Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
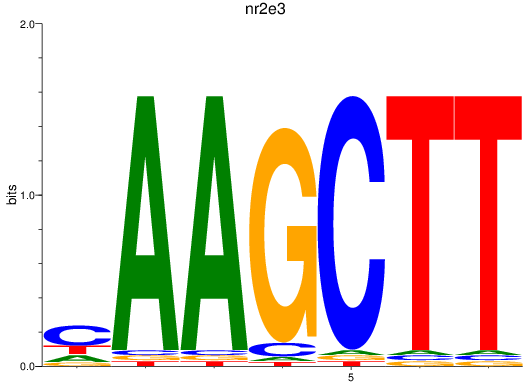
Results for nr2e3
Z-value: 1.36
Transcription factors associated with nr2e3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2e3
|
ENSDARG00000045904 | nuclear receptor subfamily 2, group E, member 3 |
|
nr2e3
|
ENSDARG00000113178 | nuclear receptor subfamily 2, group E, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2e3 | dr11_v1_chr25_+_22274642_22274642 | -0.90 | 4.1e-07 | Click! |
Activity profile of nr2e3 motif
Sorted Z-values of nr2e3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_27550768 | 3.94 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr21_-_30293224 | 3.60 |
ENSDART00000101051
|
slbp2
|
stem-loop binding protein 2 |
| chr11_-_3494964 | 3.29 |
ENSDART00000162369
|
acap3b
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3b |
| chr12_+_30789611 | 3.24 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr19_-_27564980 | 3.23 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr23_+_2740741 | 3.20 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr17_-_2596125 | 3.13 |
ENSDART00000175740
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr24_-_10014512 | 3.01 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr13_-_6252498 | 2.82 |
ENSDART00000115157
|
tuba4l
|
tubulin, alpha 4 like |
| chr25_+_22319940 | 2.77 |
ENSDART00000154065
ENSDART00000153492 ENSDART00000024866 ENSDART00000154376 |
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr10_+_3145707 | 2.76 |
ENSDART00000160046
|
hic2
|
hypermethylated in cancer 2 |
| chr15_+_38299385 | 2.76 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr19_-_27564458 | 2.71 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr17_-_2578026 | 2.69 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr20_+_33987465 | 2.69 |
ENSDART00000061751
|
zp3a.2
|
zona pellucida glycoprotein 3a, tandem duplicate 2 |
| chr20_-_29498178 | 2.67 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr22_-_17652914 | 2.59 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr15_+_38299563 | 2.53 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr9_-_34269066 | 2.49 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr9_-_8314028 | 2.47 |
ENSDART00000102739
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr5_+_57924611 | 2.45 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr23_+_27756984 | 2.44 |
ENSDART00000137103
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr20_-_44576949 | 2.33 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr9_+_33216945 | 2.33 |
ENSDART00000134029
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr12_+_13344896 | 2.29 |
ENSDART00000089017
|
rnasen
|
ribonuclease type III, nuclear |
| chr22_-_17652112 | 2.25 |
ENSDART00000189205
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr21_-_21178410 | 2.24 |
ENSDART00000185277
ENSDART00000141341 ENSDART00000145872 ENSDART00000079678 |
ftsj1
|
FtsJ RNA methyltransferase homolog 1 |
| chr23_-_22523303 | 2.23 |
ENSDART00000079019
|
spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr3_-_47235997 | 2.19 |
ENSDART00000047071
|
tmed1a
|
transmembrane p24 trafficking protein 1a |
| chr21_+_4116437 | 2.18 |
ENSDART00000167791
ENSDART00000123759 |
rapgef1b
|
Rap guanine nucleotide exchange factor (GEF) 1b |
| chr20_+_33991801 | 2.16 |
ENSDART00000061744
|
zp3a.1
|
zona pellucida glycoprotein 3a, tandem duplicate 1 |
| chr24_-_5691956 | 2.12 |
ENSDART00000189112
|
dia1b
|
deleted in autism 1b |
| chr4_-_77114795 | 2.10 |
ENSDART00000144849
|
CU467646.2
|
|
| chr14_+_30328567 | 2.10 |
ENSDART00000105889
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr14_+_21722235 | 2.10 |
ENSDART00000183667
|
stx3a
|
syntaxin 3A |
| chr3_+_51563695 | 2.09 |
ENSDART00000008607
|
ttyh2l
|
tweety homolog 2, like |
| chr4_-_1757460 | 2.08 |
ENSDART00000144074
|
tm7sf3
|
transmembrane 7 superfamily member 3 |
| chr19_-_47570672 | 2.07 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr19_+_42231431 | 1.92 |
ENSDART00000102698
|
jtb
|
jumping translocation breakpoint |
| chr17_+_5931530 | 1.91 |
ENSDART00000168326
ENSDART00000189790 |
znf513b
|
zinc finger protein 513b |
| chr12_-_25887864 | 1.90 |
ENSDART00000152983
|
si:dkey-193p11.2
|
si:dkey-193p11.2 |
| chr14_+_22129096 | 1.90 |
ENSDART00000132514
|
ccng1
|
cyclin G1 |
| chr21_-_37027252 | 1.89 |
ENSDART00000076483
|
zgc:77151
|
zgc:77151 |
| chr5_+_25733774 | 1.88 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr6_+_32338334 | 1.88 |
ENSDART00000065149
|
dock7
|
dedicator of cytokinesis 7 |
| chr10_+_15255012 | 1.88 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr5_+_37747128 | 1.87 |
ENSDART00000148795
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr10_+_15255198 | 1.86 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr10_+_3153973 | 1.86 |
ENSDART00000183223
|
hic2
|
hypermethylated in cancer 2 |
| chr5_-_20135679 | 1.86 |
ENSDART00000079402
|
usp30
|
ubiquitin specific peptidase 30 |
| chr21_+_15723069 | 1.85 |
ENSDART00000149126
ENSDART00000130628 |
p2rx4a
|
purinergic receptor P2X, ligand-gated ion channel, 4a |
| chr13_-_24717365 | 1.84 |
ENSDART00000137934
ENSDART00000003922 |
erlin1
|
ER lipid raft associated 1 |
| chr24_+_15020402 | 1.83 |
ENSDART00000148102
|
dok6
|
docking protein 6 |
| chr1_+_59314675 | 1.80 |
ENSDART00000161872
ENSDART00000160658 ENSDART00000169792 ENSDART00000160735 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr5_-_56964547 | 1.78 |
ENSDART00000074400
|
tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr19_+_4968947 | 1.76 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr10_+_16911951 | 1.74 |
ENSDART00000164933
|
UNC13B (1 of many)
|
unc-13 homolog B |
| chr1_+_2431956 | 1.73 |
ENSDART00000183832
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr8_+_26396552 | 1.72 |
ENSDART00000087151
|
amt
|
aminomethyltransferase |
| chr7_-_18554603 | 1.71 |
ENSDART00000108938
|
msantd1
|
Myb/SANT-like DNA-binding domain containing 1 |
| chr24_-_21090447 | 1.69 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr18_+_50880096 | 1.69 |
ENSDART00000169782
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr8_-_25771474 | 1.68 |
ENSDART00000193883
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr3_-_27647845 | 1.67 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr15_+_781717 | 1.65 |
ENSDART00000154903
|
znf970
|
zinc finger protein 970 |
| chr8_+_11471350 | 1.64 |
ENSDART00000092355
ENSDART00000136184 |
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr2_-_1622641 | 1.64 |
ENSDART00000082143
|
prkacbb
|
protein kinase, cAMP-dependent, catalytic, beta b |
| chr16_-_41488023 | 1.63 |
ENSDART00000169312
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr12_-_13905307 | 1.62 |
ENSDART00000152400
|
dbf4b
|
DBF4 zinc finger B |
| chr8_-_1838315 | 1.61 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr9_+_8364553 | 1.60 |
ENSDART00000190713
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr14_+_41318412 | 1.58 |
ENSDART00000064614
|
xkrx
|
XK, Kell blood group complex subunit-related, X-linked |
| chr11_-_15850794 | 1.58 |
ENSDART00000185946
|
rap1ab
|
RAP1A, member of RAS oncogene family b |
| chr6_-_1432200 | 1.56 |
ENSDART00000182901
|
LO018148.1
|
|
| chr6_+_10333920 | 1.55 |
ENSDART00000151667
ENSDART00000151477 |
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr20_-_31252809 | 1.54 |
ENSDART00000137236
|
hpcal1
|
hippocalcin-like 1 |
| chr21_+_43172506 | 1.51 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr15_-_5901514 | 1.49 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr7_+_27603211 | 1.49 |
ENSDART00000148782
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr9_-_21098413 | 1.49 |
ENSDART00000018570
|
wars2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr3_-_60589292 | 1.47 |
ENSDART00000157822
|
jmjd6
|
jumonji domain containing 6 |
| chr5_-_15494164 | 1.46 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr16_-_8132742 | 1.46 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr4_-_7876005 | 1.46 |
ENSDART00000109252
|
nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr11_+_31380495 | 1.45 |
ENSDART00000185073
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr2_-_49031303 | 1.42 |
ENSDART00000143471
|
cdc34b
|
cell division cycle 34 homolog (S. cerevisiae) b |
| chr20_-_23426339 | 1.42 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr7_+_36467796 | 1.42 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr1_+_45925150 | 1.41 |
ENSDART00000074689
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr11_+_43740949 | 1.38 |
ENSDART00000189296
|
CU862021.1
|
|
| chr17_+_13031497 | 1.37 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr9_+_22632126 | 1.37 |
ENSDART00000139434
|
etv5a
|
ets variant 5a |
| chr21_+_37477001 | 1.36 |
ENSDART00000114778
|
amot
|
angiomotin |
| chr2_-_22530969 | 1.36 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr8_-_15129573 | 1.35 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr6_+_12527725 | 1.35 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr21_+_33311622 | 1.35 |
ENSDART00000163808
|
si:ch211-151g22.1
|
si:ch211-151g22.1 |
| chr19_-_20106486 | 1.35 |
ENSDART00000043924
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr19_+_43684376 | 1.34 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr20_-_28698172 | 1.34 |
ENSDART00000190635
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr19_+_41479990 | 1.32 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr14_-_21618005 | 1.32 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr9_+_33340311 | 1.32 |
ENSDART00000140064
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr9_-_49964810 | 1.32 |
ENSDART00000167098
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr3_-_3439150 | 1.30 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr3_-_30488063 | 1.30 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr8_+_13364950 | 1.29 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr21_+_19319804 | 1.29 |
ENSDART00000063621
|
abraxas2a
|
abraxas 2a, BRISC complex subunit |
| chr7_+_71955486 | 1.29 |
ENSDART00000189349
|
CABZ01071171.1
|
Danio rerio low density lipoprotein receptor-related protein 4 (lrp4), mRNA. |
| chr14_+_5835134 | 1.29 |
ENSDART00000054867
|
aup1
|
ancient ubiquitous protein 1 |
| chr16_+_25068576 | 1.28 |
ENSDART00000125838
|
im:7147486
|
im:7147486 |
| chr5_+_60518611 | 1.28 |
ENSDART00000130565
ENSDART00000186310 |
tmem132e
|
transmembrane protein 132E |
| chr9_+_43799829 | 1.28 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr9_-_30247961 | 1.27 |
ENSDART00000131519
|
si:dkey-100n23.3
|
si:dkey-100n23.3 |
| chr13_-_6279684 | 1.26 |
ENSDART00000092105
|
agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr24_-_18809433 | 1.26 |
ENSDART00000152009
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr25_+_33033633 | 1.26 |
ENSDART00000192336
|
tln2b
|
talin 2b |
| chr16_-_8120203 | 1.25 |
ENSDART00000193430
|
snrka
|
SNF related kinase a |
| chr17_+_49637257 | 1.24 |
ENSDART00000075281
ENSDART00000065917 ENSDART00000123641 |
zgc:113372
|
zgc:113372 |
| chr23_+_27789795 | 1.24 |
ENSDART00000141458
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr17_-_10498347 | 1.23 |
ENSDART00000171324
|
mipol1
|
mirror-image polydactyly 1 |
| chr12_+_33320884 | 1.23 |
ENSDART00000188988
|
csnk1db
|
casein kinase 1, delta b |
| chr20_+_32523576 | 1.23 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr19_-_23914743 | 1.22 |
ENSDART00000139856
|
chtopa
|
chromatin target of PRMT1a |
| chr19_-_47832853 | 1.19 |
ENSDART00000170988
|
ago4
|
argonaute RISC catalytic component 4 |
| chr6_+_36821621 | 1.19 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr15_-_28587147 | 1.18 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr2_-_45135591 | 1.18 |
ENSDART00000014691
ENSDART00000123916 |
eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr4_+_25607743 | 1.16 |
ENSDART00000028297
|
acot14
|
acyl-CoA thioesterase 14 |
| chr2_+_1939391 | 1.16 |
ENSDART00000163310
|
exosc4
|
exosome component 4 |
| chr3_+_23047977 | 1.15 |
ENSDART00000155792
|
b4galnt2.2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 2 |
| chr8_-_50888806 | 1.15 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr6_+_19948043 | 1.14 |
ENSDART00000182636
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr15_-_33304133 | 1.14 |
ENSDART00000186092
|
nbeab
|
neurobeachin b |
| chr10_+_10313024 | 1.13 |
ENSDART00000142895
ENSDART00000129952 |
urm1
|
ubiquitin related modifier 1 |
| chr5_+_28260158 | 1.12 |
ENSDART00000181434
|
ncaph
|
non-SMC condensin I complex, subunit H |
| chr7_+_20917966 | 1.11 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr25_-_7670616 | 1.10 |
ENSDART00000131583
ENSDART00000142794 |
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr6_-_3982783 | 1.10 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr7_-_21905851 | 1.09 |
ENSDART00000111066
ENSDART00000020288 |
epoa
|
erythropoietin a |
| chr13_-_36525982 | 1.09 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr16_-_17316440 | 1.08 |
ENSDART00000147615
|
zyx
|
zyxin |
| chr3_-_27868183 | 1.08 |
ENSDART00000185812
|
abat
|
4-aminobutyrate aminotransferase |
| chr2_+_31942390 | 1.08 |
ENSDART00000138684
ENSDART00000146758 ENSDART00000137921 |
otulinb
|
OTU deubiquitinase with linear linkage specificity b |
| chr6_-_40778294 | 1.08 |
ENSDART00000019845
|
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr5_-_32336613 | 1.08 |
ENSDART00000139732
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_-_40959667 | 1.07 |
ENSDART00000084070
|
rbm33a
|
RNA binding motif protein 33a |
| chr5_-_22052852 | 1.07 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr23_-_31645760 | 1.06 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr14_-_46617228 | 1.06 |
ENSDART00000161010
ENSDART00000171054 |
prom1a
|
prominin 1a |
| chr6_-_29051773 | 1.04 |
ENSDART00000190508
ENSDART00000180191 ENSDART00000111682 |
evi5b
|
ecotropic viral integration site 5b |
| chr25_-_7918526 | 1.04 |
ENSDART00000104686
ENSDART00000156761 |
ambra1b
|
autophagy/beclin-1 regulator 1b |
| chr12_+_13905286 | 1.04 |
ENSDART00000147186
|
fkbp10b
|
FK506 binding protein 10b |
| chr4_-_73411863 | 1.04 |
ENSDART00000171434
|
zgc:162958
|
zgc:162958 |
| chr13_+_37656278 | 1.03 |
ENSDART00000193251
|
phf3
|
PHD finger protein 3 |
| chr22_-_6562618 | 1.03 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr9_+_29430432 | 1.03 |
ENSDART00000125632
|
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr15_+_24795473 | 1.03 |
ENSDART00000139689
ENSDART00000141033 ENSDART00000100746 ENSDART00000135677 |
gosr1
|
golgi SNAP receptor complex member 1 |
| chr6_-_40657653 | 1.02 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr15_-_31177324 | 1.02 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr23_-_27505825 | 1.01 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr9_+_6079528 | 1.01 |
ENSDART00000142167
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr13_-_4134141 | 1.01 |
ENSDART00000132354
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr6_+_40922572 | 1.01 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr9_-_12659140 | 1.01 |
ENSDART00000058565
|
pttg1ipb
|
PTTG1 interacting protein b |
| chr10_+_585719 | 1.01 |
ENSDART00000180167
|
smad4a
|
SMAD family member 4a |
| chr13_-_25750910 | 1.01 |
ENSDART00000111567
|
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr1_+_1712140 | 1.00 |
ENSDART00000081047
|
atp1a1a.1
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 1 |
| chr17_-_51262430 | 1.00 |
ENSDART00000163222
|
trappc12
|
trafficking protein particle complex 12 |
| chr19_+_8612839 | 1.00 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr15_-_14083028 | 0.99 |
ENSDART00000147796
ENSDART00000043492 ENSDART00000133080 |
trappc6bl
|
trafficking protein particle complex 6b-like |
| chr6_-_10037207 | 0.98 |
ENSDART00000179701
|
nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr2_+_49805892 | 0.98 |
ENSDART00000056248
|
wdr48b
|
WD repeat domain 48b |
| chr5_-_54481692 | 0.98 |
ENSDART00000165719
|
fbxw5
|
F-box and WD repeat domain containing 5 |
| chr8_+_16676894 | 0.98 |
ENSDART00000076586
|
si:ch211-198n5.11
|
si:ch211-198n5.11 |
| chr16_-_10223741 | 0.98 |
ENSDART00000188099
|
si:rp71-15i12.1
|
si:rp71-15i12.1 |
| chr7_-_30280934 | 0.97 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr2_+_24080694 | 0.97 |
ENSDART00000024058
|
kcnh2a
|
potassium voltage-gated channel, subfamily H (eag-related), member 2a |
| chr23_+_22785375 | 0.97 |
ENSDART00000142085
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr14_+_45028062 | 0.97 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr4_-_5019113 | 0.96 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr2_-_26596794 | 0.95 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr10_-_41397663 | 0.94 |
ENSDART00000158038
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr9_-_34260214 | 0.93 |
ENSDART00000012385
|
me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr17_-_2721336 | 0.92 |
ENSDART00000109582
ENSDART00000192691 ENSDART00000189381 |
spata7
|
spermatogenesis associated 7 |
| chr3_+_35498119 | 0.91 |
ENSDART00000178963
|
tnrc6a
|
trinucleotide repeat containing 6a |
| chr18_-_40913294 | 0.91 |
ENSDART00000059196
ENSDART00000098878 |
polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr24_+_11908833 | 0.91 |
ENSDART00000178622
|
fen1
|
flap structure-specific endonuclease 1 |
| chr19_+_20787179 | 0.90 |
ENSDART00000193204
|
adnp2b
|
ADNP homeobox 2b |
| chr15_-_14194208 | 0.89 |
ENSDART00000188237
ENSDART00000183155 ENSDART00000165520 |
pnkp
|
polynucleotide kinase 3'-phosphatase |
| chr16_-_9869056 | 0.89 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr15_-_20709289 | 0.89 |
ENSDART00000136767
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr12_+_33320504 | 0.89 |
ENSDART00000021491
|
csnk1db
|
casein kinase 1, delta b |
| chr8_-_8349653 | 0.89 |
ENSDART00000025214
|
tsr2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr2_+_55199721 | 0.88 |
ENSDART00000016143
|
zmp:0000000521
|
zmp:0000000521 |
| chr1_+_45663727 | 0.88 |
ENSDART00000038574
ENSDART00000141144 ENSDART00000149565 |
trappc5
|
trafficking protein particle complex 5 |
| chr19_+_42328423 | 0.88 |
ENSDART00000077059
|
anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr19_+_37925616 | 0.87 |
ENSDART00000148348
|
nxph1
|
neurexophilin 1 |
| chr17_-_6955082 | 0.87 |
ENSDART00000109228
|
zbtb24
|
zinc finger and BTB domain containing 24 |
| chr7_+_21752168 | 0.87 |
ENSDART00000173641
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2e3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 1.6 | GO:2000301 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.6 | 10.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.6 | 2.8 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.5 | 2.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.5 | 3.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.5 | 4.3 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.4 | 4.7 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.4 | 1.7 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.4 | 2.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.4 | 3.2 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.4 | 3.2 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 1.0 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.3 | 1.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 1.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.3 | 2.4 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.3 | 0.5 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.3 | 0.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.3 | 0.8 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.2 | 1.5 | GO:0071305 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.2 | 1.6 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.2 | 0.7 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.2 | 1.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 1.1 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.2 | 1.1 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.2 | 1.9 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.2 | 1.0 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.2 | 0.6 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.2 | 0.8 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.2 | 2.5 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 1.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 1.1 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.2 | 1.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 1.4 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.2 | 0.8 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 0.8 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.2 | 0.6 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 1.9 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 2.1 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.0 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 1.8 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 1.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.7 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 2.5 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 2.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 1.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 1.1 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 0.7 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.9 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 1.3 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 1.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 1.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 1.0 | GO:0046070 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 0.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.0 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 1.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 1.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.3 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.5 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 0.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.7 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.9 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 1.1 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 2.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.5 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.1 | 1.8 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 1.3 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 2.0 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) |
| 0.1 | 1.4 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.6 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 1.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.4 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.1 | 0.5 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 1.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.4 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.5 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 1.4 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 2.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.3 | GO:1902373 | negative regulation of RNA catabolic process(GO:1902369) negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 1.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.0 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.4 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 1.4 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.1 | GO:0009155 | purine deoxyribonucleotide catabolic process(GO:0009155) |
| 0.1 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 1.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.5 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.1 | 0.4 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.1 | 0.2 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.1 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.7 | GO:2000142 | regulation of transcription initiation from RNA polymerase II promoter(GO:0060260) regulation of DNA-templated transcription, initiation(GO:2000142) |
| 0.1 | 2.4 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 6.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.2 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 2.5 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.8 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 3.8 | GO:1902652 | cholesterol metabolic process(GO:0008203) secondary alcohol metabolic process(GO:1902652) |
| 0.1 | 0.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.6 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 5.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.0 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.8 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 0.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.5 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.6 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.4 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.4 | GO:0021634 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) optic nerve formation(GO:0021634) |
| 0.0 | 0.7 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 1.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.5 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 4.7 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 1.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.9 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0033029 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) negative regulation of T cell differentiation(GO:0045581) regulation of T-helper cell differentiation(GO:0045622) negative regulation of T-helper cell differentiation(GO:0045623) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) negative regulation of alpha-beta T cell differentiation(GO:0046639) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 1.5 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0072512 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.8 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.2 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.0 | 0.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 1.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0021985 | parturition(GO:0007567) neurohypophysis development(GO:0021985) maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.0 | 0.6 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 2.0 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.5 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.2 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 1.6 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 2.1 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.2 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:1990089 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 1.5 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 1.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.0 | 1.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 1.4 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.5 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 1.2 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 2.2 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.6 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 1.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 1.2 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.4 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0039703 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism metabolic process(GO:0044033) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 1.1 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.2 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.6 | GO:0060389 | pathway-restricted SMAD protein phosphorylation(GO:0060389) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.6 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.8 | GO:0090174 | vesicle fusion(GO:0006906) organelle membrane fusion(GO:0090174) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 1.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.4 | 1.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 2.5 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.3 | 0.9 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.3 | 1.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.3 | 3.7 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.3 | 1.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 1.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 1.8 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 0.6 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.2 | 4.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 0.8 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.2 | 1.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 2.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 1.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.5 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 1.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 2.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.3 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.6 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 1.9 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 2.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.6 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.6 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 2.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.1 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 2.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 2.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.4 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.7 | GO:0097346 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) INO80-type complex(GO:0097346) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.6 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 4.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 2.1 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.7 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 4.0 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.8 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 1.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 3.5 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 1.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 2.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 3.0 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 3.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0072686 | mitotic spindle(GO:0072686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 10.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.9 | 3.7 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.9 | 2.8 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.7 | 4.7 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.6 | 3.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.5 | 1.5 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.4 | 1.5 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.4 | 1.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 1.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.3 | 1.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 1.0 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.3 | 0.9 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.3 | 0.8 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.3 | 1.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 2.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 0.9 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 0.7 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.2 | 1.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 1.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 1.0 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.2 | 1.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 0.6 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.2 | 1.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.2 | 1.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 1.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 3.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 0.8 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 1.0 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 0.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 1.9 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 0.9 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 3.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.9 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 2.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.8 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 1.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.6 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.9 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.4 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.1 | 1.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 2.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.6 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 4.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.0 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 4.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 5.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.5 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 1.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.7 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.3 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 4.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 3.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.2 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 2.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.5 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 1.5 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.2 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.1 | 0.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.7 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 13.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 1.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.6 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 3.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 1.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 1.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.9 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.0 | 1.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 1.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 2.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 8.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 1.2 | GO:0030898 | microfilament motor activity(GO:0000146) actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.3 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 1.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 1.3 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 4.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 1.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.5 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 1.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 3.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 1.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 4.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 1.7 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.2 | 1.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 3.0 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.9 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 1.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.8 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 2.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 7.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |