Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
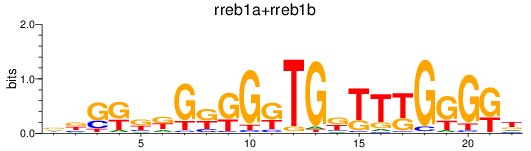
Results for rreb1a+rreb1b
Z-value: 1.60
Transcription factors associated with rreb1a+rreb1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rreb1b
|
ENSDARG00000042652 | ras responsive element binding protein 1b |
|
rreb1a
|
ENSDARG00000063701 | ras responsive element binding protein 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rreb1b | dr11_v1_chr2_+_21048661_21048661 | -0.87 | 3.2e-06 | Click! |
| rreb1a | dr11_v1_chr24_-_2381143_2381143 | -0.85 | 1.0e-05 | Click! |
Activity profile of rreb1a+rreb1b motif
Sorted Z-values of rreb1a+rreb1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_54640315 | 20.39 |
ENSDART00000101207
|
spag1b
|
sperm associated antigen 1b |
| chr25_+_258883 | 14.91 |
ENSDART00000155256
|
zgc:92481
|
zgc:92481 |
| chr2_-_44183613 | 5.89 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr16_-_54405976 | 5.32 |
ENSDART00000055395
|
osr2
|
odd-skipped related transciption factor 2 |
| chr2_-_44183451 | 5.03 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr8_+_54284961 | 4.93 |
ENSDART00000122692
|
plxnd1
|
plexin D1 |
| chr19_+_56351 | 4.57 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr17_-_12408109 | 4.32 |
ENSDART00000155509
|
ankef1b
|
ankyrin repeat and EF-hand domain containing 1b |
| chr22_-_34872533 | 4.29 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr5_-_3839285 | 4.19 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr23_-_38054 | 3.90 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr14_-_853176 | 3.81 |
ENSDART00000041988
|
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr25_-_24233884 | 3.77 |
ENSDART00000146419
|
si:dkey-11e23.9
|
si:dkey-11e23.9 |
| chr18_+_1154189 | 3.76 |
ENSDART00000135090
|
si:ch1073-75f15.2
|
si:ch1073-75f15.2 |
| chr6_+_27514465 | 3.75 |
ENSDART00000128985
ENSDART00000079397 |
ryk
|
receptor-like tyrosine kinase |
| chr3_+_60007703 | 3.75 |
ENSDART00000157351
ENSDART00000153928 ENSDART00000155876 |
si:ch211-110e21.3
|
si:ch211-110e21.3 |
| chr5_+_817016 | 3.61 |
ENSDART00000188498
|
BX537263.3
|
|
| chr5_+_828663 | 3.61 |
ENSDART00000181776
|
BX537263.4
|
|
| chr2_-_24907741 | 3.43 |
ENSDART00000155013
|
si:dkey-149i17.11
|
si:dkey-149i17.11 |
| chr25_+_37480285 | 3.39 |
ENSDART00000166187
|
CABZ01095001.1
|
|
| chr16_+_10918252 | 3.32 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr23_+_44497701 | 3.28 |
ENSDART00000149903
|
si:ch73-375g18.1
|
si:ch73-375g18.1 |
| chr18_+_17474989 | 3.27 |
ENSDART00000140565
ENSDART00000147843 |
si:dkey-102f14.7
|
si:dkey-102f14.7 |
| chr20_+_33728126 | 3.20 |
ENSDART00000038535
|
ak9
|
adenylate kinase 9 |
| chr10_-_641609 | 3.13 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr1_-_9104631 | 3.11 |
ENSDART00000146642
|
si:ch211-14k19.8
|
si:ch211-14k19.8 |
| chr10_-_5857548 | 3.10 |
ENSDART00000166933
|
si:ch211-281k19.2
|
si:ch211-281k19.2 |
| chr19_+_233143 | 3.05 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr3_-_1434135 | 2.92 |
ENSDART00000149622
|
mgp
|
matrix Gla protein |
| chr1_-_5543065 | 2.87 |
ENSDART00000103755
|
fn1b
|
fibronectin 1b |
| chr17_+_7522777 | 2.85 |
ENSDART00000184723
|
klhl10b.2
|
kelch-like family member 10b, tandem duplicate 2 |
| chr25_-_9013963 | 2.84 |
ENSDART00000073912
|
rag2
|
recombination activating gene 2 |
| chr25_+_1732838 | 2.78 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr22_+_1186866 | 2.78 |
ENSDART00000167710
|
eps8l3a
|
EPS8-like 3a |
| chr9_+_52621487 | 2.76 |
ENSDART00000166266
|
si:ch211-241j8.2
|
si:ch211-241j8.2 |
| chr25_-_1124851 | 2.76 |
ENSDART00000067558
|
spg11
|
spastic paraplegia 11 |
| chr18_+_5547185 | 2.66 |
ENSDART00000193977
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr16_-_52821023 | 2.65 |
ENSDART00000074718
|
spire1b
|
spire-type actin nucleation factor 1b |
| chr7_-_10606 | 2.63 |
ENSDART00000192650
ENSDART00000186761 |
FO704772.2
|
|
| chr3_+_37196258 | 2.56 |
ENSDART00000187944
ENSDART00000185896 |
fmnl1a
|
formin-like 1a |
| chr17_-_4395373 | 2.55 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr2_-_23479714 | 2.47 |
ENSDART00000167291
|
si:ch211-14p21.3
|
si:ch211-14p21.3 |
| chr14_-_9522364 | 2.46 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr4_-_193762 | 2.44 |
ENSDART00000169667
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr13_+_51853716 | 2.43 |
ENSDART00000175341
ENSDART00000187855 |
LT631684.1
|
|
| chr3_-_1190132 | 2.40 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr2_-_22575851 | 2.34 |
ENSDART00000182932
|
FQ377628.2
|
|
| chr6_-_24320268 | 2.26 |
ENSDART00000171733
|
si:dkeyp-85h7.1
|
si:dkeyp-85h7.1 |
| chr16_-_41059919 | 2.23 |
ENSDART00000188803
|
si:dkey-201i6.8
|
si:dkey-201i6.8 |
| chr2_+_22694382 | 2.18 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr6_-_53426773 | 2.17 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr12_+_47162456 | 2.10 |
ENSDART00000186272
ENSDART00000180811 |
RYR2
|
ryanodine receptor 2 |
| chr5_-_72125551 | 2.07 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr25_+_7571920 | 2.07 |
ENSDART00000042928
|
fuk
|
fucokinase |
| chr3_-_16289826 | 2.04 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr5_+_13359146 | 1.97 |
ENSDART00000179839
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr2_+_33747509 | 1.94 |
ENSDART00000134187
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr6_+_59967994 | 1.94 |
ENSDART00000050457
|
zgc:65895
|
zgc:65895 |
| chr12_+_2677303 | 1.94 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr17_+_53311618 | 1.92 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr8_+_4368534 | 1.92 |
ENSDART00000015214
|
pptc7a
|
PTC7 protein phosphatase homolog a |
| chr8_-_1909840 | 1.88 |
ENSDART00000147408
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr2_-_51700709 | 1.79 |
ENSDART00000188601
|
tgm1l1
|
transglutaminase 1 like 1 |
| chr2_+_33722547 | 1.79 |
ENSDART00000139193
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr21_-_44009169 | 1.79 |
ENSDART00000028960
|
ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr1_+_109798 | 1.77 |
ENSDART00000165402
|
F7 (1 of many)
|
zgc:163025 |
| chr10_-_57270 | 1.77 |
ENSDART00000058411
|
hlcs
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
| chr1_+_55002583 | 1.75 |
ENSDART00000037250
|
si:ch211-196h16.12
|
si:ch211-196h16.12 |
| chr15_-_31067589 | 1.73 |
ENSDART00000060157
|
lgals9l3
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 3 |
| chr17_+_12408188 | 1.73 |
ENSDART00000105218
|
khk
|
ketohexokinase |
| chr25_+_5288665 | 1.73 |
ENSDART00000169540
|
CABZ01039861.1
|
|
| chr5_+_480119 | 1.73 |
ENSDART00000055681
|
tek
|
TEK tyrosine kinase, endothelial |
| chr25_-_37489917 | 1.72 |
ENSDART00000160688
|
psma1
|
proteasome subunit alpha 1 |
| chr18_+_21122818 | 1.69 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr22_+_29067388 | 1.69 |
ENSDART00000133673
|
pimr100
|
Pim proto-oncogene, serine/threonine kinase, related 100 |
| chr16_+_9053275 | 1.68 |
ENSDART00000123150
|
dnah5
|
dynein, axonemal, heavy chain 5 |
| chr5_-_26181863 | 1.59 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr19_+_638339 | 1.59 |
ENSDART00000148705
|
tert
|
telomerase reverse transcriptase |
| chr12_+_47162761 | 1.59 |
ENSDART00000192339
ENSDART00000167726 |
RYR2
|
ryanodine receptor 2 |
| chr4_-_1403077 | 1.55 |
ENSDART00000189813
|
CABZ01068469.1
|
|
| chr7_-_74090168 | 1.54 |
ENSDART00000050528
|
tyrp1a
|
tyrosinase-related protein 1a |
| chr15_-_5742531 | 1.53 |
ENSDART00000045985
|
phkg1a
|
phosphorylase kinase, gamma 1a (muscle) |
| chr24_+_42131564 | 1.53 |
ENSDART00000153854
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr3_+_18398876 | 1.51 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr21_+_28445052 | 1.48 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr19_-_10081047 | 1.47 |
ENSDART00000128837
|
grin2da
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a |
| chr16_+_41826584 | 1.47 |
ENSDART00000147523
|
si:dkey-199f5.6
|
si:dkey-199f5.6 |
| chr6_+_9421279 | 1.46 |
ENSDART00000161036
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr2_+_33368414 | 1.45 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr25_-_173165 | 1.45 |
ENSDART00000193594
|
CABZ01114053.1
|
|
| chr18_-_50845804 | 1.42 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr3_-_7170661 | 1.40 |
ENSDART00000190345
|
BX005085.4
|
|
| chr21_-_41305748 | 1.40 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr20_-_5369105 | 1.39 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr3_+_1223824 | 1.36 |
ENSDART00000065922
|
wbp2nl
|
WBP2 N-terminal like |
| chr9_+_10692905 | 1.35 |
ENSDART00000061499
|
cxcr4b
|
chemokine (C-X-C motif), receptor 4b |
| chr5_+_19337108 | 1.35 |
ENSDART00000089078
|
acacb
|
acetyl-CoA carboxylase beta |
| chr7_+_74150839 | 1.32 |
ENSDART00000160195
|
ppp1cbl
|
protein phosphatase 1, catalytic subunit, beta isoform, like |
| chr4_-_9909371 | 1.31 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr13_+_41022502 | 1.31 |
ENSDART00000026808
|
dkk1a
|
dickkopf WNT signaling pathway inhibitor 1a |
| chr5_+_337215 | 1.30 |
ENSDART00000167982
|
rnf170
|
ring finger protein 170 |
| chr13_-_45523026 | 1.29 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr17_+_8925232 | 1.28 |
ENSDART00000036668
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr5_-_16351306 | 1.28 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr25_+_37348730 | 1.27 |
ENSDART00000156639
|
pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr9_-_216527 | 1.25 |
ENSDART00000163068
|
aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr23_-_9768700 | 1.25 |
ENSDART00000045126
|
lama5
|
laminin, alpha 5 |
| chr5_-_28029558 | 1.25 |
ENSDART00000078649
|
abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr8_-_17167819 | 1.24 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr17_-_52643970 | 1.24 |
ENSDART00000190594
|
spred1
|
sprouty-related, EVH1 domain containing 1 |
| chr21_+_45510448 | 1.24 |
ENSDART00000160494
ENSDART00000167914 |
fnip1
|
folliculin interacting protein 1 |
| chr9_+_56449505 | 1.22 |
ENSDART00000187725
|
CABZ01079480.1
|
|
| chr23_-_46201008 | 1.22 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr5_+_7989210 | 1.21 |
ENSDART00000168071
|
gdnfb
|
glial cell derived neurotrophic factor b |
| chr4_-_64703 | 1.20 |
ENSDART00000167851
|
CU856344.1
|
|
| chr17_+_53418445 | 1.20 |
ENSDART00000097631
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr12_+_49135755 | 1.20 |
ENSDART00000153460
|
si:zfos-911d5.4
|
si:zfos-911d5.4 |
| chr1_+_227241 | 1.20 |
ENSDART00000003317
|
tfdp1b
|
transcription factor Dp-1, b |
| chr13_-_31435137 | 1.19 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr12_-_1951233 | 1.15 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr17_+_15899039 | 1.14 |
ENSDART00000154181
|
syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr18_-_50083405 | 1.13 |
ENSDART00000098668
ENSDART00000182215 ENSDART00000165423 |
abcc8b
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8b |
| chr19_-_13923091 | 1.13 |
ENSDART00000169993
ENSDART00000158992 |
stx12
|
syntaxin 12 |
| chr9_+_36946340 | 1.12 |
ENSDART00000135281
|
si:dkey-3d4.3
|
si:dkey-3d4.3 |
| chr5_-_64865521 | 1.11 |
ENSDART00000043397
ENSDART00000145831 |
ZBTB26
|
si:ch211-236k19.4 |
| chr21_+_25625026 | 1.08 |
ENSDART00000134678
|
ovol1b
|
ovo-like zinc finger 1b |
| chr16_-_19026733 | 1.08 |
ENSDART00000104826
|
pimr98
|
Pim proto-oncogene, serine/threonine kinase, related 98 |
| chr16_-_19026414 | 1.08 |
ENSDART00000141208
|
pimr98
|
Pim proto-oncogene, serine/threonine kinase, related 98 |
| chr6_+_59818275 | 1.07 |
ENSDART00000165213
|
caskb
|
calcium/calmodulin-dependent serine protein kinase b |
| chr22_-_5252005 | 1.07 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr10_-_36825984 | 1.07 |
ENSDART00000111104
|
phf12a
|
PHD finger protein 12a |
| chr14_+_52563794 | 1.06 |
ENSDART00000168874
|
rpl26
|
ribosomal protein L26 |
| chr17_+_53311243 | 1.06 |
ENSDART00000160241
ENSDART00000160009 ENSDART00000162239 |
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr6_+_59808677 | 1.06 |
ENSDART00000164469
|
caskb
|
calcium/calmodulin-dependent serine protein kinase b |
| chr1_+_2129164 | 1.05 |
ENSDART00000074923
ENSDART00000124534 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr20_+_52595220 | 1.02 |
ENSDART00000180610
|
si:dkey-235d18.5
|
si:dkey-235d18.5 |
| chr16_+_43347966 | 1.01 |
ENSDART00000171308
|
zmp:0000000930
|
zmp:0000000930 |
| chr18_-_81280 | 0.99 |
ENSDART00000149810
ENSDART00000149321 |
hdc
|
histidine decarboxylase |
| chr5_-_417495 | 0.98 |
ENSDART00000180586
ENSDART00000189408 |
HOOK3
|
hook microtubule tethering protein 3 |
| chr6_-_442163 | 0.98 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr4_-_170120 | 0.98 |
ENSDART00000171333
|
eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr11_+_45347961 | 0.97 |
ENSDART00000172962
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr25_+_150570 | 0.97 |
ENSDART00000170892
|
adam10b
|
ADAM metallopeptidase domain 10b |
| chr1_-_8012476 | 0.96 |
ENSDART00000177051
|
CR855320.3
|
|
| chr9_+_13714379 | 0.95 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr20_+_52458765 | 0.94 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr25_-_37501371 | 0.93 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr9_-_23033818 | 0.93 |
ENSDART00000022392
|
rnd3b
|
Rho family GTPase 3b |
| chr7_-_55454406 | 0.92 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr8_+_48848200 | 0.92 |
ENSDART00000130673
|
tp73
|
tumor protein p73 |
| chr12_+_41510492 | 0.91 |
ENSDART00000170976
ENSDART00000176164 |
kif5bb
|
kinesin family member 5B, b |
| chr1_-_625875 | 0.89 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr15_+_714203 | 0.88 |
ENSDART00000153847
|
si:dkey-7i4.24
|
si:dkey-7i4.24 |
| chr4_+_90048 | 0.88 |
ENSDART00000166440
|
lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr5_-_12093618 | 0.87 |
ENSDART00000161542
|
lrrc74b
|
leucine rich repeat containing 74B |
| chr7_+_22293894 | 0.87 |
ENSDART00000056790
|
tmem256
|
transmembrane protein 256 |
| chr11_-_3334248 | 0.87 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr4_+_39742119 | 0.86 |
ENSDART00000176004
|
CR749167.2
|
|
| chr2_+_24781026 | 0.85 |
ENSDART00000145692
|
pde4ca
|
phosphodiesterase 4C, cAMP-specific a |
| chr9_+_51891 | 0.85 |
ENSDART00000163529
|
zgc:158316
|
zgc:158316 |
| chr22_+_39074688 | 0.83 |
ENSDART00000153547
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr5_-_16475374 | 0.83 |
ENSDART00000134274
ENSDART00000136004 |
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr7_+_65876335 | 0.81 |
ENSDART00000150143
|
tead1b
|
TEA domain family member 1b |
| chr8_+_46010838 | 0.79 |
ENSDART00000143126
|
si:ch211-119d14.2
|
si:ch211-119d14.2 |
| chr6_-_60031693 | 0.78 |
ENSDART00000160275
|
CABZ01079262.1
|
|
| chr22_+_6905210 | 0.77 |
ENSDART00000167530
|
CABZ01065328.2
|
|
| chr20_+_18740518 | 0.75 |
ENSDART00000142196
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr20_+_46513651 | 0.75 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr4_-_18211 | 0.73 |
ENSDART00000171737
|
ptpn12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr5_-_6508250 | 0.73 |
ENSDART00000060535
|
crybb3
|
crystallin, beta B3 |
| chr13_-_638485 | 0.72 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr8_+_8012570 | 0.72 |
ENSDART00000183429
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr14_+_94946 | 0.71 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr11_+_45461419 | 0.71 |
ENSDART00000173424
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr22_-_834106 | 0.70 |
ENSDART00000105873
|
cry4
|
cryptochrome circadian clock 4 |
| chr16_+_5908483 | 0.70 |
ENSDART00000167393
|
ulk4
|
unc-51 like kinase 4 |
| chr7_-_2039060 | 0.70 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr2_+_36007449 | 0.69 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr5_+_32009080 | 0.69 |
ENSDART00000186885
|
scai
|
suppressor of cancer cell invasion |
| chr15_-_44663712 | 0.68 |
ENSDART00000192124
|
CABZ01061894.1
|
|
| chr3_-_60027255 | 0.68 |
ENSDART00000189252
ENSDART00000154684 |
recql5
|
RecQ helicase-like 5 |
| chr12_-_28363111 | 0.68 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr19_+_32856907 | 0.67 |
ENSDART00000148232
|
rpl30
|
ribosomal protein L30 |
| chr2_+_20793982 | 0.67 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr7_+_31838320 | 0.67 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr12_-_54375 | 0.67 |
ENSDART00000152304
|
si:ch1073-357b18.4
|
si:ch1073-357b18.4 |
| chr6_-_21189295 | 0.66 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr5_-_43959972 | 0.66 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr22_+_997838 | 0.65 |
ENSDART00000149743
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr8_+_68864 | 0.65 |
ENSDART00000164574
|
prr16
|
proline rich 16 |
| chr2_+_52847049 | 0.63 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr11_+_37049105 | 0.62 |
ENSDART00000155408
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr16_-_54357200 | 0.62 |
ENSDART00000170753
|
CABZ01111454.1
|
|
| chr1_-_59571758 | 0.62 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr7_-_5191797 | 0.62 |
ENSDART00000172955
|
si:ch73-223f5.1
|
si:ch73-223f5.1 |
| chr20_-_54377933 | 0.62 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr19_-_45960191 | 0.62 |
ENSDART00000052434
ENSDART00000172732 |
eif3hb
|
eukaryotic translation initiation factor 3, subunit H, b |
| chr10_+_428269 | 0.61 |
ENSDART00000140715
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr17_+_8323348 | 0.61 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr6_-_346084 | 0.61 |
ENSDART00000162599
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
Network of associatons between targets according to the STRING database.
First level regulatory network of rreb1a+rreb1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.8 | 3.2 | GO:0009202 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) dGDP metabolic process(GO:0046066) |
| 0.8 | 22.1 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.7 | 2.7 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.6 | 3.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.6 | 1.7 | GO:0009750 | response to fructose(GO:0009750) |
| 0.6 | 2.8 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.5 | 1.6 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.5 | 1.5 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.5 | 1.5 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.5 | 2.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.4 | 2.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.4 | 1.2 | GO:0060688 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.4 | 2.8 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.4 | 3.8 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.3 | 1.7 | GO:2000561 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.3 | 1.4 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.3 | 1.3 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.3 | 1.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 1.0 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.3 | 2.6 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.3 | 1.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.3 | 1.1 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.3 | 1.4 | GO:0007343 | egg activation(GO:0007343) |
| 0.3 | 2.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.3 | 0.8 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 4.9 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.2 | 2.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 | 1.1 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.2 | 1.2 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 3.1 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 0.6 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.2 | 0.9 | GO:0010482 | ventriculo bulbo valve development(GO:0003173) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 1.5 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.2 | 1.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 2.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.4 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 3.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.7 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.4 | GO:0055026 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 1.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.2 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 3.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.0 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 1.7 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.6 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 3.6 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.1 | 0.7 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.9 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.3 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.1 | 2.1 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.1 | 1.5 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.1 | 1.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.5 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 0.5 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 1.3 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.1 | 0.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 2.4 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 1.3 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.1 | 0.7 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.5 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 0.3 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 1.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 1.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 2.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 2.0 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.6 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 1.1 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 1.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.6 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 2.2 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.8 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 2.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.7 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 13.4 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 1.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 1.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 1.2 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 0.9 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.6 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.6 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 | 0.6 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 3.9 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.5 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.9 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.7 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.1 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.2 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 1.5 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.2 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.5 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.1 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.4 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.5 | 1.6 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.5 | 2.8 | GO:0000801 | central element(GO:0000801) |
| 0.4 | 1.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.3 | 1.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 1.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.2 | 4.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 3.8 | GO:0005903 | brush border(GO:0005903) |
| 0.2 | 2.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 1.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.2 | 1.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.4 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 2.1 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.6 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 1.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 1.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 2.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.3 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 3.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.6 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.9 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 2.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.6 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:0022626 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.0 | 13.8 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.7 | 2.7 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.6 | 3.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.5 | 1.6 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.4 | 1.2 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.4 | 2.9 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.3 | 1.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.3 | 1.0 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.3 | 1.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.3 | 1.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.3 | 1.5 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 1.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 1.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 3.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 1.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.2 | 1.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 1.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 1.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 4.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 3.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 4.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.7 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 2.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 2.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.6 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.8 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 0.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 2.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 1.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.6 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 1.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.6 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.3 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.6 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.8 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 2.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 3.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 1.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 3.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 4.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 4.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 3.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 2.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 4.2 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 1.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.5 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.6 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.7 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |