
|
Multifunctional enzyme of the peroxisomal fatty acid beta-oxidation pathway; has 3-hydroxyacyl-CoA dehydrogenase and enoyl-CoA hydratase activities |
|
| YHR145C | 0.56 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJR152W | 0.55 |
DAL5
|
Allantoin permease; ureidosuccinate permease; expression is constitutive but sensitive to nitrogen catabolite repression |
|
| YPR013C | 0.55 |
Putative zinc finger protein; YPR013C is not an essential gene |
||
| YIL112W | 0.55 |
HOS4
|
Subunit of the Set3 complex, which is a meiotic-specific repressor of sporulation specific genes that contains deacetylase activity; potential Cdc28p substrate |
|
| YIL125W | 0.55 |
KGD1
|
Component of the mitochondrial alpha-ketoglutarate dehydrogenase complex, which catalyzes a key step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation of alpha-ketoglutarate to form succinyl-CoA |
|
| YOL078W | 0.54 |
AVO1
|
Component of a membrane-bound complex containing the Tor2p kinase and other proteins, which may have a role in regulation of cell growth |
|
| YFL052W | 0.53 |
Putative zinc cluster protein that contains a DNA binding domain; null mutant sensitive to calcofluor white, low osmolarity and heat, suggesting a role for YFL052Wp in cell wall integrity |
||
| YMR306W | 0.53 |
FKS3
|
Protein involved in spore wall assembly, has similarity to 1,3-beta-D-glucan synthase catalytic subunits Fks1p and Gsc2p; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YDR490C | 0.53 |
PKH1
|
Serine/threonine protein kinase involved in sphingolipid-mediated signaling pathway that controls endocytosis; activates Ypk1p and Ykr2p, components of signaling cascade required for maintenance of cell wall integrity; redundant with Pkh2p |
|
| YJL127C-B | 0.52 |
Putative protein of unknown function; identified based on homology to the filamentous fungus, Ashbya gossypii |
||
| YFL059W | 0.52 |
SNZ3
|
Member of a stationary phase-induced gene family; transcription of SNZ2 is induced prior to diauxic shift, and also in the absence of thiamin in a Thi2p-dependent manner; forms a coregulated gene pair with SNO3 |
|
| YNL142W | 0.52 |
MEP2
|
Ammonium permease involved in regulation of pseudohyphal growth; belongs to a ubiquitous family of cytoplasmic membrane proteins that transport only ammonium (NH4+); expression is under the nitrogen catabolite repression regulation |
|
| YDL221W | 0.51 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the 3' end of essential gene CDC13 |
||
| YOL058W | 0.51 |
ARG1
|
Arginosuccinate synthetase, catalyzes the formation of L-argininosuccinate from citrulline and L-aspartate in the arginine biosynthesis pathway; potential Cdc28p substrate |
|
| YBR145W | 0.50 |
ADH5
|
Alcohol dehydrogenase isoenzyme V; involved in ethanol production |
|
| YBL088C | 0.50 |
TEL1
|
Protein kinase primarily involved in telomere length regulation; contributes to cell cycle checkpoint control in response to DNA damage; functionally redundant with Mec1p; homolog of human ataxia telangiectasia (ATM) gene |
|
| YFR047C | 0.50 |
BNA6
|
Quinolinate phosphoribosyl transferase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p |
|
| YOR071C | 0.50 |
NRT1
|
High-affinity nicotinamide riboside transporter; also transports thiamine with low affinity; shares sequence similarity with Thi7p and Thi72p; proposed to be involved in 5-fluorocytosine sensitivity |
|
| YAR015W | 0.50 |
ADE1
|
N-succinyl-5-aminoimidazole-4-carboxamide ribotide (SAICAR) synthetase, required for 'de novo' purine nucleotide biosynthesis; red pigment accumulates in mutant cells deprived of adenine |
|
| YBR230W-A | 0.50 |
Putative protein of unknown function |
||
| YJL037W | 0.48 |
IRC18
|
Putative protein of unknown function; expression induced in respiratory-deficient cells and in carbon-limited chemostat cultures; similar to adjacent ORF, YJL038C; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YGR191W | 0.48 |
HIP1
|
High-affinity histidine permease, also involved in the transport of manganese ions |
|
| YNL025C | 0.47 |
SSN8
|
Cyclin-like component of the RNA polymerase II holoenzyme, involved in phosphorylation of the RNA polymerase II C-terminal domain; involved in glucose repression and telomere maintenance |
|
| YIL100W | 0.47 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps the dubious ORF YIL100C-A |
||
| YIR017C | 0.46 |
MET28
|
Transcriptional activator in the Cbf1p-Met4p-Met28p complex, participates in the regulation of sulfur metabolism |
|
| YNL220W | 0.46 |
ADE12
|
Adenylosuccinate synthase, catalyzes the first step in synthesis of adenosine monophosphate from inosine 5'monophosphate during purine nucleotide biosynthesis; exhibits binding to single-stranded autonomously replicating (ARS) core sequence |
|
| YLR228C | 0.45 |
ECM22
|
Sterol regulatory element binding protein, regulates transcription of the sterol biosynthetic genes ERG2 and ERG3; member of the fungus-specific Zn[2]-Cys[6] binuclear cluster family of transcription factors; homologous to Upc2p |
|
| YJR039W | 0.45 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YDL233W | 0.45 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YDL233W is not an essential gene |
||
| YLR431C | 0.44 |
ATG23
|
Peripheral membrane protein required for the cytoplasm-to-vacuole targeting (Cvt) pathway; cycles between the pre-autophagosome (PAS) and non-PAS locations; forms a complex with Atg9p and Atg27p |
|
| YDR441C | 0.44 |
APT2
|
Apparent pseudogene, not transcribed or translated under normal conditions; encodes a protein with similarity to adenine phosphoribosyltransferase, but artificially expressed protein exhibits no enzymatic activity |
|
| YDL210W | 0.43 |
UGA4
|
Permease that serves as a gamma-aminobutyrate (GABA) transport protein involved in the utilization of GABA as a nitrogen source; catalyzes the transport of putrescine and delta-aminolevulinic acid (ALA); localized to the vacuolar membrane |
|
| YBR182C | 0.42 |
SMP1
|
Putative transcription factor involved in regulating the response to osmotic stress; member of the MADS-box family of transcription factors |
|
| YBL102W | 0.42 |
SFT2
|
Non-essential tetra-spanning membrane protein found mostly in the late Golgi, can suppress some sed5 alleles; may be part of the transport machinery, but precise function is unknown; similar to mammalian syntaxin 5 |
|
| YLR053C | 0.42 |
Putative protein of unknown function |
||
| YNL334C | 0.42 |
SNO2
|
Protein of unknown function, nearly identical to Sno3p; expression is induced before the diauxic shift and also in the absence of thiamin |
|
| YBL101C | 0.41 |
ECM21
|
Non-essential protein of unknown function; promoter contains several Gcn4p binding elements |
|
| YFR034C | 0.41 |
PHO4
|
Basic helix-loop-helix (bHLH) transcription factor of the myc-family; binds cooperatively with Pho2p to the PHO5 promoter; function is regulated by phosphorylation at multiple sites and by phosphate availability |
|
| YJR037W | 0.41 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified gene HUL4/YJR036C; YJR037W is not an essential gene. |
||
| YDL194W | 0.41 |
SNF3
|
Plasma membrane glucose sensor that regulates glucose transport; has 12 predicted transmembrane segments; long cytoplasmic C-terminal tail is required for low glucose induction of hexose transporter genes HXT2 and HXT4 |
|
| YBR250W | 0.41 |
SPO23
|
Protein of unknown function; associates with meiosis-specific protein Spo1p |
|
| YIL100C-A | 0.40 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL186C | 0.40 |
TPN1
|
Plasma membrane pyridoxine (vitamin B6) transporter; member of the purine-cytosine permease subfamily within the major facilitator superfamily; proton symporter with similarity to Fcy21p, Fcy2p, and Fcy22p |
|
| YGL219C | 0.40 |
MDM34
|
Mitochondrial outer membrane protein, required for normal mitochondrial morphology and inheritance; localizes to dots on the mitochondrial surface near mtDNA nucleoids; interacts genetically with MDM31 and MDM32 |
|
| YGL056C | 0.39 |
SDS23
|
One of two S. cerevisiae homologs (Sds23p and Sds24p) of the Schizosaccharomyces pombe Sds23 protein, which genetic studies have implicated in APC/cyclosome regulation |
|
| YLR311C | 0.39 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YAR050W | 0.39 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YHR078W | 0.38 |
High osmolarity-regulated gene of unknown function |
||
| YPR009W | 0.38 |
SUT2
|
Putative transcription factor; multicopy suppressor of mutations that cause low activity of the cAMP/protein kinase A pathway; highly similar to Sut1p |
|
| YKR058W | 0.38 |
GLG1
|
Self-glucosylating initiator of glycogen synthesis, also glucosylates n-dodecyl-beta-D-maltoside; similar to mammalian glycogenin |
|
| YNL311C | 0.38 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; putative F-box protein; analysis of integrated high-throughput datasets predicts involvement in ubiquitin-dependent protein catabolism |
||
| YOL147C | 0.38 |
PEX11
|
Peroxisomal membrane protein required for peroxisome proliferation and medium-chain fatty acid oxidation, most abundant protein in the peroxisomal membrane, regulated by Adr1p and Pip2p-Oaf1p, promoter contains ORE and UAS1-like elements |
|
| YHL016C | 0.38 |
DUR3
|
Plasma membrane transporter for both urea and polyamines, expression is highly sensitive to nitrogen catabolite repression and induced by allophanate, the last intermediate of the allantoin degradative pathway |
|
| YNR059W | 0.37 |
MNT4
|
Putative alpha-1,3-mannosyltransferase, not required for protein O-glycosylation |
|
| YFL060C | 0.37 |
SNO3
|
Protein of unknown function, nearly identical to Sno2p; expression is induced before the diauxic shift and also in the absence of thiamin |
|
| YLR124W | 0.37 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YNR001C | 0.36 |
CIT1
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate; the rate-limiting enzyme of the TCA cycle; nuclear encoded mitochondrial protein |
|
| YMR169C | 0.35 |
ALD3
|
Cytoplasmic aldehyde dehydrogenase, involved in beta-alanine synthesis; uses NAD+ as the preferred coenzyme; very similar to Ald2p; expression is induced by stress and repressed by glucose |
|
| YNR063W | 0.34 |
Putative zinc-cluster protein of unknown function |
||
| YGR190C | 0.33 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene HIP1/YGR191W |
||
| YGR204W | 0.33 |
ADE3
|
Cytoplasmic trifunctional enzyme C1-tetrahydrofolate synthase, involved in single carbon metabolism and required for biosynthesis of purines, thymidylate, methionine, and histidine |
|
| YDR273W | 0.33 |
DON1
|
Meiosis-specific component of the spindle pole body, part of the leading edge protein (LEP) coat, forms a ring-like structure at the leading edge of the prospore membrane during meiosis II |
|
| YGL197W | 0.32 |
MDS3
|
Protein with an N-terminal kelch-like domain, putative negative regulator of early meiotic gene expression; required, with Pmd1p, for growth under alkaline conditions |
|
| YMR280C | 0.32 |
CAT8
|
Zinc cluster transcriptional activator necessary for derepression of a variety of genes under non-fermentative growth conditions, active after diauxic shift, binds carbon source responsive elements |
|
| YPL107W | 0.32 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YPL107W is not an essential gene |
||
| YMR195W | 0.31 |
ICY1
|
Protein of unknown function, required for viability in rich media of cells lacking mitochondrial DNA; mutants have an invasive growth defect with elongated morphology; induced by amino acid starvation |
|
| YDL054C | 0.31 |
MCH1
|
Protein with similarity to mammalian monocarboxylate permeases, which are involved in transport of monocarboxylic acids across the plasma membrane; mutant is not deficient in monocarboxylate transport |
|
| YDR216W | 0.31 |
ADR1
|
Carbon source-responsive zinc-finger transcription factor, required for transcription of the glucose-repressed gene ADH2, of peroxisomal protein genes, and of genes required for ethanol, glycerol, and fatty acid utilization |
|
| YPR184W | 0.31 |
GDB1
|
Glycogen debranching enzyme containing glucanotranferase and alpha-1,6-amyloglucosidase activities, required for glycogen degradation; phosphorylated in mitochondria |
|
| YMR058W | 0.30 |
FET3
|
Ferro-O2-oxidoreductase required for high-affinity iron uptake and involved in mediating resistance to copper ion toxicity, belongs to class of integral membrane multicopper oxidases |
|
| YHL037C | 0.30 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNL104C | 0.30 |
LEU4
|
Alpha-isopropylmalate synthase (2-isopropylmalate synthase); the main isozyme responsible for the first step in the leucine biosynthesis pathway |
|
| YJR036C | 0.29 |
HUL4
|
Protein with similarity to hect domain E3 ubiquitin-protein ligases, not essential for viability |
|
| YDR525W-A | 0.29 |
SNA2
|
Protein of unknown function, has similarity to Pmp3p, which is involved in cation transport; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YIL023C | 0.29 |
YKE4
|
Zinc transporter; localizes to the ER; null mutant is sensitive to calcofluor white, leads to zinc accumulation in cytosol; ortholog of the mouse KE4 and member of the ZIP (ZRT, IRT-like Protein) family |
|
| YLR092W | 0.29 |
SUL2
|
High affinity sulfate permease; sulfate uptake is mediated by specific sulfate transporters Sul1p and Sul2p, which control the concentration of endogenous activated sulfate intermediates |
|
| YDR171W | 0.29 |
HSP42
|
Small heat shock protein (sHSP) with chaperone activity; forms barrel-shaped oligomers that suppress unfolded protein aggregation; involved in cytoskeleton reorganization after heat shock |
|
| YOR178C | 0.29 |
GAC1
|
Regulatory subunit for Glc7p type-1 protein phosphatase (PP1), tethers Glc7p to Gsy2p glycogen synthase, binds Hsf1p heat shock transcription factor, required for induction of some HSF-regulated genes under heat shock |
|
| YMR179W | 0.28 |
SPT21
|
Protein required for normal transcription at several loci including HTA2-HTB2 and HHF2-HHT2, but not required at the other histone loci; functionally related to Spt10p; involved in telomere maintenance |
|
| YDR409W | 0.28 |
SIZ1
|
SUMO/Smt3 ligase that promotes the attachment of sumo (Smt3p; small ubiquitin-related modifier) to proteins; binds Ubc9p and may bind septins; specifically required for sumoylation of septins in vivo; localized to the septin ring |
|
| YEL021W | 0.28 |
URA3
|
Orotidine-5'-phosphate (OMP) decarboxylase, catalyzes the sixth enzymatic step in the de novo biosynthesis of pyrimidines, converting OMP into uridine monophosphate (UMP); converts 5-FOA into 5-fluorouracil, a toxic compound |
|
| YNL143C | 0.28 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YER084W-A | 0.28 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDL079C | 0.27 |
MRK1
|
Glycogen synthase kinase 3 (GSK-3) homolog; one of four GSK-3 homologs in S. cerevisiae that function to activate Msn2p-dependent transcription of stress responsive genes and that function in protein degradation |
|
| YDR258C | 0.27 |
HSP78
|
Oligomeric mitochondrial matrix chaperone that cooperates with Ssc1p in mitochondrial thermotolerance after heat shock; prevents the aggregation of misfolded matrix proteins; component of the mitochondrial proteolysis system |
|
| YAR047C | 0.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR028W | 0.27 |
YOP1
|
Membrane protein that interacts with Yip1p to mediate membrane traffic; overexpression results in cell death and accumulation of internal cell membranes; regulates vesicular traffic in stressed cells |
|
| YPL027W | 0.27 |
SMA1
|
Protein of unknown function involved in the assembly of the prospore membrane during sporulation |
|
| YNL250W | 0.27 |
RAD50
|
Subunit of MRX complex, with Mre11p and Xrs2p, involved in processing double-strand DNA breaks in vegetative cells, initiation of meiotic DSBs, telomere maintenance, and nonhomologous end joining |
|
| YKL171W | 0.27 |
Putative protein of unknown function; predicted protein kinase; implicated in proteasome function; epitope-tagged protein localizes to the cytoplasm |
||
| YGR088W | 0.27 |
CTT1
|
Cytosolic catalase T, has a role in protection from oxidative damage by hydrogen peroxide |
|
| YJR153W | 0.26 |
PGU1
|
Endo-polygalacturonase, pectolytic enzyme that hydrolyzes the alpha-1,4-glycosidic bonds in the rhamnogalacturonan chains in pectins |
|
| YDR533C | 0.26 |
HSP31
|
Possible chaperone and cysteine protease with similarity to E. coli Hsp31; member of the DJ-1/ThiJ/PfpI superfamily, which includes human DJ-1 involved in Parkinson's disease; exists as a dimer and contains a putative metal-binding site |
|
| YOL081W | 0.26 |
IRA2
|
GTPase-activating protein that negatively regulates RAS by converting it from the GTP- to the GDP-bound inactive form, required for reducing cAMP levels under nutrient limiting conditions, has similarity to Ira1p and human neurofibromin |
|
| YML116W-A | 0.26 |
Putative protein of unknown function |
||
| YMR271C | 0.26 |
URA10
|
Minor orotate phosphoribosyltransferase (OPRTase) isozyme that catalyzes the fifth enzymatic step in the de novo biosynthesis of pyrimidines, converting orotate into orotidine-5'-phosphate; major OPRTase encoded by URA5 |
|
| YIL117C | 0.26 |
PRM5
|
Pheromone-regulated protein, predicted to have 1 transmembrane segment; induced during cell integrity signaling |
Network of associatons between targets according to the STRING database.
First level regulatory network of BAS1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 12.9 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 1.6 | 6.3 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.9 | 3.7 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.9 | 2.8 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.8 | 4.2 | GO:0034354 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.7 | 2.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.6 | 3.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.6 | 1.7 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.5 | 2.3 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.4 | 1.7 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.4 | 2.0 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.4 | 4.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.4 | 3.0 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.3 | 1.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.3 | 1.0 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.3 | 0.9 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 0.2 | 0.7 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.2 | 1.0 | GO:0019346 | transsulfuration(GO:0019346) |
| 0.2 | 1.0 | GO:0045980 | negative regulation of nucleotide metabolic process(GO:0045980) |
| 0.2 | 0.9 | GO:0009450 | gamma-aminobutyric acid metabolic process(GO:0009448) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.2 | 0.9 | GO:0046033 | AMP biosynthetic process(GO:0006167) AMP metabolic process(GO:0046033) |
| 0.2 | 0.4 | GO:0043419 | urea catabolic process(GO:0043419) |
| 0.2 | 1.7 | GO:0015758 | glucose transport(GO:0015758) |
| 0.2 | 0.8 | GO:0034764 | positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) positive regulation of sterol transport(GO:0032373) positive regulation of transmembrane transport(GO:0034764) regulation of sterol import by regulation of transcription from RNA polymerase II promoter(GO:0035968) positive regulation of sterol import by positive regulation of transcription from RNA polymerase II promoter(GO:0035969) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.2 | 0.8 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.2 | 0.6 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.2 | 1.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 0.5 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 1.9 | GO:0009228 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.2 | 0.5 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.2 | 1.0 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.2 | 0.5 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 0.2 | 0.5 | GO:0045117 | azole transport(GO:0045117) |
| 0.2 | 0.5 | GO:0043388 | regulation of sulfur amino acid metabolic process(GO:0031335) positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.1 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.7 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.5 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.1 | 1.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.0 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.6 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.3 | GO:1904951 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 0.1 | 1.1 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.1 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.3 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) |
| 0.1 | 0.5 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) |
| 0.1 | 1.3 | GO:0034440 | fatty acid oxidation(GO:0019395) lipid oxidation(GO:0034440) |
| 0.1 | 2.1 | GO:0072350 | tricarboxylic acid metabolic process(GO:0072350) |
| 0.1 | 0.9 | GO:0015688 | iron chelate transport(GO:0015688) |
| 0.1 | 0.1 | GO:0034225 | cellular response to zinc ion starvation(GO:0034224) regulation of transcription from RNA polymerase II promoter in response to zinc ion starvation(GO:0034225) |
| 0.1 | 0.7 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.1 | GO:0051038 | negative regulation of transcription involved in meiotic cell cycle(GO:0051038) |
| 0.1 | 0.5 | GO:0010672 | regulation of transcription from RNA polymerase II promoter involved in meiotic cell cycle(GO:0010672) |
| 0.1 | 1.2 | GO:0015749 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.1 | 0.3 | GO:0042744 | hydrogen peroxide metabolic process(GO:0042743) hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.3 | GO:0072530 | cytosine transport(GO:0015856) purine-containing compound transmembrane transport(GO:0072530) |
| 0.1 | 0.3 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) |
| 0.1 | 0.5 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 0.2 | GO:0043966 | histone H3 acetylation(GO:0043966) histone H3-K9 acetylation(GO:0043970) histone H3-K14 acetylation(GO:0044154) histone H3-K9 modification(GO:0061647) |
| 0.1 | 0.4 | GO:0044205 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.2 | GO:0018209 | peptidyl-serine phosphorylation(GO:0018105) peptidyl-serine modification(GO:0018209) |
| 0.1 | 0.3 | GO:0032071 | regulation of deoxyribonuclease activity(GO:0032070) regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.1 | 0.9 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.1 | 0.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.4 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.1 | 0.2 | GO:0010039 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.1 | 2.7 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 1.0 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.4 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 1.0 | GO:0015918 | sterol transport(GO:0015918) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) regulation of vacuole organization(GO:0044088) |
| 0.1 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.2 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 1.4 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.1 | 0.4 | GO:0051274 | (1->6)-beta-D-glucan biosynthetic process(GO:0006078) beta-glucan biosynthetic process(GO:0051274) |
| 0.1 | 0.2 | GO:0009107 | lipoate metabolic process(GO:0009106) lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.1 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0034091 | regulation of maintenance of sister chromatid cohesion(GO:0034091) positive regulation of mitotic nuclear division(GO:0045840) positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of sister chromatid cohesion(GO:0045876) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.6 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 3.8 | GO:0048468 | ascospore formation(GO:0030437) cell development(GO:0048468) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.0 | 0.2 | GO:0051601 | exocyst assembly(GO:0001927) exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.0 | 0.2 | GO:0060237 | regulation of fungal-type cell wall organization(GO:0060237) |
| 0.0 | 0.2 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0000435 | regulation of transcription from RNA polymerase II promoter by galactose(GO:0000431) positive regulation of transcription from RNA polymerase II promoter by galactose(GO:0000435) |
| 0.0 | 0.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.0 | 0.1 | GO:0031138 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.0 | 0.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0045033 | peroxisome inheritance(GO:0045033) |
| 0.0 | 0.2 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.3 | GO:0007118 | budding cell apical bud growth(GO:0007118) |
| 0.0 | 0.1 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0055078 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.2 | GO:0061393 | positive regulation of transcription from RNA polymerase II promoter in response to osmotic stress(GO:0061393) |
| 0.0 | 1.3 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.0 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.0 | 0.1 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.3 | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress(GO:0036003) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.1 | GO:0016577 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 0.0 | 1.0 | GO:0030435 | sporulation resulting in formation of a cellular spore(GO:0030435) |
| 0.0 | 0.1 | GO:0046686 | response to arsenic-containing substance(GO:0046685) response to cadmium ion(GO:0046686) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:0048209 | regulation of COPII vesicle coating(GO:0003400) regulation of vesicle targeting, to, from or within Golgi(GO:0048209) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.2 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.2 | GO:0010324 | membrane invagination(GO:0010324) lysosomal microautophagy(GO:0016237) single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.7 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.0 | 0.1 | GO:0001101 | response to acid chemical(GO:0001101) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.1 | GO:0043207 | immune effector process(GO:0002252) immune system process(GO:0002376) response to virus(GO:0009615) response to external biotic stimulus(GO:0043207) defense response to virus(GO:0051607) response to other organism(GO:0051707) defense response to other organism(GO:0098542) |
| 0.0 | 0.0 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 0.1 | GO:0042766 | nucleosome mobilization(GO:0042766) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.5 | 1.6 | GO:0045265 | mitochondrial proton-transporting ATP synthase, stator stalk(GO:0000274) proton-transporting ATP synthase, stator stalk(GO:0045265) |
| 0.5 | 2.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.4 | 1.2 | GO:0000111 | nucleotide-excision repair factor 2 complex(GO:0000111) |
| 0.3 | 0.5 | GO:0032126 | eisosome(GO:0032126) |
| 0.2 | 0.9 | GO:0030287 | cell wall-bounded periplasmic space(GO:0030287) |
| 0.1 | 0.7 | GO:0072380 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 0.1 | 0.5 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.1 | 0.7 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 1.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.6 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.1 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.4 | GO:0044233 | ERMES complex(GO:0032865) ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.9 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.1 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.1 | GO:0031307 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.2 | GO:0030874 | nucleolar chromatin(GO:0030874) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.3 | GO:0031499 | TRAMP complex(GO:0031499) |
| 0.0 | 0.2 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.1 | GO:0001400 | mating projection base(GO:0001400) |
| 0.0 | 0.7 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 13.2 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.0 | GO:0009277 | fungal-type cell wall(GO:0009277) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.4 | GO:0005628 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.0 | 0.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0098552 | side of membrane(GO:0098552) cytoplasmic side of membrane(GO:0098562) |
| 0.0 | 2.1 | GO:0000323 | storage vacuole(GO:0000322) lytic vacuole(GO:0000323) fungal-type vacuole(GO:0000324) |
| 0.0 | 0.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0070210 | Rpd3L-Expanded complex(GO:0070210) |
| 0.0 | 0.1 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 1.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0000799 | transcription factor TFIIIC complex(GO:0000127) condensin complex(GO:0000796) nuclear condensin complex(GO:0000799) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.6 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 1.1 | 5.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 1.0 | 3.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.9 | 3.7 | GO:0004375 | glycine dehydrogenase (decarboxylating) activity(GO:0004375) oxidoreductase activity, acting on the CH-NH2 group of donors, disulfide as acceptor(GO:0016642) |
| 0.9 | 2.8 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.6 | 0.6 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.5 | 0.5 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.5 | 3.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.5 | 3.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.4 | 1.7 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.4 | 2.5 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.4 | 1.6 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.4 | 2.0 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.4 | 1.9 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.4 | 1.1 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.3 | 11.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.3 | 1.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 0.9 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.3 | 0.6 | GO:0042947 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.3 | 0.5 | GO:0015489 | putrescine transmembrane transporter activity(GO:0015489) |
| 0.2 | 0.9 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.2 | 2.9 | GO:0015578 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.2 | 0.8 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 0.2 | 0.9 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.2 | 0.5 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 0.5 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.2 | 1.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.6 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.1 | 0.6 | GO:0016885 | biotin carboxylase activity(GO:0004075) ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.1 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.1 | 0.8 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.4 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 1.0 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.1 | 0.4 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.1 | 0.8 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 0.6 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 0.6 | GO:0042887 | amide transmembrane transporter activity(GO:0042887) |
| 0.1 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.1 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.1 | 0.8 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.2 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.1 | 0.2 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.6 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions(GO:0016722) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 3.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.1 | GO:0016769 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.3 | GO:0004030 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.2 | GO:0016405 | CoA-ligase activity(GO:0016405) acid-thiol ligase activity(GO:0016878) |
| 0.1 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.2 | GO:0015603 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.0 | 0.2 | GO:0001128 | RNA polymerase II transcription coactivator activity involved in preinitiation complex assembly(GO:0001128) |
| 0.0 | 0.8 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.6 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0052742 | phosphatidylinositol kinase activity(GO:0052742) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0004601 | peroxidase activity(GO:0004601) oxidoreductase activity, acting on peroxide as acceptor(GO:0016684) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.0 | 1.3 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0022838 | substrate-specific channel activity(GO:0022838) |
| 0.0 | 0.2 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.5 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.0 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 0.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.0 | GO:0000772 | mating pheromone activity(GO:0000772) pheromone activity(GO:0005186) |
| 0.0 | 0.0 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.0 | 0.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.0 | GO:0030332 | cyclin binding(GO:0030332) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 0.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 16.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.2 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 0.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 16.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |