Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
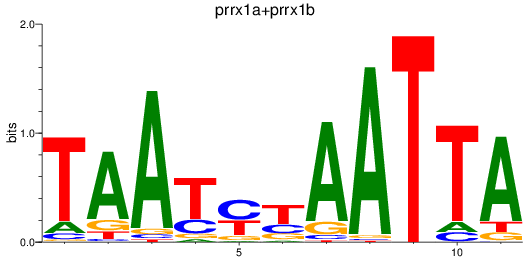
Results for prrx1a+prrx1b
Z-value: 1.59
Transcription factors associated with prrx1a+prrx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prrx1a
|
ENSDARG00000033971 | paired related homeobox 1a |
|
prrx1b
|
ENSDARG00000042027 | paired related homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prrx1a | dr11_v1_chr2_-_23172708_23172708 | 0.86 | 4.9e-06 | Click! |
| prrx1b | dr11_v1_chr20_+_34511678_34511678 | 0.56 | 1.7e-02 | Click! |
Activity profile of prrx1a+prrx1b motif
Sorted Z-values of prrx1a+prrx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_48397702 | 5.78 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr9_-_48407408 | 5.12 |
ENSDART00000058248
|
zgc:172182
|
zgc:172182 |
| chr8_-_21372446 | 4.91 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr22_-_15562933 | 4.44 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr10_-_17083180 | 4.21 |
ENSDART00000170083
|
fam166b
|
family with sequence similarity 166, member B |
| chr20_-_7072487 | 4.11 |
ENSDART00000145954
|
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr22_-_910926 | 4.01 |
ENSDART00000180075
|
FP016205.1
|
|
| chr9_+_48219111 | 3.98 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr12_-_990149 | 3.97 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr21_+_43404945 | 3.79 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr18_+_25752592 | 3.73 |
ENSDART00000111767
|
si:ch211-39k3.2
|
si:ch211-39k3.2 |
| chr7_-_30367650 | 3.73 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr7_+_35075847 | 3.72 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr14_-_49063157 | 3.69 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr6_+_40354424 | 3.49 |
ENSDART00000047416
|
slc4a8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr4_+_9669717 | 3.42 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr18_+_5490668 | 3.40 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr24_-_17067284 | 3.38 |
ENSDART00000111237
|
armc3
|
armadillo repeat containing 3 |
| chr16_-_45058919 | 3.26 |
ENSDART00000177134
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr19_+_43978814 | 3.24 |
ENSDART00000102314
|
nkd3
|
naked cuticle homolog 3 |
| chr7_-_28148310 | 3.20 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr21_+_6556635 | 3.19 |
ENSDART00000139598
|
col5a1
|
procollagen, type V, alpha 1 |
| chr4_-_9909371 | 3.19 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr25_-_16146851 | 3.19 |
ENSDART00000104043
|
dkk3b
|
dickkopf WNT signaling pathway inhibitor 3b |
| chr5_+_42400777 | 3.02 |
ENSDART00000183114
|
BX548073.8
|
|
| chr3_+_28953274 | 2.96 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr14_-_21123551 | 2.96 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr5_+_42386705 | 2.95 |
ENSDART00000143034
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr5_+_42407962 | 2.95 |
ENSDART00000188489
|
BX548073.11
|
|
| chr25_+_26895394 | 2.92 |
ENSDART00000155820
|
si:dkey-42p14.3
|
si:dkey-42p14.3 |
| chr4_-_16001118 | 2.91 |
ENSDART00000041070
ENSDART00000125389 |
mest
|
mesoderm specific transcript |
| chr21_-_43022048 | 2.86 |
ENSDART00000138329
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr5_+_42379517 | 2.85 |
ENSDART00000103325
|
pimr59
|
Pim proto-oncogene, serine/threonine kinase, related 59 |
| chr17_-_39772999 | 2.85 |
ENSDART00000155727
|
pimr60
|
Pim proto-oncogene, serine/threonine kinase, related 60 |
| chr5_+_42393896 | 2.79 |
ENSDART00000189550
|
BX548073.13
|
|
| chr4_+_72723304 | 2.77 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr23_+_39695827 | 2.76 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr7_+_72003301 | 2.74 |
ENSDART00000012918
ENSDART00000182268 ENSDART00000185750 |
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr23_+_42810055 | 2.70 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr4_-_390431 | 2.67 |
ENSDART00000067482
ENSDART00000138500 |
dynlt1
|
dynein, light chain, Tctex-type 1 |
| chr7_-_20758825 | 2.66 |
ENSDART00000156717
ENSDART00000182629 ENSDART00000179801 |
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr1_-_42289704 | 2.64 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr16_-_21140097 | 2.63 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr8_-_13046089 | 2.60 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr7_+_20344222 | 2.59 |
ENSDART00000141186
ENSDART00000139274 |
ponzr1
|
plac8 onzin related protein 1 |
| chr7_+_20344032 | 2.54 |
ENSDART00000144948
ENSDART00000138786 |
ponzr1
|
plac8 onzin related protein 1 |
| chr7_+_26629084 | 2.54 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr8_-_48675775 | 2.51 |
ENSDART00000060785
|
pimr183
|
Pim proto-oncogene, serine/threonine kinase, related 183 |
| chr6_-_8311044 | 2.49 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr17_+_31592191 | 2.49 |
ENSDART00000153765
|
si:dkey-13p1.3
|
si:dkey-13p1.3 |
| chr5_+_40299568 | 2.49 |
ENSDART00000142157
|
arl15a
|
ADP-ribosylation factor-like 15a |
| chr12_+_18663154 | 2.48 |
ENSDART00000057918
|
si:ch211-147h1.4
|
si:ch211-147h1.4 |
| chr15_-_14884332 | 2.47 |
ENSDART00000165237
|
si:ch211-24o8.4
|
si:ch211-24o8.4 |
| chr15_-_45110011 | 2.38 |
ENSDART00000182047
ENSDART00000188662 |
CABZ01072607.1
|
|
| chr24_-_38657683 | 2.35 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr15_-_35246742 | 2.33 |
ENSDART00000131479
|
mff
|
mitochondrial fission factor |
| chr12_-_20795867 | 2.31 |
ENSDART00000152835
ENSDART00000153424 |
nme2a
|
NME/NM23 nucleoside diphosphate kinase 2a |
| chr10_-_28835771 | 2.27 |
ENSDART00000192220
ENSDART00000188436 |
alcama
|
activated leukocyte cell adhesion molecule a |
| chr11_+_12744575 | 2.21 |
ENSDART00000131059
ENSDART00000081335 ENSDART00000142481 |
rbb4l
|
retinoblastoma binding protein 4, like |
| chr8_+_21406769 | 2.21 |
ENSDART00000135766
|
si:dkey-163f12.6
|
si:dkey-163f12.6 |
| chr12_+_3871452 | 2.21 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr20_-_44496245 | 2.19 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr24_+_35827766 | 2.17 |
ENSDART00000144700
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr1_+_44003654 | 2.14 |
ENSDART00000131337
|
si:dkey-22i16.10
|
si:dkey-22i16.10 |
| chr7_-_48667056 | 2.13 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr6_+_52350443 | 2.06 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr10_-_34107919 | 2.04 |
ENSDART00000167428
ENSDART00000145279 |
pimr144
pimr146
|
Pim proto-oncogene, serine/threonine kinase, related 144 Pim proto-oncogene, serine/threonine kinase, related 146 |
| chr10_+_6641514 | 2.02 |
ENSDART00000150003
|
si:ch211-57m13.5
|
si:ch211-57m13.5 |
| chr1_-_56080112 | 2.00 |
ENSDART00000075469
ENSDART00000161473 |
c3a.6
|
complement component c3a, duplicate 6 |
| chr4_+_2267641 | 1.98 |
ENSDART00000165503
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr6_+_6924637 | 1.98 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr18_-_36909773 | 1.97 |
ENSDART00000141694
|
si:ch211-160d20.5
|
si:ch211-160d20.5 |
| chr3_-_37759147 | 1.97 |
ENSDART00000151067
|
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr4_-_5831522 | 1.95 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr17_-_8268406 | 1.94 |
ENSDART00000149873
ENSDART00000064668 ENSDART00000148403 |
ahi1
|
Abelson helper integration site 1 |
| chr25_+_34641536 | 1.92 |
ENSDART00000167033
|
CABZ01079011.1
|
|
| chr2_+_56463167 | 1.91 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr20_+_2134816 | 1.90 |
ENSDART00000039249
|
l3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr21_-_8420830 | 1.88 |
ENSDART00000138040
|
lhx2a
|
LIM homeobox 2a |
| chr21_+_26720803 | 1.87 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr1_-_46244523 | 1.80 |
ENSDART00000143908
|
si:ch211-138g9.3
|
si:ch211-138g9.3 |
| chr11_-_45138857 | 1.78 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr23_-_18913032 | 1.75 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr17_-_26926577 | 1.73 |
ENSDART00000050202
|
rcan3
|
regulator of calcineurin 3 |
| chr21_+_27382893 | 1.72 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr23_+_20689255 | 1.71 |
ENSDART00000182420
|
usp21
|
ubiquitin specific peptidase 21 |
| chr12_+_28856151 | 1.71 |
ENSDART00000152969
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr10_-_34089779 | 1.68 |
ENSDART00000140070
|
pimr144
|
Pim proto-oncogene, serine/threonine kinase, related 144 |
| chr8_-_48675411 | 1.65 |
ENSDART00000165081
|
pimr183
|
Pim proto-oncogene, serine/threonine kinase, related 183 |
| chr10_-_34772211 | 1.63 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr11_-_3860534 | 1.61 |
ENSDART00000082425
|
gata2a
|
GATA binding protein 2a |
| chr20_-_52338782 | 1.61 |
ENSDART00000109735
ENSDART00000132941 |
si:ch1073-287p18.1
|
si:ch1073-287p18.1 |
| chr22_-_32507966 | 1.57 |
ENSDART00000104693
|
pcbp4
|
poly(rC) binding protein 4 |
| chr12_+_34378733 | 1.56 |
ENSDART00000018939
|
tha1
|
threonine aldolase 1 |
| chr6_+_9130989 | 1.56 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr20_+_34717403 | 1.55 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr23_-_16485190 | 1.55 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr24_+_26997798 | 1.55 |
ENSDART00000089506
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr1_+_25801648 | 1.54 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr4_+_78008298 | 1.53 |
ENSDART00000174237
|
cct2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr17_-_19345521 | 1.52 |
ENSDART00000082085
|
gsc
|
goosecoid |
| chr25_+_3327071 | 1.52 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr9_+_35876927 | 1.49 |
ENSDART00000138834
|
mab21l3
|
mab-21-like 3 |
| chr3_-_24980067 | 1.48 |
ENSDART00000048871
|
desi1a
|
desumoylating isopeptidase 1a |
| chr10_-_26744131 | 1.48 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr16_+_31921812 | 1.48 |
ENSDART00000176928
ENSDART00000193733 |
rps9
|
ribosomal protein S9 |
| chr13_-_9496141 | 1.47 |
ENSDART00000131472
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr1_-_49250490 | 1.47 |
ENSDART00000150386
|
si:ch73-6k14.2
|
si:ch73-6k14.2 |
| chr14_-_46173265 | 1.46 |
ENSDART00000164321
|
zmp:0000000758
|
zmp:0000000758 |
| chr5_-_72105216 | 1.44 |
ENSDART00000044786
|
thnsl2
|
threonine synthase-like 2 |
| chr10_-_34243269 | 1.44 |
ENSDART00000191406
ENSDART00000131549 |
pimr167
|
Pim proto-oncogene, serine/threonine kinase, related 167 |
| chr6_-_3573716 | 1.43 |
ENSDART00000026693
|
dmbx1b
|
diencephalon/mesencephalon homeobox 1b |
| chr16_+_23984755 | 1.43 |
ENSDART00000145328
|
apoc2
|
apolipoprotein C-II |
| chr10_-_34150577 | 1.42 |
ENSDART00000191166
ENSDART00000137763 |
pimr150
pimr147
|
Pim proto-oncogene, serine/threonine kinase, related 150 Pim proto-oncogene, serine/threonine kinase, related 147 |
| chr12_+_2428247 | 1.42 |
ENSDART00000152529
|
lrrc18b
|
leucine rich repeat containing 18b |
| chr9_-_34300707 | 1.42 |
ENSDART00000049805
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr13_-_9546366 | 1.41 |
ENSDART00000140408
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr1_-_669717 | 1.40 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr21_-_4468216 | 1.40 |
ENSDART00000193272
|
pimr160
|
Pim proto-oncogene, serine/threonine kinase, related 160 |
| chr1_-_44899287 | 1.38 |
ENSDART00000187522
|
tcirg1a
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3a |
| chr10_-_34123733 | 1.38 |
ENSDART00000157708
ENSDART00000143954 |
pimr150
|
Pim proto-oncogene, serine/threonine kinase, related 150 |
| chr15_-_44512461 | 1.38 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr21_-_4327738 | 1.38 |
ENSDART00000151457
|
pimr166
|
Pim proto-oncogene, serine/threonine kinase, related 166 |
| chr20_-_2134620 | 1.38 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr10_-_3332362 | 1.38 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr3_+_33918510 | 1.37 |
ENSDART00000055248
|
alkbh7
|
alkB homolog 7 |
| chr10_-_34200750 | 1.36 |
ENSDART00000137279
|
pimr148
|
Pim proto-oncogene, serine/threonine kinase, related 148 |
| chr21_-_4347983 | 1.36 |
ENSDART00000151809
|
pimr164
|
Pim proto-oncogene, serine/threonine kinase, related 164 |
| chr21_-_4410134 | 1.36 |
ENSDART00000151498
|
pimr162
|
Pim proto-oncogene, serine/threonine kinase, related 162 |
| chr13_+_9046473 | 1.36 |
ENSDART00000136557
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr10_-_34223567 | 1.35 |
ENSDART00000184889
|
pimr145
|
Pim proto-oncogene, serine/threonine kinase, related 145 |
| chr14_-_46070802 | 1.34 |
ENSDART00000038670
|
elf2a
|
E74-like factor 2a (ets domain transcription factor) |
| chr13_+_9091958 | 1.34 |
ENSDART00000171741
ENSDART00000147051 |
pimr155
|
Pim proto-oncogene, serine/threonine kinase, related 155 |
| chr1_-_45049603 | 1.34 |
ENSDART00000023336
|
rps6
|
ribosomal protein S6 |
| chr23_+_45282858 | 1.33 |
ENSDART00000162353
|
CABZ01073265.1
|
|
| chr21_-_4487752 | 1.32 |
ENSDART00000168410
|
pimr161
|
Pim proto-oncogene, serine/threonine kinase, related 161 |
| chr18_-_15932704 | 1.32 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr13_+_30903816 | 1.31 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr20_-_46534179 | 1.31 |
ENSDART00000060689
|
eif2b2
|
eukaryotic translation initiation factor 2B, subunit 2 beta |
| chr21_-_5393125 | 1.31 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr10_-_34174858 | 1.30 |
ENSDART00000131346
|
pimr145
|
Pim proto-oncogene, serine/threonine kinase, related 145 |
| chr14_-_24410673 | 1.30 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr21_-_4368035 | 1.30 |
ENSDART00000151126
|
pimr163
|
Pim proto-oncogene, serine/threonine kinase, related 163 |
| chr4_-_28373909 | 1.30 |
ENSDART00000056132
|
tprkb
|
Tp53rk binding protein |
| chr7_+_66822229 | 1.29 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr23_-_12345764 | 1.29 |
ENSDART00000133956
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr8_+_1118328 | 1.28 |
ENSDART00000081445
|
ubox5
|
U-box domain containing 5 |
| chr2_-_30135446 | 1.27 |
ENSDART00000141906
|
trpa1a
|
transient receptor potential cation channel, subfamily A, member 1a |
| chr13_+_9112544 | 1.26 |
ENSDART00000143138
ENSDART00000131495 |
pimr153
|
Pim proto-oncogene, serine/threonine kinase, related 153 |
| chr6_+_43234213 | 1.26 |
ENSDART00000112474
|
arl6ip5a
|
ADP-ribosylation factor-like 6 interacting protein 5a |
| chr10_-_17222083 | 1.25 |
ENSDART00000134059
|
depdc5
|
DEP domain containing 5 |
| chr12_-_32013125 | 1.24 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr24_-_9960290 | 1.24 |
ENSDART00000143390
ENSDART00000092975 ENSDART00000184953 |
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr10_-_31440500 | 1.24 |
ENSDART00000024778
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr16_-_31717851 | 1.24 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr1_+_53954230 | 1.24 |
ENSDART00000037729
ENSDART00000159900 |
ccsapa
|
centriole, cilia and spindle-associated protein a |
| chr1_+_12009673 | 1.23 |
ENSDART00000080100
|
slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr15_-_21702317 | 1.23 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr6_+_49081141 | 1.21 |
ENSDART00000131080
ENSDART00000050000 |
tshba
|
thyroid stimulating hormone, beta subunit, a |
| chr1_-_46981134 | 1.21 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr15_-_17869115 | 1.21 |
ENSDART00000112838
|
atf5b
|
activating transcription factor 5b |
| chr20_+_11731039 | 1.20 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr23_-_44226556 | 1.20 |
ENSDART00000149115
|
zgc:158659
|
zgc:158659 |
| chr9_-_30555725 | 1.19 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr6_+_40661703 | 1.19 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr13_+_9162465 | 1.18 |
ENSDART00000102108
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr21_-_4428354 | 1.17 |
ENSDART00000151174
|
CT027677.2
|
|
| chr19_+_9174166 | 1.17 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr10_+_29698467 | 1.17 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr25_+_31267268 | 1.16 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr22_-_38274188 | 1.16 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr3_+_12440099 | 1.14 |
ENSDART00000158060
|
vasnb
|
vasorin b |
| chr19_-_7495006 | 1.14 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr4_+_74141400 | 1.14 |
ENSDART00000166994
|
trim24
|
tripartite motif containing 24 |
| chr15_+_42285643 | 1.13 |
ENSDART00000152731
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr5_+_25074648 | 1.12 |
ENSDART00000188951
|
mrpl41
|
mitochondrial ribosomal protein L41 |
| chr23_+_26733232 | 1.12 |
ENSDART00000035080
|
zgc:158263
|
zgc:158263 |
| chr20_-_30920356 | 1.11 |
ENSDART00000022951
|
kif25
|
kinesin family member 25 |
| chr14_+_48964628 | 1.11 |
ENSDART00000105427
|
mif
|
macrophage migration inhibitory factor |
| chr13_+_9184801 | 1.10 |
ENSDART00000133916
ENSDART00000144483 |
pimr156
|
Pim proto-oncogene, serine/threonine kinase, related 156 |
| chr17_+_30545895 | 1.10 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr5_+_41322783 | 1.09 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr22_+_14051894 | 1.08 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr18_-_21271373 | 1.08 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr20_+_24448007 | 1.08 |
ENSDART00000139866
|
si:dkey-273g18.1
|
si:dkey-273g18.1 |
| chr25_+_18964782 | 1.07 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr16_-_41944765 | 1.07 |
ENSDART00000142648
|
gramd1a
|
GRAM domain containing 1A |
| chr13_-_36535128 | 1.06 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr14_+_8475007 | 1.06 |
ENSDART00000148210
|
dnajc4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr1_-_43727418 | 1.05 |
ENSDART00000133715
ENSDART00000074597 ENSDART00000132542 ENSDART00000181792 |
bdh2
SLC9B2
|
3-hydroxybutyrate dehydrogenase, type 2 si:dkey-162b23.4 |
| chr5_-_11943750 | 1.05 |
ENSDART00000074979
|
rnft2
|
ring finger protein, transmembrane 2 |
| chr10_-_34741738 | 1.05 |
ENSDART00000163072
|
dclk1a
|
doublecortin-like kinase 1a |
| chr6_+_41191482 | 1.05 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr20_+_46371458 | 1.05 |
ENSDART00000152912
|
adgrg11
|
adhesion G protein-coupled receptor G11 |
| chr25_+_22730490 | 1.04 |
ENSDART00000149455
|
abcc8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr23_+_25893020 | 1.04 |
ENSDART00000144769
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr2_+_16781015 | 1.03 |
ENSDART00000155147
ENSDART00000003845 |
tfa
|
transferrin-a |
| chr14_-_1355544 | 1.02 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr7_+_20030888 | 1.02 |
ENSDART00000192808
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr1_-_9195629 | 1.02 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of prrx1a+prrx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.9 | 2.6 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.6 | 2.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.6 | 4.0 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.5 | 1.6 | GO:1902893 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.5 | 1.6 | GO:0042364 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.5 | 5.1 | GO:0072088 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.4 | 1.8 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 1.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.4 | 1.5 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.4 | 1.5 | GO:0050955 | thermoception(GO:0050955) detection of temperature stimulus involved in thermoception(GO:0050960) detection of temperature stimulus involved in sensory perception(GO:0050961) |
| 0.4 | 1.5 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.3 | 1.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.3 | 1.3 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.3 | 2.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 0.3 | GO:0021559 | trigeminal nerve development(GO:0021559) glossopharyngeal nerve development(GO:0021563) |
| 0.3 | 1.7 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 4.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 1.6 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.3 | 1.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.3 | 4.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 2.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 0.9 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 0.6 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 1.0 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.2 | 0.6 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.2 | 1.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 0.8 | GO:0015868 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.2 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 2.6 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.2 | 1.2 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.2 | 1.1 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.2 | 0.9 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.2 | 1.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 1.0 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.2 | 0.5 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.2 | 2.0 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.2 | 1.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.2 | 1.4 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.2 | 0.6 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.2 | 1.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 3.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 2.3 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 2.5 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.8 | GO:0021767 | mammillary body development(GO:0021767) social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 0.5 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.9 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.5 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.9 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.8 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.6 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 3.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 2.9 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.1 | 0.5 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.1 | 43.4 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 0.8 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.8 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.8 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 1.5 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 0.7 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.9 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 1.8 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 0.3 | GO:0046333 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 1.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.8 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.7 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 | 1.5 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 2.0 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 1.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.4 | GO:0097300 | necrotic cell death(GO:0070265) programmed necrotic cell death(GO:0097300) |
| 0.1 | 1.2 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.1 | 0.1 | GO:0048331 | axial mesoderm structural organization(GO:0048331) mesoderm structural organization(GO:0048338) |
| 0.1 | 0.2 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.8 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.9 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 3.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.8 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 1.0 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 1.0 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.7 | GO:0046959 | learning(GO:0007612) nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.6 | GO:0044550 | melanin biosynthetic process(GO:0042438) secondary metabolite biosynthetic process(GO:0044550) |
| 0.1 | 0.7 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 7.5 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.1 | 4.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 2.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.7 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 1.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.6 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.3 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 0.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 2.3 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.5 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) presynaptic active zone organization(GO:1990709) |
| 0.0 | 0.4 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.6 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 1.0 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.3 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.3 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0046635 | positive regulation of alpha-beta T cell activation(GO:0046635) |
| 0.0 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 1.3 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.6 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.5 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.6 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 2.8 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.5 | GO:0071696 | ectodermal placode development(GO:0071696) |
| 0.0 | 0.4 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.5 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0071632 | optomotor response(GO:0071632) |
| 0.0 | 0.1 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.3 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 2.1 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.7 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.0 | 1.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 1.0 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.0 | 1.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.9 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.9 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.2 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 1.2 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 1.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 1.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.5 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 1.1 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.4 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.8 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 1.1 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 2.2 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.1 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.1 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 1.7 | GO:0019932 | second-messenger-mediated signaling(GO:0019932) |
| 0.0 | 1.2 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.0 | 0.4 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 1.3 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 0.3 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.3 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 1.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 1.5 | GO:0070646 | protein modification by small protein removal(GO:0070646) |
| 0.0 | 0.2 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 1.4 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 2.8 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.3 | 1.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.3 | 1.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 0.8 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 4.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 0.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 1.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 1.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.2 | 1.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 1.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 1.6 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 1.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 1.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.5 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 1.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 3.6 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 1.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 4.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 3.0 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 0.6 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 2.8 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.6 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 2.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 3.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.1 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.5 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 1.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 1.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.0 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.8 | 3.3 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.6 | 3.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.6 | 2.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.5 | 3.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.4 | 1.8 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.4 | 1.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.4 | 1.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.3 | 2.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.3 | 1.4 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.3 | 1.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 1.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.3 | 3.7 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.3 | 2.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 1.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 3.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 2.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 2.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 1.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 1.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.2 | 0.8 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.2 | 1.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 1.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 1.0 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.2 | 0.6 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.3 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 2.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 1.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.5 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 1.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 1.6 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 0.8 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 1.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.9 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.1 | 0.4 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 1.0 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 1.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.5 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 0.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.9 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 2.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.6 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.1 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 1.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.9 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.2 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.6 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 3.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 2.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.1 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 4.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 38.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.0 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.7 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.4 | GO:0010851 | cyclase regulator activity(GO:0010851) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 2.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 1.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 4.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 4.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 1.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.8 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 2.0 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.6 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.7 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 3.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 2.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 3.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 1.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 3.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.8 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 4.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.7 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 1.3 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |