Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
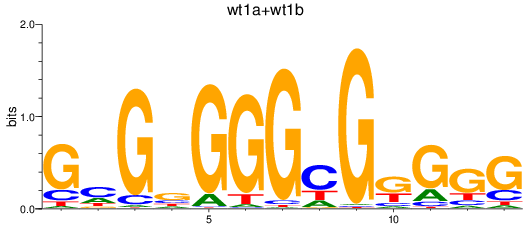
Results for wt1a+wt1b
Z-value: 1.90
Transcription factors associated with wt1a+wt1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
wt1b
|
ENSDARG00000007990 | WT1 transcription factor b |
|
wt1a
|
ENSDARG00000031420 | WT1 transcription factor a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| wt1a | dr11_v1_chr25_-_15214161_15214161 | 0.88 | 1.5e-06 | Click! |
| wt1b | dr11_v1_chr18_-_45617146_45617146 | 0.39 | 1.1e-01 | Click! |
Activity profile of wt1a+wt1b motif
Sorted Z-values of wt1a+wt1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_147574 | 7.05 |
ENSDART00000104762
ENSDART00000131635 |
slc16a10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr16_-_54405976 | 6.64 |
ENSDART00000055395
|
osr2
|
odd-skipped related transciption factor 2 |
| chr15_+_31526225 | 6.48 |
ENSDART00000154456
|
wdr95
|
WD40 repeat domain 95 |
| chr2_-_44183613 | 5.98 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr6_-_60104628 | 5.78 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr3_-_22829710 | 5.40 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr7_-_10560964 | 5.30 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr2_-_44183451 | 5.00 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr1_-_59422880 | 4.98 |
ENSDART00000167244
|
si:ch211-188p14.2
|
si:ch211-188p14.2 |
| chr7_+_16166558 | 4.90 |
ENSDART00000173785
|
si:ch73-367j5.3
|
si:ch73-367j5.3 |
| chr16_+_10918252 | 4.83 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr12_-_11649690 | 4.78 |
ENSDART00000149713
|
btbd16
|
BTB (POZ) domain containing 16 |
| chr10_-_5857548 | 4.57 |
ENSDART00000166933
|
si:ch211-281k19.2
|
si:ch211-281k19.2 |
| chr24_+_42131564 | 4.40 |
ENSDART00000153854
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr20_-_9580446 | 4.10 |
ENSDART00000014168
|
zfp36l1b
|
zinc finger protein 36, C3H type-like 1b |
| chr24_+_33622769 | 4.05 |
ENSDART00000079342
|
dnah5l
|
dynein, axonemal, heavy chain 5 like |
| chr13_-_36703164 | 4.05 |
ENSDART00000044357
|
cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr20_+_27242595 | 4.04 |
ENSDART00000149300
|
si:dkey-85n7.6
|
si:dkey-85n7.6 |
| chr25_+_1732838 | 3.96 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr14_+_80685 | 3.87 |
ENSDART00000188443
|
stag3
|
stromal antigen 3 |
| chr24_+_42127983 | 3.87 |
ENSDART00000190157
ENSDART00000176032 ENSDART00000175790 |
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr5_-_68022631 | 3.82 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr3_+_7617353 | 3.72 |
ENSDART00000165551
|
zgc:109949
|
zgc:109949 |
| chr21_-_308852 | 3.67 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr19_+_636886 | 3.65 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr19_-_31802296 | 3.64 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr16_+_10963602 | 3.63 |
ENSDART00000141032
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr22_+_18187857 | 3.63 |
ENSDART00000166300
|
mef2b
|
myocyte enhancer factor 2b |
| chr3_+_14388010 | 3.60 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr1_-_59571758 | 3.55 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr3_-_1190132 | 3.55 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr19_+_63567 | 3.37 |
ENSDART00000165657
ENSDART00000165183 |
zhx2b
|
zinc fingers and homeoboxes 2b |
| chr16_+_4838808 | 3.31 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr19_+_233143 | 3.27 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr1_-_411331 | 3.26 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr5_-_46273938 | 3.25 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr1_-_58562129 | 3.23 |
ENSDART00000159070
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr24_+_42132962 | 3.15 |
ENSDART00000187739
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr19_+_48111285 | 3.13 |
ENSDART00000169420
|
nme2b.2
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2 |
| chr20_+_54356272 | 3.07 |
ENSDART00000145735
|
znf410
|
zinc finger protein 410 |
| chr1_-_51734524 | 3.07 |
ENSDART00000109640
ENSDART00000122628 |
junba
|
JunB proto-oncogene, AP-1 transcription factor subunit a |
| chr3_-_16289826 | 3.05 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr5_-_22619879 | 3.05 |
ENSDART00000051623
|
zgc:113208
|
zgc:113208 |
| chr4_-_22472653 | 3.04 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr17_-_31695217 | 3.03 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr23_-_19140781 | 3.03 |
ENSDART00000143580
|
si:ch73-381f5.2
|
si:ch73-381f5.2 |
| chr22_-_34872533 | 2.97 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr22_+_18188045 | 2.97 |
ENSDART00000140106
|
mef2b
|
myocyte enhancer factor 2b |
| chr23_+_19182819 | 2.90 |
ENSDART00000131804
|
si:dkey-93l1.4
|
si:dkey-93l1.4 |
| chr7_-_10606 | 2.87 |
ENSDART00000192650
ENSDART00000186761 |
FO704772.2
|
|
| chr9_+_2507526 | 2.85 |
ENSDART00000166579
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr8_+_2456854 | 2.85 |
ENSDART00000133938
ENSDART00000002764 |
polb
|
polymerase (DNA directed), beta |
| chr8_+_54284961 | 2.85 |
ENSDART00000122692
|
plxnd1
|
plexin D1 |
| chr25_+_35774544 | 2.85 |
ENSDART00000034737
ENSDART00000188162 |
cpne8
|
copine VIII |
| chr3_+_1211242 | 2.80 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr8_-_34762163 | 2.78 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr24_-_4450238 | 2.71 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr5_-_2689753 | 2.71 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr5_+_32924669 | 2.71 |
ENSDART00000085219
|
lmo4a
|
LIM domain only 4a |
| chr10_+_439692 | 2.70 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr2_+_33747509 | 2.67 |
ENSDART00000134187
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr18_+_5549672 | 2.67 |
ENSDART00000184970
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr7_-_18168493 | 2.64 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr23_+_19691146 | 2.60 |
ENSDART00000143001
|
si:dkey-93l1.6
|
si:dkey-93l1.6 |
| chr1_-_58561963 | 2.56 |
ENSDART00000165040
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr16_+_4839078 | 2.53 |
ENSDART00000150111
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr6_-_9922266 | 2.51 |
ENSDART00000151549
|
pimr73
|
Pim proto-oncogene, serine/threonine kinase, related 73 |
| chr25_+_150570 | 2.51 |
ENSDART00000170892
|
adam10b
|
ADAM metallopeptidase domain 10b |
| chr3_+_22079219 | 2.50 |
ENSDART00000122782
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr25_+_192116 | 2.48 |
ENSDART00000153983
|
zgc:114188
|
zgc:114188 |
| chr2_+_33722547 | 2.46 |
ENSDART00000139193
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr4_-_16001118 | 2.43 |
ENSDART00000041070
ENSDART00000125389 |
mest
|
mesoderm specific transcript |
| chr19_+_935565 | 2.41 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr4_+_41602 | 2.41 |
ENSDART00000159640
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr21_-_43117327 | 2.38 |
ENSDART00000122352
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr7_+_73630751 | 2.38 |
ENSDART00000159745
|
PCP4L1
|
si:dkey-46i9.1 |
| chr10_+_45345574 | 2.36 |
ENSDART00000166085
|
ppiab
|
peptidylprolyl isomerase Ab (cyclophilin A) |
| chr6_-_39160422 | 2.33 |
ENSDART00000148661
|
stat2
|
signal transducer and activator of transcription 2 |
| chr23_+_9560991 | 2.30 |
ENSDART00000081433
ENSDART00000131594 ENSDART00000130069 ENSDART00000138601 |
adrm1
|
adhesion regulating molecule 1 |
| chr18_+_33788340 | 2.30 |
ENSDART00000136950
|
si:dkey-145c18.2
|
si:dkey-145c18.2 |
| chr1_+_54908895 | 2.27 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr14_-_9522364 | 2.27 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr10_+_10351685 | 2.26 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr22_-_16180849 | 2.26 |
ENSDART00000090390
|
vcam1b
|
vascular cell adhesion molecule 1b |
| chr24_+_24831727 | 2.24 |
ENSDART00000080969
|
trim55b
|
tripartite motif containing 55b |
| chr4_-_68563862 | 2.24 |
ENSDART00000182970
|
BX548011.2
|
|
| chr23_+_9560797 | 2.23 |
ENSDART00000180014
|
adrm1
|
adhesion regulating molecule 1 |
| chr8_-_29719393 | 2.22 |
ENSDART00000077635
|
si:ch211-103n10.5
|
si:ch211-103n10.5 |
| chr19_-_10043142 | 2.20 |
ENSDART00000193016
|
grin2da
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a |
| chr11_+_25459697 | 2.17 |
ENSDART00000161481
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr23_-_46201008 | 2.15 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr21_-_22114625 | 2.14 |
ENSDART00000177426
ENSDART00000135410 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr10_-_28835771 | 2.12 |
ENSDART00000192220
ENSDART00000188436 |
alcama
|
activated leukocyte cell adhesion molecule a |
| chr22_+_38310957 | 2.10 |
ENSDART00000040550
|
traf5
|
Tnf receptor-associated factor 5 |
| chr22_+_15898221 | 2.09 |
ENSDART00000062587
|
klf2a
|
Kruppel-like factor 2a |
| chr6_-_442163 | 2.06 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr12_+_2522642 | 2.06 |
ENSDART00000152567
|
frmpd2
|
FERM and PDZ domain containing 2 |
| chr21_-_45878872 | 2.04 |
ENSDART00000029763
|
sap30l
|
sap30-like |
| chr19_-_19442983 | 2.03 |
ENSDART00000052649
|
sb:cb649
|
sb:cb649 |
| chr20_+_18209895 | 2.00 |
ENSDART00000111063
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr1_+_57371447 | 2.00 |
ENSDART00000152229
ENSDART00000181077 |
si:dkey-27j5.3
|
si:dkey-27j5.3 |
| chr11_+_41560792 | 2.00 |
ENSDART00000127292
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr3_+_32492467 | 1.97 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr25_+_24250247 | 1.96 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr15_+_1397811 | 1.95 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr20_-_26420939 | 1.95 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr12_+_695619 | 1.93 |
ENSDART00000161691
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr12_+_9880493 | 1.92 |
ENSDART00000055019
|
ndufa4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 |
| chr21_-_5056812 | 1.91 |
ENSDART00000139713
ENSDART00000140859 |
zgc:77838
|
zgc:77838 |
| chr14_+_42172257 | 1.90 |
ENSDART00000074362
|
pcdh18b
|
protocadherin 18b |
| chr14_-_2933185 | 1.88 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr2_+_26288301 | 1.88 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr13_-_31435137 | 1.88 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr11_-_4302251 | 1.85 |
ENSDART00000182554
|
CT033790.1
|
|
| chr6_-_59942335 | 1.85 |
ENSDART00000168416
|
fbxl3b
|
F-box and leucine-rich repeat protein 3b |
| chr16_-_1757521 | 1.80 |
ENSDART00000124660
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr22_-_11714104 | 1.80 |
ENSDART00000145265
ENSDART00000063127 ENSDART00000183743 |
crygs1
|
crystallin, gamma S1 |
| chr21_-_39058490 | 1.79 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr14_+_24283915 | 1.78 |
ENSDART00000172868
|
klhl3
|
kelch-like family member 3 |
| chr17_+_53311618 | 1.77 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr1_-_51157454 | 1.76 |
ENSDART00000047851
|
jag1a
|
jagged 1a |
| chr20_-_26421112 | 1.76 |
ENSDART00000183767
ENSDART00000182330 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr1_-_59232267 | 1.75 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr14_+_42172677 | 1.72 |
ENSDART00000148544
|
pcdh18b
|
protocadherin 18b |
| chr9_+_19489514 | 1.72 |
ENSDART00000152032
ENSDART00000114256 ENSDART00000190572 ENSDART00000147571 ENSDART00000151918 ENSDART00000152034 |
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr1_-_54107321 | 1.71 |
ENSDART00000148382
ENSDART00000150357 |
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr20_+_13894123 | 1.69 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr4_+_1600034 | 1.69 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr9_+_1654284 | 1.69 |
ENSDART00000062854
|
nfe2l2a
|
nuclear factor, erythroid 2-like 2a |
| chr2_+_24304854 | 1.66 |
ENSDART00000078972
|
fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr14_-_12822 | 1.65 |
ENSDART00000180650
ENSDART00000188819 |
msx1a
|
muscle segment homeobox 1a |
| chr4_-_68569527 | 1.65 |
ENSDART00000192091
|
BX548011.5
|
|
| chr25_+_22683640 | 1.64 |
ENSDART00000131993
|
ush1c
|
Usher syndrome 1C |
| chr1_-_51474974 | 1.63 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr12_+_41510492 | 1.62 |
ENSDART00000170976
ENSDART00000176164 |
kif5bb
|
kinesin family member 5B, b |
| chr15_-_47822597 | 1.62 |
ENSDART00000193236
ENSDART00000161391 |
CZQB01095947.1
|
|
| chr19_-_617246 | 1.61 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr8_-_52715911 | 1.60 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr7_-_51102479 | 1.59 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr22_-_367569 | 1.57 |
ENSDART00000041895
|
ssu72
|
SSU72 homolog, RNA polymerase II CTD phosphatase |
| chr5_+_51443009 | 1.57 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr14_-_45346558 | 1.56 |
ENSDART00000090844
|
zgc:153018
|
zgc:153018 |
| chr6_+_31684 | 1.56 |
ENSDART00000188853
ENSDART00000184553 |
CZQB01141835.2
|
|
| chr22_+_2860260 | 1.55 |
ENSDART00000106221
|
si:dkey-20i20.2
|
si:dkey-20i20.2 |
| chr20_-_54381034 | 1.55 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr1_+_22174251 | 1.54 |
ENSDART00000137429
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr20_+_16173618 | 1.53 |
ENSDART00000192109
ENSDART00000104112 ENSDART00000129633 |
zyg11
|
zyg-11 homolog (C. elegans) |
| chr19_+_33701734 | 1.51 |
ENSDART00000123270
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr9_-_39005317 | 1.51 |
ENSDART00000014207
|
myl1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr16_-_13613475 | 1.50 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr20_+_18740518 | 1.49 |
ENSDART00000142196
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr25_-_37501371 | 1.48 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr22_+_10752511 | 1.46 |
ENSDART00000081188
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr16_-_54810464 | 1.46 |
ENSDART00000030658
|
zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr14_+_30910114 | 1.43 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr21_+_5801105 | 1.42 |
ENSDART00000151225
ENSDART00000184487 |
ccng2
|
cyclin G2 |
| chr10_+_10210455 | 1.42 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr5_-_16351306 | 1.41 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr8_-_1884955 | 1.41 |
ENSDART00000081563
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr22_+_10752787 | 1.41 |
ENSDART00000186542
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr8_+_8298439 | 1.40 |
ENSDART00000170566
|
srpk3
|
SRSF protein kinase 3 |
| chr17_-_43466317 | 1.40 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr20_-_5369105 | 1.40 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr22_-_17729778 | 1.39 |
ENSDART00000192132
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr22_+_1006573 | 1.37 |
ENSDART00000123458
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr8_-_9118958 | 1.36 |
ENSDART00000037922
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr18_-_15467446 | 1.36 |
ENSDART00000187847
|
endouc
|
endonuclease, polyU-specific C |
| chr4_+_39742119 | 1.33 |
ENSDART00000176004
|
CR749167.2
|
|
| chr1_-_625875 | 1.33 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr11_-_236766 | 1.32 |
ENSDART00000163978
|
dusp7
|
dual specificity phosphatase 7 |
| chr5_+_11840905 | 1.31 |
ENSDART00000030444
|
tesca
|
tescalcin a |
| chr16_-_44399335 | 1.31 |
ENSDART00000165058
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr1_-_49225890 | 1.30 |
ENSDART00000111598
|
cxcl18b
|
chemokine (C-X-C motif) ligand 18b |
| chr9_-_8454060 | 1.29 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr11_+_40649412 | 1.28 |
ENSDART00000043016
ENSDART00000134560 |
slc45a1
|
solute carrier family 45, member 1 |
| chr6_+_59808677 | 1.28 |
ENSDART00000164469
|
caskb
|
calcium/calmodulin-dependent serine protein kinase b |
| chr1_-_51157660 | 1.27 |
ENSDART00000137172
|
jag1a
|
jagged 1a |
| chr1_+_55293424 | 1.27 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr9_-_105135 | 1.24 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr21_+_103194 | 1.24 |
ENSDART00000162755
|
zer1
|
zyg-11 related, cell cycle regulator |
| chr5_+_22969651 | 1.22 |
ENSDART00000089992
ENSDART00000145477 |
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr1_-_53907092 | 1.22 |
ENSDART00000007732
|
capn9
|
calpain 9 |
| chr11_+_6456146 | 1.21 |
ENSDART00000036939
|
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr11_+_6819050 | 1.21 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr5_-_38777852 | 1.19 |
ENSDART00000131603
|
si:dkey-58f10.4
|
si:dkey-58f10.4 |
| chr19_+_1964005 | 1.18 |
ENSDART00000172049
|
sh3bp5a
|
SH3-domain binding protein 5a (BTK-associated) |
| chr5_-_12093618 | 1.17 |
ENSDART00000161542
|
lrrc74b
|
leucine rich repeat containing 74B |
| chr10_+_32561317 | 1.17 |
ENSDART00000109029
|
map6a
|
microtubule-associated protein 6a |
| chr5_+_43870389 | 1.17 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr11_-_236984 | 1.17 |
ENSDART00000170778
|
dusp7
|
dual specificity phosphatase 7 |
| chr21_-_22325124 | 1.16 |
ENSDART00000142100
|
gdpd4b
|
glycerophosphodiester phosphodiesterase domain containing 4b |
| chr12_+_5189776 | 1.15 |
ENSDART00000081298
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr16_-_44349845 | 1.15 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr2_+_24781026 | 1.14 |
ENSDART00000145692
|
pde4ca
|
phosphodiesterase 4C, cAMP-specific a |
| chr22_-_188102 | 1.14 |
ENSDART00000125391
ENSDART00000170463 |
CABZ01052268.1
|
|
| chr2_+_20793982 | 1.13 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr23_-_40779334 | 1.13 |
ENSDART00000141432
|
DUPD1 (1 of many)
|
si:dkeyp-27c8.1 |
| chr4_+_16710001 | 1.11 |
ENSDART00000035899
|
pkp2
|
plakophilin 2 |
| chr19_+_33701366 | 1.10 |
ENSDART00000162517
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr17_+_24615091 | 1.10 |
ENSDART00000064739
|
rpl13a
|
ribosomal protein L13a |
Network of associatons between targets according to the STRING database.
First level regulatory network of wt1a+wt1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 1.2 | 3.6 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 1.2 | 5.8 | GO:0071071 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.9 | 2.7 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.8 | 5.8 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.8 | 2.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.8 | 5.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.7 | 2.1 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.7 | 2.7 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.7 | 2.0 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.6 | 1.8 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.6 | 1.7 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.5 | 2.6 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.5 | 1.6 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.5 | 1.9 | GO:0060468 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.4 | 0.9 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.4 | 3.7 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.4 | 1.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.4 | 3.3 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.3 | 1.0 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.3 | 1.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.3 | 1.9 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.3 | 4.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 2.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.3 | 0.9 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.3 | 4.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 1.6 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.3 | 1.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.3 | 3.8 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 0.9 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.2 | 1.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 3.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 0.6 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.2 | 3.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.2 | 0.8 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 0.6 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.2 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.6 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 0.9 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.2 | 0.9 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 | 2.0 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.2 | 2.3 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.2 | 2.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 0.9 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 1.2 | GO:1904019 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.2 | 1.7 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.2 | 1.7 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.2 | 3.0 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.2 | 1.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 1.4 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 1.0 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.1 | 0.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.4 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.6 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 1.0 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 2.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 1.4 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.4 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 0.8 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 2.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.9 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.2 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.7 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.1 | 0.9 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 2.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.5 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 2.9 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.1 | 0.2 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.1 | 1.0 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 0.6 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 1.6 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.5 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.8 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 2.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 2.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 2.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 2.9 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 2.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.5 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 8.8 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.1 | 1.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 1.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.9 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 2.8 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.1 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 2.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.3 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.1 | 0.4 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 5.4 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 1.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.3 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 1.7 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 5.5 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 2.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.4 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.7 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 0.2 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 2.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 20.9 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 5.7 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 0.6 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 0.3 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 1.2 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 2.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.5 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 1.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.6 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.6 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.3 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.6 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 1.4 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.0 | 0.1 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.0 | 1.1 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 1.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.0 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 1.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.8 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 1.8 | GO:0031076 | embryonic camera-type eye development(GO:0031076) |
| 0.0 | 0.4 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.3 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.0 | 0.9 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.3 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.6 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 1.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.3 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 1.3 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.2 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 0.4 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.8 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.0 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.0 | 0.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 2.4 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.7 | GO:0045197 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.0 | 0.1 | GO:0015893 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.5 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 2.2 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.4 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 1.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.9 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.6 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.3 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 2.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.1 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 3.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 1.0 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 1.5 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 1.9 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.8 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.6 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.4 | 4.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.4 | 2.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.3 | 4.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.3 | 2.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.3 | 3.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 1.6 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 3.7 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.3 | 1.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 3.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 3.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 0.9 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 2.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 1.6 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.2 | 0.9 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 2.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.2 | 3.9 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.2 | 2.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 2.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 2.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 2.6 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 2.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 2.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.6 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 1.9 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 0.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 5.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 2.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 4.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 4.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 2.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.2 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 1.8 | 7.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.2 | 3.6 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 1.2 | 5.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.8 | 2.5 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.8 | 4.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.7 | 5.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.7 | 2.7 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.6 | 2.9 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.6 | 2.3 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.4 | 3.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 2.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.3 | 1.0 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.3 | 1.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.3 | 1.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.3 | 0.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 3.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 2.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 2.0 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.2 | 2.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 0.8 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 1.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.8 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 0.8 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.2 | 4.7 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.2 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 1.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 3.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 1.8 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 2.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 1.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 5.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 2.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.3 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 3.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 0.9 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 3.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 2.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.7 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.9 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 6.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 2.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 3.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.6 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.1 | 1.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 3.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 2.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.5 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 1.8 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.7 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 1.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 2.7 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 0.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 2.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.6 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 1.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 2.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 2.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.7 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 3.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.0 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 1.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 12.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 5.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 3.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.1 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 3.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 5.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.3 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.3 | 17.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 5.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 3.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 5.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.5 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 1.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 5.8 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.3 | 2.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 2.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 2.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 1.9 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.2 | 2.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 12.8 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.2 | 2.7 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 3.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 3.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.6 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.1 | 3.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.9 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |