Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for ikzf1
Z-value: 1.03
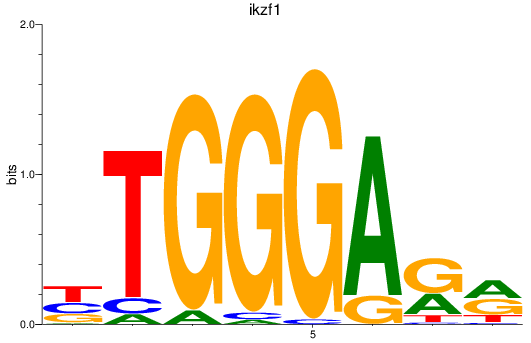
Transcription factors associated with ikzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ikzf1
|
ENSDARG00000013539 | IKAROS family zinc finger 1 (Ikaros) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ikzf1 | dr11_v1_chr13_-_15982707_15982707 | -0.79 | 1.1e-04 | Click! |
Activity profile of ikzf1 motif
Sorted Z-values of ikzf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_1009831 | 4.09 |
ENSDART00000172414
|
fabp1b.2
|
fatty acid binding protein 1b, liver, tandem duplicate 2 |
| chr25_+_30196039 | 3.43 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr7_+_67699009 | 3.29 |
ENSDART00000192810
|
zgc:162592
|
zgc:162592 |
| chr11_-_1550709 | 2.53 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr1_-_33645967 | 2.26 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr21_+_13383413 | 2.20 |
ENSDART00000151345
|
zgc:113162
|
zgc:113162 |
| chr10_-_25217347 | 2.20 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr23_-_45955177 | 2.15 |
ENSDART00000165963
ENSDART00000186649 ENSDART00000185773 |
LO017850.1
|
|
| chr5_+_39099172 | 2.14 |
ENSDART00000006079
|
bmp2k
|
BMP2 inducible kinase |
| chr24_-_34680956 | 2.09 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr18_-_16953978 | 2.06 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr2_+_35603637 | 2.00 |
ENSDART00000147278
|
plk3
|
polo-like kinase 3 (Drosophila) |
| chr6_+_21005725 | 1.97 |
ENSDART00000041370
|
cx44.2
|
connexin 44.2 |
| chr7_+_61764040 | 1.95 |
ENSDART00000056745
|
acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr7_+_39410180 | 1.87 |
ENSDART00000168641
|
CT030188.1
|
|
| chr6_+_54680730 | 1.86 |
ENSDART00000074602
|
smpd2b
|
sphingomyelin phosphodiesterase 2b, neutral membrane (neutral sphingomyelinase) |
| chr12_-_1034383 | 1.85 |
ENSDART00000152455
ENSDART00000152346 |
polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr7_+_39410393 | 1.85 |
ENSDART00000158561
ENSDART00000185173 |
CT030188.1
|
|
| chr13_-_15929402 | 1.84 |
ENSDART00000090273
|
ttl
|
tubulin tyrosine ligase |
| chr24_-_26622423 | 1.83 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr13_-_24825691 | 1.66 |
ENSDART00000142745
|
slka
|
STE20-like kinase a |
| chr2_-_15324837 | 1.61 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr8_+_52442785 | 1.61 |
ENSDART00000189958
|
zgc:77112
|
zgc:77112 |
| chr8_-_1730686 | 1.59 |
ENSDART00000183802
|
CABZ01065416.1
|
|
| chr15_+_30126971 | 1.58 |
ENSDART00000100214
|
nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr7_-_59159253 | 1.57 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr3_+_27665160 | 1.56 |
ENSDART00000103660
|
clcn7
|
chloride channel 7 |
| chr5_+_44805269 | 1.54 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr15_-_47193564 | 1.53 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr5_+_9037650 | 1.52 |
ENSDART00000158226
|
si:ch211-155m12.1
|
si:ch211-155m12.1 |
| chr7_+_67699178 | 1.51 |
ENSDART00000160086
|
zgc:162592
|
zgc:162592 |
| chr21_+_13387965 | 1.49 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr5_-_54712159 | 1.47 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr16_+_31511739 | 1.44 |
ENSDART00000049420
|
ndrg1b
|
N-myc downstream regulated 1b |
| chr11_+_11303458 | 1.44 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr19_-_27830818 | 1.42 |
ENSDART00000131767
|
papd7
|
PAP associated domain containing 7 |
| chr20_-_13623882 | 1.42 |
ENSDART00000125218
ENSDART00000152499 |
sytl3
|
synaptotagmin-like 3 |
| chr19_+_34230108 | 1.41 |
ENSDART00000141950
|
galnt12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 |
| chr13_-_36525982 | 1.40 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr18_-_16907436 | 1.40 |
ENSDART00000146464
|
swap70b
|
switching B cell complex subunit SWAP70b |
| chr21_-_4849029 | 1.40 |
ENSDART00000168930
ENSDART00000151019 |
notch1a
|
notch 1a |
| chr1_+_40613297 | 1.39 |
ENSDART00000040798
ENSDART00000168067 ENSDART00000130490 |
naa15b
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit b |
| chr17_+_6793001 | 1.32 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr20_-_15161669 | 1.31 |
ENSDART00000080333
ENSDART00000063882 |
plpp6
|
phospholipid phosphatase 6 |
| chr3_+_46762703 | 1.31 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr9_-_11676491 | 1.28 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr17_+_33415319 | 1.28 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr17_+_2549503 | 1.28 |
ENSDART00000156843
|
si:dkey-248g15.3
|
si:dkey-248g15.3 |
| chr16_+_33142734 | 1.23 |
ENSDART00000138244
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr20_+_42246948 | 1.22 |
ENSDART00000061135
|
gopc
|
golgi-associated PDZ and coiled-coil motif containing |
| chr21_-_34951265 | 1.21 |
ENSDART00000135222
|
lipia
|
lipase, member Ia |
| chr2_-_42558549 | 1.21 |
ENSDART00000025997
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr24_-_9989634 | 1.20 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr15_+_29025090 | 1.19 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr19_+_20201254 | 1.19 |
ENSDART00000010140
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr8_-_43716897 | 1.18 |
ENSDART00000163237
|
ep400
|
E1A binding protein p400 |
| chr21_+_43172506 | 1.18 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr20_-_48898560 | 1.18 |
ENSDART00000163071
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr17_+_33415542 | 1.16 |
ENSDART00000183169
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr10_-_44008241 | 1.14 |
ENSDART00000137686
|
acads
|
acyl-CoA dehydrogenase short chain |
| chr6_+_36821621 | 1.14 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr1_-_43712120 | 1.13 |
ENSDART00000074600
|
SLC9B2
|
si:dkey-162b23.4 |
| chr17_+_28102487 | 1.13 |
ENSDART00000131638
ENSDART00000076265 |
zgc:91908
|
zgc:91908 |
| chr22_-_17671348 | 1.12 |
ENSDART00000137995
|
tjp3
|
tight junction protein 3 |
| chr23_+_26026383 | 1.12 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr14_+_28492990 | 1.11 |
ENSDART00000160347
ENSDART00000187176 ENSDART00000088094 |
stag2b
|
stromal antigen 2b |
| chr7_+_25003313 | 1.10 |
ENSDART00000131935
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr15_+_11693624 | 1.08 |
ENSDART00000193630
ENSDART00000161930 |
strn4
|
striatin, calmodulin binding protein 4 |
| chr19_+_1658698 | 1.07 |
ENSDART00000158311
|
klhl7
|
kelch-like family member 7 |
| chr6_+_1724889 | 1.06 |
ENSDART00000157415
|
acvr2ab
|
activin A receptor type 2Ab |
| chr23_+_43668756 | 1.05 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr7_-_58098814 | 1.05 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr17_-_25331439 | 1.04 |
ENSDART00000155422
ENSDART00000082324 |
zpcx
|
zona pellucida protein C |
| chr2_-_49031303 | 1.03 |
ENSDART00000143471
|
cdc34b
|
cell division cycle 34 homolog (S. cerevisiae) b |
| chr13_+_15816573 | 1.03 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr11_-_27953135 | 1.02 |
ENSDART00000168338
|
ece1
|
endothelin converting enzyme 1 |
| chr8_+_14959587 | 1.02 |
ENSDART00000138548
ENSDART00000041697 |
abl2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr19_+_20201593 | 1.01 |
ENSDART00000163026
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr17_-_8592824 | 1.01 |
ENSDART00000127022
|
CU462878.1
|
|
| chr8_-_39822917 | 1.00 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr6_+_2195625 | 1.00 |
ENSDART00000155659
|
acvr1bb
|
activin A receptor type 1Bb |
| chr17_+_38295847 | 0.99 |
ENSDART00000008532
|
mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr20_-_4049862 | 0.98 |
ENSDART00000158057
|
sprtn
|
SprT-like N-terminal domain |
| chr17_+_25187670 | 0.98 |
ENSDART00000190873
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr17_-_6613458 | 0.96 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr5_-_23675222 | 0.96 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr3_+_17653784 | 0.94 |
ENSDART00000159984
ENSDART00000157682 ENSDART00000187937 |
kat2a
|
K(lysine) acetyltransferase 2A |
| chr17_-_24879003 | 0.94 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr24_-_31090948 | 0.94 |
ENSDART00000176799
|
hccsb
|
holocytochrome c synthase b |
| chr14_+_7048930 | 0.93 |
ENSDART00000109138
|
hbegfa
|
heparin-binding EGF-like growth factor a |
| chr2_+_243778 | 0.92 |
ENSDART00000182262
|
CABZ01085887.1
|
|
| chr9_-_28255029 | 0.91 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr7_+_34549377 | 0.91 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr17_-_30521043 | 0.90 |
ENSDART00000087111
|
itsn2b
|
intersectin 2b |
| chr21_-_25295087 | 0.90 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr2_+_54844755 | 0.90 |
ENSDART00000170428
|
rnf126
|
ring finger protein 126 |
| chr3_+_35005730 | 0.89 |
ENSDART00000029451
|
prkcbb
|
protein kinase C, beta b |
| chr25_-_6049339 | 0.89 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr20_+_36806398 | 0.89 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr2_-_42492445 | 0.88 |
ENSDART00000139929
|
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr23_-_31967554 | 0.88 |
ENSDART00000183973
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr9_-_34260214 | 0.88 |
ENSDART00000012385
|
me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr5_+_48666485 | 0.88 |
ENSDART00000158000
ENSDART00000031141 |
polr3g
|
polymerase (RNA) III (DNA directed) polypeptide G |
| chr5_+_39099380 | 0.87 |
ENSDART00000166657
|
bmp2k
|
BMP2 inducible kinase |
| chr2_-_5723786 | 0.87 |
ENSDART00000100924
|
cwc25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae) |
| chr20_-_31238313 | 0.87 |
ENSDART00000028471
|
hpcal1
|
hippocalcin-like 1 |
| chr22_-_15704704 | 0.86 |
ENSDART00000017838
ENSDART00000130238 |
safb
|
scaffold attachment factor B |
| chr19_-_17864213 | 0.86 |
ENSDART00000151043
|
ints8
|
integrator complex subunit 8 |
| chr6_-_30485009 | 0.85 |
ENSDART00000025698
|
zgc:153311
|
zgc:153311 |
| chr14_+_20918701 | 0.84 |
ENSDART00000144736
ENSDART00000148204 |
zgc:66433
|
zgc:66433 |
| chr11_-_11791718 | 0.84 |
ENSDART00000180476
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr8_+_20488322 | 0.84 |
ENSDART00000036630
|
zgc:101100
|
zgc:101100 |
| chr2_+_23701613 | 0.84 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr11_-_26701611 | 0.82 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr20_-_15161502 | 0.82 |
ENSDART00000187072
|
plpp6
|
phospholipid phosphatase 6 |
| chr9_+_21402863 | 0.80 |
ENSDART00000125357
|
cx30.3
|
connexin 30.3 |
| chr16_-_22585289 | 0.80 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr12_-_13549538 | 0.79 |
ENSDART00000133895
|
ghdc
|
GH3 domain containing |
| chr16_-_31791165 | 0.79 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr7_+_62080456 | 0.78 |
ENSDART00000092580
|
rbpjb
|
recombination signal binding protein for immunoglobulin kappa J region b |
| chr20_-_53078607 | 0.77 |
ENSDART00000163494
ENSDART00000191730 |
CABZ01066813.1
|
|
| chr6_-_12135741 | 0.77 |
ENSDART00000155090
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr17_-_25861787 | 0.76 |
ENSDART00000182503
|
BX000364.5
|
|
| chr3_+_31925067 | 0.75 |
ENSDART00000127330
ENSDART00000055279 |
snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr5_+_24087035 | 0.75 |
ENSDART00000183644
|
tp53
|
tumor protein p53 |
| chr1_+_51066671 | 0.75 |
ENSDART00000064007
|
srd5a2a
|
steroid-5-alpha-reductase, alpha polypeptide 2a |
| chr7_+_24006875 | 0.75 |
ENSDART00000033755
|
homezb
|
homeobox and leucine zipper encoding b |
| chr6_+_19933763 | 0.75 |
ENSDART00000166192
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr5_-_26247215 | 0.74 |
ENSDART00000136806
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr6_-_41135215 | 0.73 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr17_+_25331576 | 0.73 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr23_-_270847 | 0.72 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr9_+_38888025 | 0.72 |
ENSDART00000148306
|
map2
|
microtubule-associated protein 2 |
| chr17_+_23462972 | 0.72 |
ENSDART00000112959
ENSDART00000192168 |
ankrd1a
|
ankyrin repeat domain 1a (cardiac muscle) |
| chr13_+_15580758 | 0.71 |
ENSDART00000087194
ENSDART00000013525 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr19_+_9050852 | 0.71 |
ENSDART00000151031
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr8_-_15182232 | 0.71 |
ENSDART00000138855
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr5_+_61657702 | 0.71 |
ENSDART00000134387
ENSDART00000171248 |
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr22_+_4443689 | 0.69 |
ENSDART00000185490
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr3_+_50172452 | 0.68 |
ENSDART00000191224
|
epn3a
|
epsin 3a |
| chr9_-_25366541 | 0.68 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr17_-_33416020 | 0.68 |
ENSDART00000140149
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr6_+_41554794 | 0.67 |
ENSDART00000165424
|
srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr20_+_22799857 | 0.67 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr7_+_54259657 | 0.67 |
ENSDART00000170174
|
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr10_-_35177257 | 0.67 |
ENSDART00000077426
|
pom121
|
POM121 transmembrane nucleoporin |
| chr21_+_39100289 | 0.67 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr17_-_11357851 | 0.67 |
ENSDART00000153915
|
si:ch211-185a18.2
|
si:ch211-185a18.2 |
| chr5_-_23705828 | 0.67 |
ENSDART00000189419
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr6_+_58289335 | 0.66 |
ENSDART00000177399
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr9_+_55857193 | 0.65 |
ENSDART00000160980
|
sept10
|
septin 10 |
| chr6_-_11792152 | 0.64 |
ENSDART00000183403
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr6_-_54433995 | 0.64 |
ENSDART00000017230
|
snrpc
|
small nuclear ribonucleoprotein polypeptide C |
| chr8_-_23776399 | 0.64 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr8_-_50888806 | 0.63 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr2_+_5300405 | 0.63 |
ENSDART00000019925
|
GNB4
|
G protein subunit beta 4 |
| chr23_+_1661743 | 0.63 |
ENSDART00000044776
|
stxbp3
|
syntaxin binding protein 3 |
| chr13_+_38302665 | 0.62 |
ENSDART00000145777
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr17_-_14774117 | 0.62 |
ENSDART00000080401
|
si:ch211-266o15.1
|
si:ch211-266o15.1 |
| chr23_+_33963619 | 0.61 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr15_-_1001177 | 0.60 |
ENSDART00000160730
|
zgc:162936
|
zgc:162936 |
| chr13_-_31389661 | 0.60 |
ENSDART00000134630
|
zdhhc16a
|
zinc finger, DHHC-type containing 16a |
| chr6_-_10740365 | 0.60 |
ENSDART00000150942
|
sp3b
|
Sp3b transcription factor |
| chr20_+_22799641 | 0.60 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr15_-_1835189 | 0.60 |
ENSDART00000154369
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr2_+_58377395 | 0.60 |
ENSDART00000193511
|
vapal
|
VAMP (vesicle-associated membrane protein)-associated protein A, like |
| chr6_-_10725847 | 0.59 |
ENSDART00000184567
|
sp3b
|
Sp3b transcription factor |
| chr2_-_22659450 | 0.59 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr6_-_16456093 | 0.59 |
ENSDART00000083305
ENSDART00000181640 |
slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr18_+_7543347 | 0.59 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr2_-_30784502 | 0.59 |
ENSDART00000056735
|
rgs20
|
regulator of G protein signaling 20 |
| chr3_+_17806213 | 0.59 |
ENSDART00000055890
|
znf385c
|
zinc finger protein 385C |
| chr5_+_20319519 | 0.59 |
ENSDART00000004217
|
coro1ca
|
coronin, actin binding protein, 1Ca |
| chr16_-_13680692 | 0.58 |
ENSDART00000047452
|
ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr15_-_20709289 | 0.57 |
ENSDART00000136767
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr25_-_31763897 | 0.57 |
ENSDART00000041740
|
ubl7a
|
ubiquitin-like 7a (bone marrow stromal cell-derived) |
| chr7_+_36898622 | 0.57 |
ENSDART00000190773
|
tox3
|
TOX high mobility group box family member 3 |
| chr10_+_35347993 | 0.57 |
ENSDART00000131350
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr5_+_26686639 | 0.57 |
ENSDART00000079064
|
tango2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr12_+_23991276 | 0.56 |
ENSDART00000153136
|
psme4b
|
proteasome activator subunit 4b |
| chr12_+_28955766 | 0.56 |
ENSDART00000123417
ENSDART00000139347 |
znf668
|
zinc finger protein 668 |
| chr10_-_38468847 | 0.55 |
ENSDART00000133914
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr6_-_53204808 | 0.55 |
ENSDART00000160955
|
slc38a3b
|
solute carrier family 38, member 3b |
| chr15_-_47895200 | 0.55 |
ENSDART00000027060
|
DMWD
|
zmp:0000000529 |
| chr5_+_32817688 | 0.55 |
ENSDART00000139472
|
crata
|
carnitine O-acetyltransferase a |
| chr7_+_31130667 | 0.53 |
ENSDART00000173937
|
tjp1a
|
tight junction protein 1a |
| chr18_+_35861930 | 0.53 |
ENSDART00000185223
|
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr23_-_25135046 | 0.53 |
ENSDART00000184844
ENSDART00000103989 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr12_+_27285994 | 0.53 |
ENSDART00000087204
|
dusp3a
|
dual specificity phosphatase 3a |
| chr2_-_21847935 | 0.52 |
ENSDART00000003940
|
rab2a
|
RAB2A, member RAS oncogene family |
| chr9_+_6997861 | 0.52 |
ENSDART00000190491
|
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr22_-_14255659 | 0.52 |
ENSDART00000167088
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr9_+_38458193 | 0.52 |
ENSDART00000008053
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr20_+_54336137 | 0.52 |
ENSDART00000113792
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr4_-_9822519 | 0.51 |
ENSDART00000131357
|
parp12b
|
poly (ADP-ribose) polymerase family, member 12b |
| chr2_+_23062085 | 0.51 |
ENSDART00000153745
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr2_-_33645411 | 0.51 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr25_+_33063762 | 0.51 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr3_-_38777553 | 0.49 |
ENSDART00000193045
|
znf281a
|
zinc finger protein 281a |
| chr15_-_2857961 | 0.49 |
ENSDART00000033263
|
ankrd49
|
ankyrin repeat domain 49 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ikzf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.5 | 1.4 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.5 | 2.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 3.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.3 | 2.0 | GO:0044819 | mitotic G1/S transition checkpoint(GO:0044819) |
| 0.3 | 1.2 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.3 | 1.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.3 | 1.0 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.3 | 0.8 | GO:1901216 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.2 | 0.7 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 | 1.4 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.2 | 0.7 | GO:0032816 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.2 | 0.9 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 1.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 1.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 0.9 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.2 | 2.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 0.9 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.2 | 3.0 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 1.0 | GO:0097065 | anterior head development(GO:0097065) |
| 0.2 | 1.0 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.2 | 1.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 0.5 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.2 | 1.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.4 | GO:1901004 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.1 | 0.5 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.1 | 2.1 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 0.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.5 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.6 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 4.8 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 0.5 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.1 | 1.1 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.5 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.4 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 0.8 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.7 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 1.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.9 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of protein tyrosine kinase activity(GO:0061098) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.6 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.6 | GO:0045117 | azole transport(GO:0045117) |
| 0.1 | 0.6 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.6 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 1.9 | GO:0006684 | sphingomyelin metabolic process(GO:0006684) |
| 0.1 | 2.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.9 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.1 | 0.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.6 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.1 | 1.6 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 0.6 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 1.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.5 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.6 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 1.0 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.4 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 1.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.2 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.1 | 0.6 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.5 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.4 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 2.2 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.0 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.7 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 1.9 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.5 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.5 | GO:0046620 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.0 | 0.4 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 3.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.5 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.9 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 1.3 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 0.2 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.3 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.0 | 0.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 1.1 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 1.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.8 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.3 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 1.6 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 1.0 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.6 | GO:0070972 | protein localization to endoplasmic reticulum(GO:0070972) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.9 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.2 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.4 | GO:0048264 | positive regulation of BMP signaling pathway(GO:0030513) determination of ventral identity(GO:0048264) |
| 0.0 | 0.2 | GO:0051402 | neuron apoptotic process(GO:0051402) |
| 0.0 | 0.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.6 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.5 | 1.5 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.3 | 1.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.3 | 1.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.3 | 1.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 0.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 0.9 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 0.5 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 1.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 2.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 2.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.5 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 1.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.7 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.7 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 1.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 4.7 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 1.4 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.4 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 1.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 2.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0071565 | nBAF complex(GO:0071565) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.8 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.5 | 2.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.4 | 3.4 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.4 | 6.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 1.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.3 | 1.1 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.3 | 0.8 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.3 | 1.1 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 3.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 0.9 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.2 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 0.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.4 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.2 | 1.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 1.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.6 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 1.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.5 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 2.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.5 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 1.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.9 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.4 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.1 | 0.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.5 | GO:0070883 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.1 | 1.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.0 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.1 | 1.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.0 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 1.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 1.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.3 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.6 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 4.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.0 | 1.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 2.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 2.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 2.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 0.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.5 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 1.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 9.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 3.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 0.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 0.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 0.8 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |