Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
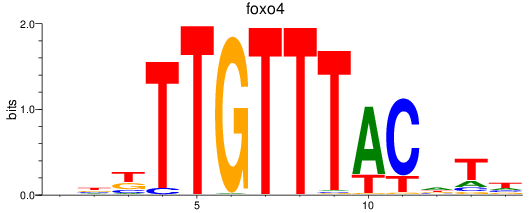
Results for foxo4
Z-value: 2.03
Transcription factors associated with foxo4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxo4
|
ENSDARG00000055792 | forkhead box O4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxo4 | dr11_v1_chr14_+_30910114_30910114 | -0.55 | 1.7e-02 | Click! |
Activity profile of foxo4 motif
Sorted Z-values of foxo4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_31511739 | 7.11 |
ENSDART00000049420
|
ndrg1b
|
N-myc downstream regulated 1b |
| chr15_+_19990068 | 6.17 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr4_-_1720648 | 5.56 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr16_-_26820634 | 5.24 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr13_+_22675802 | 5.08 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr7_+_46019780 | 4.82 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr7_+_33372680 | 4.71 |
ENSDART00000193436
ENSDART00000099988 |
glceb
|
glucuronic acid epimerase b |
| chr1_+_45969240 | 4.64 |
ENSDART00000042086
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr5_-_33236637 | 4.58 |
ENSDART00000085512
ENSDART00000144694 |
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr4_-_4256300 | 4.40 |
ENSDART00000103319
ENSDART00000150279 |
cd9b
|
CD9 molecule b |
| chr14_+_15155684 | 4.07 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr23_+_13533885 | 4.06 |
ENSDART00000144386
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr11_+_2710530 | 4.06 |
ENSDART00000132768
ENSDART00000030921 ENSDART00000040147 |
mapk14b
|
mitogen-activated protein kinase 14b |
| chr16_-_41714988 | 3.99 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr22_-_28777557 | 3.93 |
ENSDART00000135214
ENSDART00000131761 ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr5_+_44846434 | 3.83 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr12_+_22404108 | 3.78 |
ENSDART00000153055
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr6_-_39700965 | 3.75 |
ENSDART00000156645
|
espl1
|
extra spindle pole bodies like 1, separase |
| chr9_+_2041535 | 3.72 |
ENSDART00000093187
|
lnpa
|
limb and neural patterns a |
| chr22_-_28777374 | 3.72 |
ENSDART00000188206
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr3_-_15475067 | 3.67 |
ENSDART00000025324
ENSDART00000139575 |
spns1
|
spinster homolog 1 (Drosophila) |
| chr23_-_3758637 | 3.67 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr9_+_24088062 | 3.62 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr15_+_46313082 | 3.61 |
ENSDART00000153830
|
si:ch1073-190k2.1
|
si:ch1073-190k2.1 |
| chr5_+_3891485 | 3.55 |
ENSDART00000129329
ENSDART00000091711 |
rpain
|
RPA interacting protein |
| chr3_+_32411343 | 3.50 |
ENSDART00000186287
ENSDART00000141793 |
rras
|
RAS related |
| chr17_+_33158350 | 3.47 |
ENSDART00000104476
|
snx9a
|
sorting nexin 9a |
| chr5_-_65121747 | 3.46 |
ENSDART00000165556
|
tor2a
|
torsin family 2, member A |
| chr15_-_44052927 | 3.46 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr14_+_35424539 | 3.42 |
ENSDART00000171809
ENSDART00000162185 |
sytl4
|
synaptotagmin-like 4 |
| chr11_-_25257045 | 3.41 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr7_-_51749683 | 3.31 |
ENSDART00000083190
|
hdac8
|
histone deacetylase 8 |
| chr8_+_12951155 | 3.27 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr17_+_14965570 | 3.23 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr19_-_12965020 | 3.22 |
ENSDART00000128975
|
slc25a32a
|
solute carrier family 25 (mitochondrial folate carrier), member 32a |
| chr11_-_25257595 | 3.21 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr5_+_28271412 | 3.19 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr3_-_26191960 | 3.19 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr5_+_44805269 | 3.14 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr3_+_10152092 | 3.12 |
ENSDART00000066053
|
cbx2
|
chromobox homolog 2 (Drosophila Pc class) |
| chr9_-_12659140 | 3.10 |
ENSDART00000058565
|
pttg1ipb
|
PTTG1 interacting protein b |
| chr11_-_34577034 | 3.09 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr5_-_69212184 | 3.05 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr11_+_11120532 | 3.03 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr17_+_25856671 | 3.03 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr16_+_42667560 | 3.01 |
ENSDART00000023452
|
dpy19l1l
|
dpy-19-like 1, like (H. sapiens) |
| chr15_-_28587490 | 3.00 |
ENSDART00000186196
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr6_-_13022166 | 2.95 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr15_-_28587147 | 2.93 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr15_-_30984557 | 2.93 |
ENSDART00000080328
|
nf1a
|
neurofibromin 1a |
| chr5_+_44846280 | 2.92 |
ENSDART00000084370
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr10_-_38243579 | 2.91 |
ENSDART00000150159
|
usp25
|
ubiquitin specific peptidase 25 |
| chr17_-_7218481 | 2.84 |
ENSDART00000181967
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr6_+_38626926 | 2.82 |
ENSDART00000190339
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr5_+_22579975 | 2.73 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr10_-_6588793 | 2.72 |
ENSDART00000163788
|
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr20_+_53368611 | 2.71 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr3_-_29962345 | 2.67 |
ENSDART00000136819
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr14_-_32876280 | 2.66 |
ENSDART00000173168
|
si:rp71-46j2.7
|
si:rp71-46j2.7 |
| chr22_-_18240968 | 2.65 |
ENSDART00000027605
|
tmem161a
|
transmembrane protein 161A |
| chr19_-_3821678 | 2.63 |
ENSDART00000169639
|
si:dkey-206d17.12
|
si:dkey-206d17.12 |
| chr19_-_868187 | 2.62 |
ENSDART00000186626
|
eomesa
|
eomesodermin homolog a |
| chr17_+_32622933 | 2.61 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr25_+_7494181 | 2.59 |
ENSDART00000165005
|
cat
|
catalase |
| chr15_-_37589600 | 2.58 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr7_-_51749895 | 2.55 |
ENSDART00000175523
ENSDART00000189639 |
hdac8
|
histone deacetylase 8 |
| chr20_-_52928541 | 2.53 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr6_-_32045951 | 2.51 |
ENSDART00000016629
ENSDART00000139055 |
efcab7
|
EF-hand calcium binding domain 7 |
| chr10_+_32050906 | 2.49 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr16_-_39267185 | 2.49 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr5_-_22602979 | 2.47 |
ENSDART00000146287
|
nono
|
non-POU domain containing, octamer-binding |
| chr13_+_9368621 | 2.44 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr2_-_42492201 | 2.42 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr9_+_41459759 | 2.41 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr2_-_4787566 | 2.40 |
ENSDART00000160663
ENSDART00000157808 |
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr2_+_16696052 | 2.40 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr11_+_18612421 | 2.37 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr21_-_13662237 | 2.36 |
ENSDART00000091647
ENSDART00000151547 |
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr13_+_11828516 | 2.36 |
ENSDART00000110141
|
sufu
|
suppressor of fused homolog (Drosophila) |
| chr22_-_5171362 | 2.36 |
ENSDART00000124889
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr6_-_8244474 | 2.36 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr13_-_33207367 | 2.35 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr20_+_25904199 | 2.34 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr5_-_22602780 | 2.33 |
ENSDART00000011699
|
nono
|
non-POU domain containing, octamer-binding |
| chr10_+_28160265 | 2.32 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr12_+_8822717 | 2.30 |
ENSDART00000021628
|
reep3b
|
receptor accessory protein 3b |
| chr24_-_21343982 | 2.29 |
ENSDART00000012653
|
spice1
|
spindle and centriole associated protein 1 |
| chr4_-_16876281 | 2.24 |
ENSDART00000016690
ENSDART00000044005 ENSDART00000042874 ENSDART00000125762 ENSDART00000185974 |
tmpoa
|
thymopoietin a |
| chr23_+_31596441 | 2.23 |
ENSDART00000053534
|
tbpl1
|
TBP-like 1 |
| chr14_-_15155384 | 2.21 |
ENSDART00000172666
|
uvssa
|
UV-stimulated scaffold protein A |
| chr23_-_36449111 | 2.20 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr17_-_9962578 | 2.20 |
ENSDART00000021942
|
eapp
|
e2f-associated phosphoprotein |
| chr17_-_15546862 | 2.20 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr11_+_18873619 | 2.19 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr9_+_37366973 | 2.17 |
ENSDART00000016370
|
dirc2
|
disrupted in renal carcinoma 2 |
| chr13_-_21650404 | 2.17 |
ENSDART00000078352
|
tspan14
|
tetraspanin 14 |
| chr20_+_1385674 | 2.16 |
ENSDART00000145981
|
mmachc
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr3_-_33113879 | 2.15 |
ENSDART00000044677
|
rarab
|
retinoic acid receptor, alpha b |
| chr10_-_15879569 | 2.14 |
ENSDART00000136789
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr15_+_34069746 | 2.11 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr3_-_25369557 | 2.06 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr3_-_13546610 | 2.05 |
ENSDART00000159647
|
amdhd2
|
amidohydrolase domain containing 2 |
| chr17_+_44030692 | 2.04 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr17_+_38295847 | 2.04 |
ENSDART00000008532
|
mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr15_+_39977461 | 2.04 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr24_-_25004553 | 2.03 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr22_+_38276024 | 2.03 |
ENSDART00000143792
|
rcor3
|
REST corepressor 3 |
| chr21_-_43398122 | 2.03 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr3_+_46762703 | 2.03 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr19_+_46222918 | 2.02 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr19_-_25119443 | 2.01 |
ENSDART00000148953
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr19_+_31585341 | 2.01 |
ENSDART00000052185
|
gmnn
|
geminin, DNA replication inhibitor |
| chr4_-_14954029 | 1.99 |
ENSDART00000038642
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr22_-_5171829 | 1.97 |
ENSDART00000140313
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr23_+_31942040 | 1.96 |
ENSDART00000088607
|
nemp1
|
nuclear envelope integral membrane protein 1 |
| chr23_-_17450746 | 1.95 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr6_-_40922971 | 1.94 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr3_+_32403758 | 1.93 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr5_-_51998708 | 1.93 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr23_-_10722664 | 1.93 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr19_+_46222428 | 1.93 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr8_+_16990120 | 1.92 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr22_+_14117078 | 1.91 |
ENSDART00000013575
|
bzw1a
|
basic leucine zipper and W2 domains 1a |
| chr21_+_19547806 | 1.90 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr25_-_25142387 | 1.90 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr21_-_13661631 | 1.89 |
ENSDART00000184408
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr16_+_5612547 | 1.89 |
ENSDART00000140226
ENSDART00000189352 |
CYTH2
|
si:dkey-283b15.4 |
| chr7_+_38808027 | 1.89 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr5_-_56924747 | 1.88 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr24_+_39211288 | 1.87 |
ENSDART00000061540
|
im:7160594
|
im:7160594 |
| chr17_-_23709347 | 1.87 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr18_+_16749091 | 1.87 |
ENSDART00000061265
|
rnf141
|
ring finger protein 141 |
| chr3_+_35498119 | 1.86 |
ENSDART00000178963
|
tnrc6a
|
trinucleotide repeat containing 6a |
| chr13_-_13030851 | 1.85 |
ENSDART00000009499
|
nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr10_-_36633882 | 1.85 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr5_+_41477526 | 1.84 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr7_-_13906409 | 1.81 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr23_-_10723009 | 1.80 |
ENSDART00000189721
|
foxp1a
|
forkhead box P1a |
| chr5_+_55934129 | 1.79 |
ENSDART00000050969
|
tmem150ab
|
transmembrane protein 150Ab |
| chr2_+_1988036 | 1.79 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr24_-_19718077 | 1.77 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr12_-_25380028 | 1.76 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr17_-_14966384 | 1.75 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr7_+_56472585 | 1.73 |
ENSDART00000135259
ENSDART00000073596 |
ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr8_-_25716074 | 1.71 |
ENSDART00000007482
|
tspy
|
testis specific protein, Y-linked |
| chr10_-_35186310 | 1.70 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr17_+_24111392 | 1.65 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr6_-_42336987 | 1.63 |
ENSDART00000128777
ENSDART00000075601 |
fancd2
|
Fanconi anemia, complementation group D2 |
| chr8_+_20488322 | 1.63 |
ENSDART00000036630
|
zgc:101100
|
zgc:101100 |
| chr1_+_53321878 | 1.62 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr25_+_22017182 | 1.62 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr1_-_23596391 | 1.59 |
ENSDART00000155184
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr21_+_20386865 | 1.58 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr17_-_22573311 | 1.58 |
ENSDART00000141523
ENSDART00000140022 ENSDART00000079390 ENSDART00000188644 |
exo1
|
exonuclease 1 |
| chr24_+_37709191 | 1.57 |
ENSDART00000066558
|
decr2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr17_+_32360673 | 1.56 |
ENSDART00000155519
|
si:ch211-139d20.3
|
si:ch211-139d20.3 |
| chr17_-_37474689 | 1.56 |
ENSDART00000103980
|
crip2
|
cysteine-rich protein 2 |
| chr20_-_35470891 | 1.56 |
ENSDART00000152993
ENSDART00000016090 |
pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr13_+_42602406 | 1.56 |
ENSDART00000133388
ENSDART00000147996 |
mlh1
|
mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) |
| chr14_-_30490465 | 1.56 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr14_+_26439227 | 1.55 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr19_-_34063567 | 1.54 |
ENSDART00000157815
ENSDART00000183907 |
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr18_+_6857071 | 1.53 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr20_+_23742574 | 1.52 |
ENSDART00000050559
|
sh3rf1
|
SH3 domain containing ring finger 1 |
| chr5_+_53009083 | 1.52 |
ENSDART00000157836
|
hint2
|
histidine triad nucleotide binding protein 2 |
| chr3_+_19685873 | 1.52 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr5_-_40190949 | 1.51 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr3_-_15679107 | 1.50 |
ENSDART00000080441
|
zgc:66443
|
zgc:66443 |
| chr13_+_1100197 | 1.50 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr11_+_21586335 | 1.50 |
ENSDART00000091182
|
FAM72B
|
zgc:101564 |
| chr22_-_11520405 | 1.49 |
ENSDART00000063157
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr5_+_1965296 | 1.49 |
ENSDART00000156224
|
dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr21_+_34088377 | 1.48 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr11_-_26701611 | 1.48 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr1_-_23595779 | 1.48 |
ENSDART00000134860
ENSDART00000138852 |
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr8_+_8643901 | 1.48 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr11_+_24313931 | 1.47 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr24_+_14240196 | 1.46 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr17_-_11417904 | 1.45 |
ENSDART00000103228
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr21_+_34088110 | 1.44 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr16_+_6944311 | 1.43 |
ENSDART00000144763
|
eaf1
|
ELL associated factor 1 |
| chr21_-_9782502 | 1.41 |
ENSDART00000158836
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr16_+_6944564 | 1.40 |
ENSDART00000104252
|
eaf1
|
ELL associated factor 1 |
| chr10_+_11260170 | 1.38 |
ENSDART00000155742
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr6_-_35309661 | 1.35 |
ENSDART00000114960
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr21_-_27272657 | 1.35 |
ENSDART00000040754
ENSDART00000175009 |
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr21_+_17051478 | 1.35 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr8_-_18879905 | 1.34 |
ENSDART00000100491
|
sh3gl1b
|
SH3-domain GRB2-like 1b |
| chr7_-_38570299 | 1.33 |
ENSDART00000143815
|
celf1
|
cugbp, Elav-like family member 1 |
| chr6_-_40657653 | 1.30 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr9_+_2343096 | 1.30 |
ENSDART00000062292
ENSDART00000191722 ENSDART00000135180 |
atf2
|
activating transcription factor 2 |
| chr18_+_41561285 | 1.30 |
ENSDART00000169621
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr14_-_7885707 | 1.28 |
ENSDART00000029981
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr7_+_24528866 | 1.27 |
ENSDART00000180552
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr5_-_32074534 | 1.27 |
ENSDART00000112342
ENSDART00000135855 |
arpc5lb
|
actin related protein 2/3 complex, subunit 5-like, b |
| chr24_-_5932982 | 1.26 |
ENSDART00000138412
ENSDART00000135124 ENSDART00000007373 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr20_+_22799857 | 1.26 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr15_-_5720583 | 1.25 |
ENSDART00000158034
ENSDART00000190332 ENSDART00000109599 |
urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr18_-_20466061 | 1.23 |
ENSDART00000060311
|
paqr5a
|
progestin and adipoQ receptor family member Va |
| chr5_-_66749535 | 1.23 |
ENSDART00000132183
|
kat5b
|
K(lysine) acetyltransferase 5b |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxo4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.3 | 5.2 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 1.2 | 3.7 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 1.1 | 7.9 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 1.0 | 3.0 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 1.0 | 2.9 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.9 | 2.6 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.8 | 5.6 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.8 | 1.5 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.7 | 5.9 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.7 | 2.7 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.7 | 2.6 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.6 | 3.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.6 | 2.5 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.6 | 1.9 | GO:0034036 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.6 | 1.9 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.6 | 2.4 | GO:1904590 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.6 | 4.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.5 | 1.6 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.5 | 1.6 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.5 | 2.0 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.5 | 4.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.5 | 4.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.5 | 2.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.5 | 2.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.5 | 2.9 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.4 | 1.3 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.4 | 3.9 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.4 | 2.0 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.4 | 4.8 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.4 | 4.7 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.4 | 1.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.4 | 1.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.3 | 3.5 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 | 2.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 | 1.3 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.3 | 2.5 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.3 | 2.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.3 | 2.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.3 | 6.6 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.3 | 1.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.3 | 3.7 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.3 | 1.5 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 3.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 0.8 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.2 | 1.4 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 0.9 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.2 | 2.8 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.2 | 0.9 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.2 | 2.1 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.2 | 1.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 1.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.2 | 1.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 17.2 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.2 | 2.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.5 | GO:0036316 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.2 | 0.7 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 2.5 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.2 | 1.0 | GO:0051883 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.2 | 1.0 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 3.5 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.2 | 2.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 0.7 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 1.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 2.2 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 8.3 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 1.9 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 6.3 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 1.0 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 1.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 3.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 2.7 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 2.0 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 2.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.8 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 1.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 2.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.7 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 1.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.0 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 2.0 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 3.4 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 3.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 1.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.7 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 4.8 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.1 | 3.4 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 1.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 2.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 2.0 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 3.0 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 4.7 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 1.2 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.4 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 1.1 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 3.3 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 1.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 1.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 1.5 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.3 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 1.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.7 | GO:0044273 | glycosaminoglycan catabolic process(GO:0006027) sulfur compound catabolic process(GO:0044273) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.5 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 1.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0051984 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of chromosome segregation(GO:0051984) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.9 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 2.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.7 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 3.4 | GO:0009953 | dorsal/ventral pattern formation(GO:0009953) |
| 0.0 | 0.4 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.2 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.5 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.7 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 2.0 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.4 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.6 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 1.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 2.9 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 1.2 | 4.8 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.6 | 1.8 | GO:0031213 | RSF complex(GO:0031213) |
| 0.5 | 2.0 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.5 | 1.9 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.4 | 2.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.4 | 2.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.3 | 1.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.3 | 1.0 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.3 | 1.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 3.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 2.3 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 1.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 1.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 3.1 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.2 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 1.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 7.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 1.6 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.2 | 7.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 1.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 3.8 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 9.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 2.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.2 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 6.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 3.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 6.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 5.0 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 4.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 4.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 5.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.3 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 0.7 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 4.3 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 1.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 2.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.1 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 5.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 5.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 3.2 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.1 | GO:0034704 | voltage-gated calcium channel complex(GO:0005891) calcium channel complex(GO:0034704) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 3.8 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 6.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 4.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.4 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 1.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 1.3 | 5.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.2 | 4.7 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 1.1 | 3.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.8 | 2.5 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.8 | 3.0 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.7 | 2.7 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.7 | 3.3 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.6 | 2.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.6 | 1.9 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.5 | 1.6 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.5 | 4.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.5 | 1.5 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.5 | 2.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.4 | 3.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.4 | 5.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.4 | 1.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.4 | 5.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 0.9 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.3 | 1.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 2.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 2.5 | GO:0004096 | catalase activity(GO:0004096) |
| 0.3 | 1.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.3 | 2.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 1.9 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 4.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.2 | 1.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 2.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 1.1 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 1.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 3.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 0.6 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 0.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 0.7 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 1.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 0.8 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 1.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 1.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 5.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 3.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 3.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 2.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 3.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 7.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 3.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 6.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 13.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 3.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 1.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 2.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 1.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 1.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.8 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 1.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 5.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 4.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 2.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.8 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 5.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 2.8 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.0 | 1.7 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 1.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.4 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 3.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 3.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 3.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 5.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 2.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.4 | 5.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 3.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.4 | 1.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 1.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 2.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 2.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.2 | 2.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.2 | 2.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 3.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 3.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 4.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 2.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 1.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 2.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 5.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 3.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 2.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.2 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 1.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.8 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 1.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 2.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.7 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |