Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
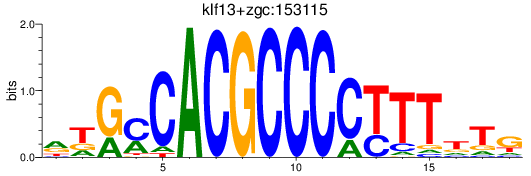
Results for klf13+zgc:153115
Z-value: 1.33
Transcription factors associated with klf13+zgc:153115
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf13
|
ENSDARG00000061368 | Kruppel-like factor 13 |
|
zgc
|
ENSDARG00000069342 | 153115 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf13 | dr11_v1_chr25_-_34512102_34512102 | -0.86 | 5.0e-06 | Click! |
| zgc:153115 | dr11_v1_chr2_-_7246338_7246338 | 0.63 | 4.7e-03 | Click! |
Activity profile of klf13+zgc:153115 motif
Sorted Z-values of klf13+zgc:153115 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_17197546 | 7.52 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr9_+_8380728 | 5.27 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr5_-_69482891 | 4.89 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr10_-_34002185 | 4.69 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr10_-_44017642 | 4.50 |
ENSDART00000135240
ENSDART00000014669 |
acads
|
acyl-CoA dehydrogenase short chain |
| chr4_-_20177868 | 4.12 |
ENSDART00000003621
|
sinup
|
siaz-interacting nuclear protein |
| chr1_-_34450784 | 3.72 |
ENSDART00000140515
|
lmo7b
|
LIM domain 7b |
| chr23_-_31645760 | 3.70 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr7_+_1473929 | 3.62 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr1_-_34450622 | 3.50 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr3_-_40054615 | 3.45 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr8_+_47219107 | 3.20 |
ENSDART00000146018
ENSDART00000075068 |
mthfr
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr5_+_16117871 | 3.09 |
ENSDART00000090657
|
znrf3
|
zinc and ring finger 3 |
| chr14_+_7048930 | 2.89 |
ENSDART00000109138
|
hbegfa
|
heparin-binding EGF-like growth factor a |
| chr21_+_34167178 | 2.70 |
ENSDART00000158308
|
trpc5b
|
transient receptor potential cation channel, subfamily C, member 5b |
| chr17_-_18898115 | 2.66 |
ENSDART00000028044
|
galcb
|
galactosylceramidase b |
| chr8_-_2616326 | 2.58 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr23_-_45398622 | 2.53 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr22_+_835728 | 2.50 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr21_-_32060993 | 2.44 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr7_+_38349667 | 2.39 |
ENSDART00000010046
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr6_+_7414215 | 2.34 |
ENSDART00000049339
|
sox21a
|
SRY (sex determining region Y)-box 21a |
| chr12_+_46634736 | 2.34 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr11_+_5499661 | 2.33 |
ENSDART00000027850
|
slc35e1
|
solute carrier family 35, member E1 |
| chr9_+_30421489 | 2.22 |
ENSDART00000145025
ENSDART00000132058 |
zgc:113314
|
zgc:113314 |
| chr15_-_1001177 | 2.19 |
ENSDART00000160730
|
zgc:162936
|
zgc:162936 |
| chr23_+_12160900 | 2.19 |
ENSDART00000136046
|
ppp1r3da
|
protein phosphatase 1, regulatory subunit 3Da |
| chr2_+_59015878 | 2.18 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr16_+_43077909 | 2.17 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr22_-_20924747 | 2.09 |
ENSDART00000185845
ENSDART00000179672 |
ell
|
elongation factor RNA polymerase II |
| chr9_+_19489304 | 2.06 |
ENSDART00000151920
|
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr25_+_6122823 | 2.06 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr2_-_39675829 | 2.06 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr9_-_19489264 | 2.05 |
ENSDART00000122894
|
wdr4
|
WD repeat domain 4 |
| chr10_+_6383270 | 2.00 |
ENSDART00000170548
|
zgc:114200
|
zgc:114200 |
| chr12_+_46869271 | 1.99 |
ENSDART00000166560
|
hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr17_-_37795865 | 1.93 |
ENSDART00000025853
|
zfp36l1a
|
zinc finger protein 36, C3H type-like 1a |
| chr8_+_10862353 | 1.90 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr25_+_3788443 | 1.90 |
ENSDART00000189747
|
chid1
|
chitinase domain containing 1 |
| chr1_+_604127 | 1.84 |
ENSDART00000133165
|
jam2a
|
junctional adhesion molecule 2a |
| chr14_-_24101897 | 1.74 |
ENSDART00000143695
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr12_-_3053873 | 1.62 |
ENSDART00000023796
ENSDART00000137148 |
dcxr
|
dicarbonyl/L-xylulose reductase |
| chr12_-_3053699 | 1.61 |
ENSDART00000139721
|
dcxr
|
dicarbonyl/L-xylulose reductase |
| chr18_+_7543347 | 1.60 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr24_-_36238054 | 1.56 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr3_-_52899394 | 1.54 |
ENSDART00000128223
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr1_-_52201266 | 1.51 |
ENSDART00000143805
ENSDART00000023757 |
rab3da
|
RAB3D, member RAS oncogene family, a |
| chr7_-_32895668 | 1.46 |
ENSDART00000141828
|
ano5b
|
anoctamin 5b |
| chr13_-_42673978 | 1.46 |
ENSDART00000133848
ENSDART00000099738 ENSDART00000099729 ENSDART00000169083 |
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr12_+_48340133 | 1.45 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr20_+_29565906 | 1.45 |
ENSDART00000062383
|
ywhaqa
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide a |
| chr12_-_48312647 | 1.43 |
ENSDART00000114415
|
ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr23_-_5818992 | 1.38 |
ENSDART00000148730
|
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr23_-_10745288 | 1.38 |
ENSDART00000140745
ENSDART00000013768 |
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr10_+_22034477 | 1.36 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr16_-_17713859 | 1.36 |
ENSDART00000149275
|
zgc:174935
|
zgc:174935 |
| chr20_-_51656512 | 1.35 |
ENSDART00000129965
|
LO018154.1
|
|
| chr2_-_47957673 | 1.34 |
ENSDART00000056305
|
fzd8b
|
frizzled class receptor 8b |
| chr19_+_46113828 | 1.26 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr7_+_29044888 | 1.26 |
ENSDART00000086871
|
gfod2
|
glucose-fructose oxidoreductase domain containing 2 |
| chr23_-_17429775 | 1.24 |
ENSDART00000043076
|
ppdpfb
|
pancreatic progenitor cell differentiation and proliferation factor b |
| chr2_+_43894368 | 1.19 |
ENSDART00000143885
|
gbp3
|
guanylate binding protein 3 |
| chr21_+_40685895 | 1.14 |
ENSDART00000017709
|
ccdc82
|
coiled-coil domain containing 82 |
| chr21_+_6114709 | 1.13 |
ENSDART00000065858
|
fpgs
|
folylpolyglutamate synthase |
| chr10_+_76864 | 1.13 |
ENSDART00000036375
|
vps26c
|
Down syndrome critical region 3 |
| chr20_+_23501535 | 1.09 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr16_+_10329701 | 1.08 |
ENSDART00000172845
|
mdc1
|
mediator of DNA damage checkpoint 1 |
| chr6_-_10034145 | 1.08 |
ENSDART00000185999
|
nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr21_+_6197223 | 1.07 |
ENSDART00000147716
|
si:dkey-93m18.3
|
si:dkey-93m18.3 |
| chr6_-_25165693 | 1.06 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr17_-_51260476 | 1.06 |
ENSDART00000084348
|
trappc12
|
trafficking protein particle complex 12 |
| chr10_+_29204581 | 1.06 |
ENSDART00000148503
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr22_-_3182965 | 1.05 |
ENSDART00000158009
|
lonp1
|
lon peptidase 1, mitochondrial |
| chr8_+_104114 | 1.04 |
ENSDART00000172101
|
sncaip
|
synuclein, alpha interacting protein |
| chr25_+_186583 | 1.03 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr3_-_50998577 | 1.02 |
ENSDART00000157735
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr10_+_21444654 | 1.02 |
ENSDART00000140113
ENSDART00000184386 ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr22_-_38224315 | 1.00 |
ENSDART00000165430
ENSDART00000140968 |
hps3
|
Hermansky-Pudlak syndrome 3 |
| chr5_+_43458304 | 0.99 |
ENSDART00000051114
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr6_+_23122789 | 0.97 |
ENSDART00000049226
ENSDART00000067560 |
acox1
|
acyl-CoA oxidase 1, palmitoyl |
| chr15_-_19443997 | 0.94 |
ENSDART00000114936
|
esamb
|
endothelial cell adhesion molecule b |
| chr18_+_6558338 | 0.93 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr7_+_20260172 | 0.92 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr3_-_31924643 | 0.88 |
ENSDART00000122589
|
rnf113a
|
ring finger protein 113A |
| chr13_+_21676235 | 0.87 |
ENSDART00000137804
ENSDART00000134950 ENSDART00000129653 |
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr5_+_11812089 | 0.85 |
ENSDART00000111359
|
fbxo21
|
F-box protein 21 |
| chr23_-_18707418 | 0.84 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr1_-_493218 | 0.83 |
ENSDART00000031635
|
ercc5
|
excision repair cross-complementation group 5 |
| chr23_+_10396842 | 0.83 |
ENSDART00000167593
ENSDART00000125103 |
eif4ba
|
eukaryotic translation initiation factor 4Ba |
| chr21_+_6114305 | 0.79 |
ENSDART00000141607
|
fpgs
|
folylpolyglutamate synthase |
| chr21_-_41070182 | 0.79 |
ENSDART00000026064
|
larsb
|
leucyl-tRNA synthetase b |
| chr1_-_34335752 | 0.76 |
ENSDART00000140157
|
si:dkey-24h22.5
|
si:dkey-24h22.5 |
| chr20_-_25748407 | 0.76 |
ENSDART00000063152
|
chst14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr11_+_13071645 | 0.75 |
ENSDART00000162259
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr24_+_35387517 | 0.75 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr19_+_47290287 | 0.74 |
ENSDART00000078382
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr14_+_22172047 | 0.73 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr21_-_11657043 | 0.71 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr8_-_40183197 | 0.71 |
ENSDART00000005118
|
gpx8
|
glutathione peroxidase 8 (putative) |
| chr11_+_11152214 | 0.69 |
ENSDART00000148030
|
ly75
|
lymphocyte antigen 75 |
| chr6_+_54576520 | 0.69 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr18_+_27077853 | 0.68 |
ENSDART00000125326
ENSDART00000192660 ENSDART00000098334 |
ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr16_-_26132122 | 0.67 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr20_+_35058634 | 0.67 |
ENSDART00000122696
|
hnrnpub
|
heterogeneous nuclear ribonucleoprotein Ub |
| chr5_+_61657702 | 0.67 |
ENSDART00000134387
ENSDART00000171248 |
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr22_-_6562618 | 0.66 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr23_-_4019699 | 0.64 |
ENSDART00000159780
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr23_-_4019928 | 0.63 |
ENSDART00000021062
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr10_+_3299829 | 0.62 |
ENSDART00000183684
|
zgc:56235
|
zgc:56235 |
| chr21_-_13230925 | 0.61 |
ENSDART00000023834
|
setb
|
SET nuclear proto-oncogene b |
| chr19_-_32944050 | 0.61 |
ENSDART00000137611
|
azin1b
|
antizyme inhibitor 1b |
| chr7_+_41146560 | 0.59 |
ENSDART00000143285
ENSDART00000173852 ENSDART00000174003 ENSDART00000038487 ENSDART00000173463 ENSDART00000166448 ENSDART00000052274 |
puf60b
|
poly-U binding splicing factor b |
| chr13_-_36582341 | 0.59 |
ENSDART00000137335
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr25_-_37284370 | 0.59 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr3_+_17806213 | 0.58 |
ENSDART00000055890
|
znf385c
|
zinc finger protein 385C |
| chr20_-_54564018 | 0.58 |
ENSDART00000099832
|
zgc:153012
|
zgc:153012 |
| chr18_+_18104235 | 0.57 |
ENSDART00000145342
|
cbln1
|
cerebellin 1 precursor |
| chr16_-_7228276 | 0.57 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr9_+_4378153 | 0.56 |
ENSDART00000191264
ENSDART00000182384 |
kalrna
|
kalirin RhoGEF kinase a |
| chr7_-_73815262 | 0.56 |
ENSDART00000185351
|
zgc:165555
|
zgc:165555 |
| chr24_-_33780387 | 0.55 |
ENSDART00000079210
|
cdk5
|
cyclin-dependent kinase 5 |
| chr13_-_44808783 | 0.54 |
ENSDART00000099984
|
glo1
|
glyoxalase 1 |
| chr7_-_5396154 | 0.53 |
ENSDART00000172980
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr2_-_3614005 | 0.52 |
ENSDART00000110399
|
pter
|
phosphotriesterase related |
| chr18_+_6558146 | 0.51 |
ENSDART00000169401
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr21_+_37436907 | 0.51 |
ENSDART00000182611
ENSDART00000076328 |
pgrmc1
|
progesterone receptor membrane component 1 |
| chr14_+_6963312 | 0.49 |
ENSDART00000150050
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr16_+_39271123 | 0.47 |
ENSDART00000043823
ENSDART00000141801 |
osbpl10b
|
oxysterol binding protein-like 10b |
| chr5_+_68807170 | 0.46 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr1_+_7189856 | 0.46 |
ENSDART00000092114
|
si:ch73-383l1.1
|
si:ch73-383l1.1 |
| chr19_-_2421793 | 0.45 |
ENSDART00000180238
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr25_-_4235037 | 0.44 |
ENSDART00000093003
|
syt7a
|
synaptotagmin VIIa |
| chr21_-_13231101 | 0.43 |
ENSDART00000190943
|
setb
|
SET nuclear proto-oncogene b |
| chr11_+_34921492 | 0.40 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr25_+_35913614 | 0.39 |
ENSDART00000022437
|
gpia
|
glucose-6-phosphate isomerase a |
| chr7_+_4474880 | 0.38 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr22_+_1421212 | 0.37 |
ENSDART00000161813
|
zgc:101130
|
zgc:101130 |
| chr3_+_17951790 | 0.37 |
ENSDART00000164663
|
aclya
|
ATP citrate lyase a |
| chr24_-_37688398 | 0.37 |
ENSDART00000141414
|
zgc:112185
|
zgc:112185 |
| chr1_-_55196103 | 0.36 |
ENSDART00000140153
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr4_-_5455506 | 0.36 |
ENSDART00000156593
ENSDART00000154676 |
si:dkey-14d8.22
|
si:dkey-14d8.22 |
| chr3_+_31925067 | 0.36 |
ENSDART00000127330
ENSDART00000055279 |
snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr9_-_28399071 | 0.35 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr19_+_29854223 | 0.35 |
ENSDART00000109729
ENSDART00000157419 |
si:ch73-130a3.4
|
si:ch73-130a3.4 |
| chr21_+_15704556 | 0.34 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr3_-_47876427 | 0.34 |
ENSDART00000180844
ENSDART00000124480 |
adgrl1a
|
adhesion G protein-coupled receptor L1a |
| chr7_+_69528850 | 0.33 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr25_+_25123385 | 0.33 |
ENSDART00000163892
|
ldha
|
lactate dehydrogenase A4 |
| chr21_+_45386033 | 0.32 |
ENSDART00000151773
|
jade2
|
jade family PHD finger 2 |
| chr17_+_6765621 | 0.30 |
ENSDART00000156637
ENSDART00000007622 |
afg1la
|
AFG1 like ATPase a |
| chr3_+_726000 | 0.30 |
ENSDART00000158510
|
dicp1.17
|
diverse immunoglobulin domain-containing protein 1.17 |
| chr3_+_62327332 | 0.28 |
ENSDART00000158414
|
BX470259.1
|
|
| chr7_-_73846995 | 0.27 |
ENSDART00000188079
|
FP236812.4
|
|
| chr24_+_17005647 | 0.26 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr23_+_30006206 | 0.26 |
ENSDART00000150153
|
tnfrsf9a
|
tumor necrosis factor receptor superfamily, member 9a |
| chr10_+_44940693 | 0.25 |
ENSDART00000157515
|
cnnm4a
|
cyclin and CBS domain divalent metal cation transport mediator 4a |
| chr15_-_9031996 | 0.25 |
ENSDART00000124998
|
rtn2a
|
reticulon 2a |
| chr12_-_22670279 | 0.25 |
ENSDART00000164888
|
zcchc4
|
zinc finger, CCHC domain containing 4 |
| chr7_-_18601206 | 0.24 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr11_-_36341028 | 0.23 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr7_+_58730201 | 0.23 |
ENSDART00000073640
|
plag1
|
pleiomorphic adenoma gene 1 |
| chr22_+_2217939 | 0.22 |
ENSDART00000145394
ENSDART00000146694 |
znf1172
|
zinc finger protein 1172 |
| chr7_+_9880811 | 0.22 |
ENSDART00000128376
|
lins1
|
lines homolog 1 |
| chr22_-_27622854 | 0.22 |
ENSDART00000190874
|
CR388079.3
|
|
| chr22_-_6968756 | 0.22 |
ENSDART00000136636
|
fgfr1bl
|
fibroblast growth factor receptor 1b, like |
| chr12_-_4540564 | 0.21 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr19_-_205104 | 0.19 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr8_-_21268303 | 0.19 |
ENSDART00000067211
|
gpr37l1b
|
G protein-coupled receptor 37 like 1b |
| chr3_-_33934788 | 0.19 |
ENSDART00000151411
|
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr1_+_56755536 | 0.19 |
ENSDART00000157727
|
CABZ01049361.1
|
|
| chr4_-_39448829 | 0.19 |
ENSDART00000168526
|
si:dkey-261o4.9
|
si:dkey-261o4.9 |
| chr12_+_11080776 | 0.18 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr10_+_10728870 | 0.18 |
ENSDART00000109282
|
swi5
|
SWI5 homologous recombination repair protein |
| chr25_-_18730697 | 0.17 |
ENSDART00000182475
|
si:dkeyp-93a5.2
|
si:dkeyp-93a5.2 |
| chr12_+_6391243 | 0.17 |
ENSDART00000152765
|
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr15_-_3078600 | 0.17 |
ENSDART00000172842
|
fndc3a
|
fibronectin type III domain containing 3A |
| chr23_-_790620 | 0.17 |
ENSDART00000166818
|
CU927920.1
|
|
| chr9_-_37749973 | 0.17 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr21_+_28535203 | 0.16 |
ENSDART00000184950
|
CR749775.2
|
|
| chr21_+_43854594 | 0.16 |
ENSDART00000142718
|
csf1rb
|
colony stimulating factor 1 receptor, b |
| chr6_-_54111928 | 0.15 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr4_+_14957360 | 0.14 |
ENSDART00000002770
ENSDART00000111882 ENSDART00000148292 |
tspan33a
|
tetraspanin 33a |
| chr18_+_41549764 | 0.14 |
ENSDART00000059135
|
bcl7bb
|
BCL tumor suppressor 7Bb |
| chr1_-_57839070 | 0.12 |
ENSDART00000152571
|
si:dkey-1c7.3
|
si:dkey-1c7.3 |
| chr25_-_20268027 | 0.12 |
ENSDART00000138763
|
dnajb9a
|
DnaJ (Hsp40) homolog, subfamily B, member 9a |
| chr16_-_24561354 | 0.12 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr18_+_40584288 | 0.11 |
ENSDART00000087692
|
si:ch211-132b12.3
|
si:ch211-132b12.3 |
| chr21_+_1382078 | 0.11 |
ENSDART00000188463
|
TCF4
|
transcription factor 4 |
| chr3_+_59117136 | 0.10 |
ENSDART00000182745
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr15_-_5563551 | 0.10 |
ENSDART00000099520
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr19_+_22727940 | 0.10 |
ENSDART00000052509
|
trhrb
|
thyrotropin-releasing hormone receptor b |
| chr11_-_4235811 | 0.09 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr20_+_572037 | 0.09 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr22_-_16275236 | 0.08 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr10_+_38643304 | 0.07 |
ENSDART00000067447
|
mmp30
|
matrix metallopeptidase 30 |
| chr9_-_1984604 | 0.07 |
ENSDART00000082339
|
hoxd12a
|
homeobox D12a |
| chr19_+_46237665 | 0.06 |
ENSDART00000159391
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr18_-_6943577 | 0.06 |
ENSDART00000132399
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr20_+_46311242 | 0.06 |
ENSDART00000038696
|
flvcr2b
|
feline leukemia virus subgroup C cellular receptor family, member 2b |
| chr10_+_1899595 | 0.04 |
ENSDART00000040271
|
CU861476.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of klf13+zgc:153115
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 1.1 | 3.2 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.8 | 3.1 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.5 | 2.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.5 | 1.9 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.4 | 1.3 | GO:2001014 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.4 | 2.1 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.3 | 2.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 | 2.7 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.3 | 2.9 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.3 | 4.1 | GO:0001840 | neural plate development(GO:0001840) |
| 0.3 | 0.8 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.3 | 3.4 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.2 | 1.7 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 1.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 3.6 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.2 | 2.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 7.5 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.2 | 0.5 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 0.7 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.2 | 1.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 1.7 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.1 | 1.1 | GO:0009146 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 0.9 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.8 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.1 | 0.8 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.6 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.9 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 1.0 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.6 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.7 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 2.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 2.2 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 1.4 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 1.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.6 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.1 | 1.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 2.0 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 | 0.7 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.4 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 1.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.0 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.3 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.4 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 4.1 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 1.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 1.3 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 2.3 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.1 | GO:0019079 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 3.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 1.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.5 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 4.7 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.3 | 1.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 2.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 1.4 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 2.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 0.7 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.1 | 1.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.1 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 3.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.8 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 2.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.2 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 4.6 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.7 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 2.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.7 | 2.0 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.4 | 1.3 | GO:1990715 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.4 | 1.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.3 | 4.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.3 | 1.4 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.3 | 0.8 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.2 | 1.0 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 3.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 1.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 0.5 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 2.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.9 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.1 | 3.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.9 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 0.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.8 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.6 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 2.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.3 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 3.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 2.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 1.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.6 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 3.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.7 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 1.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 1.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 2.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 2.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.4 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.9 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.2 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 2.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.6 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.7 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 6.7 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 5.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 1.5 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.8 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 2.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 5.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 0.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.4 | 7.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.3 | 4.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 5.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 0.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.8 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 1.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |