Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
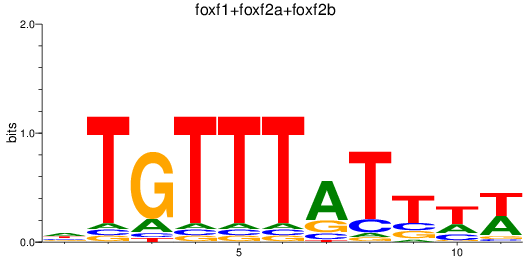
Results for foxf1+foxf2a+foxf2b
Z-value: 1.42
Transcription factors associated with foxf1+foxf2a+foxf2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxf1
|
ENSDARG00000015399 | forkhead box F1 |
|
foxf2a
|
ENSDARG00000017195 | forkhead box F2a |
|
foxf2b
|
ENSDARG00000070389 | forkhead box F2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxf2b | dr11_v1_chr20_+_26690036_26690036 | -0.35 | 1.5e-01 | Click! |
| foxf2a | dr11_v1_chr2_-_716426_716426 | 0.24 | 3.3e-01 | Click! |
| foxf1 | dr11_v1_chr18_+_30847237_30847237 | 0.21 | 4.0e-01 | Click! |
Activity profile of foxf1+foxf2a+foxf2b motif
Sorted Z-values of foxf1+foxf2a+foxf2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_11756040 | 2.75 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr4_-_1720648 | 1.89 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr13_+_22675802 | 1.88 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr11_-_30508843 | 1.53 |
ENSDART00000101667
ENSDART00000179930 |
map4k3a
|
mitogen-activated protein kinase kinase kinase kinase 3a |
| chr23_-_33558161 | 1.52 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr1_-_6028876 | 1.50 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr1_-_50611031 | 1.46 |
ENSDART00000148285
|
ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr8_-_53044300 | 1.41 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr1_+_496268 | 1.40 |
ENSDART00000109415
|
blzf1
|
basic leucine zipper nuclear factor 1 |
| chr2_+_27652386 | 1.36 |
ENSDART00000188261
|
tmem68
|
transmembrane protein 68 |
| chr4_-_6373735 | 1.35 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr2_+_44512324 | 1.35 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr19_+_43669122 | 1.30 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr24_-_17023392 | 1.28 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr1_+_51386649 | 1.27 |
ENSDART00000152289
|
atg4da
|
autophagy related 4D, cysteine peptidase a |
| chr5_-_3960161 | 1.24 |
ENSDART00000111453
|
myo19
|
myosin XIX |
| chr9_+_41459759 | 1.22 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr2_+_27651984 | 1.22 |
ENSDART00000126762
|
tmem68
|
transmembrane protein 68 |
| chr1_-_43920371 | 1.21 |
ENSDART00000109283
|
scpp7
|
secretory calcium-binding phosphoprotein 7 |
| chr8_-_33154677 | 1.20 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr1_+_45969240 | 1.18 |
ENSDART00000042086
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr8_-_39859688 | 1.13 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr3_+_24511959 | 1.11 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr11_+_24046179 | 1.09 |
ENSDART00000006703
|
maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr8_+_49092077 | 1.08 |
ENSDART00000032355
|
zgc:56525
|
zgc:56525 |
| chr15_-_33807758 | 1.08 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr6_+_2174082 | 1.06 |
ENSDART00000073936
|
acvr1bb
|
activin A receptor type 1Bb |
| chr3_-_4501026 | 1.05 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr6_+_22104857 | 1.04 |
ENSDART00000129202
|
si:dkey-156n14.5
|
si:dkey-156n14.5 |
| chr11_-_11890001 | 1.02 |
ENSDART00000081544
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr8_+_29636431 | 1.00 |
ENSDART00000133047
|
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr23_-_22523303 | 1.00 |
ENSDART00000079019
|
spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr19_+_25971000 | 1.00 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr22_+_12361317 | 1.00 |
ENSDART00000189963
ENSDART00000159614 |
r3hdm1
|
R3H domain containing 1 |
| chr7_-_60351537 | 0.99 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr15_-_29354020 | 0.98 |
ENSDART00000127795
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr16_+_18535618 | 0.98 |
ENSDART00000021596
|
rxrbb
|
retinoid x receptor, beta b |
| chr7_-_53117131 | 0.98 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr8_-_53044089 | 0.98 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr18_-_44847855 | 0.97 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr6_+_23809501 | 0.97 |
ENSDART00000168701
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr15_+_19990068 | 0.96 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr2_+_1988036 | 0.95 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr6_+_20954400 | 0.95 |
ENSDART00000143248
ENSDART00000165806 |
stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr7_+_13830052 | 0.94 |
ENSDART00000191360
|
abhd2a
|
abhydrolase domain containing 2a |
| chr7_+_24889783 | 0.94 |
ENSDART00000005329
ENSDART00000159955 |
mark2b
|
MAP/microtubule affinity-regulating kinase 2b |
| chr15_+_23657051 | 0.93 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr1_+_19538299 | 0.93 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr6_+_4255319 | 0.93 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr8_+_23147218 | 0.93 |
ENSDART00000030920
ENSDART00000141175 ENSDART00000146264 |
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr7_-_60351876 | 0.92 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr24_+_17068724 | 0.92 |
ENSDART00000191137
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr21_+_25802190 | 0.92 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr11_+_18612421 | 0.91 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr12_-_18961491 | 0.91 |
ENSDART00000172574
|
ep300a
|
E1A binding protein p300 a |
| chr15_+_44283723 | 0.91 |
ENSDART00000167722
|
cwf19l2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr20_+_34390196 | 0.90 |
ENSDART00000183596
|
trmt1l
|
tRNA methyltransferase 1-like |
| chr13_-_12667220 | 0.90 |
ENSDART00000079594
|
fam241a
|
family with sequence similarity 241 member A |
| chr21_-_25295087 | 0.90 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr24_+_17069420 | 0.89 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr7_-_13906409 | 0.89 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr12_+_27096835 | 0.87 |
ENSDART00000149475
|
ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr14_-_21660548 | 0.87 |
ENSDART00000161713
ENSDART00000089845 |
kdm3b
|
lysine (K)-specific demethylase 3B |
| chr2_+_31437547 | 0.87 |
ENSDART00000141170
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr23_-_18017946 | 0.86 |
ENSDART00000104592
|
pm20d1.2
|
peptidase M20 domain containing 1, tandem duplicate 2 |
| chr9_+_22780901 | 0.86 |
ENSDART00000110992
ENSDART00000143972 |
rif1
|
replication timing regulatory factor 1 |
| chr1_+_26676758 | 0.85 |
ENSDART00000152299
|
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr1_-_43920576 | 0.85 |
ENSDART00000191914
|
scpp7
|
secretory calcium-binding phosphoprotein 7 |
| chr22_+_30195257 | 0.85 |
ENSDART00000027803
ENSDART00000172496 |
add3a
|
adducin 3 (gamma) a |
| chr23_+_27778670 | 0.84 |
ENSDART00000053863
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr6_-_37745508 | 0.84 |
ENSDART00000078316
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr12_-_35582521 | 0.84 |
ENSDART00000162175
ENSDART00000168958 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr14_-_30960470 | 0.84 |
ENSDART00000129989
|
si:ch211-191o15.6
|
si:ch211-191o15.6 |
| chr5_-_22573624 | 0.84 |
ENSDART00000131889
ENSDART00000080886 ENSDART00000147513 ENSDART00000080882 |
aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr25_+_16646113 | 0.82 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr22_-_10752471 | 0.81 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr14_-_16082806 | 0.81 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr7_-_50367642 | 0.81 |
ENSDART00000134941
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr21_-_26490186 | 0.80 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr8_-_30424182 | 0.80 |
ENSDART00000099021
|
dock8
|
dedicator of cytokinesis 8 |
| chr13_-_32626247 | 0.78 |
ENSDART00000100663
|
wdr35
|
WD repeat domain 35 |
| chr17_-_5352924 | 0.78 |
ENSDART00000167275
|
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr12_-_35582683 | 0.78 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr25_-_25142387 | 0.78 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr12_+_26467847 | 0.78 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr17_+_10748366 | 0.78 |
ENSDART00000018683
ENSDART00000097274 |
ATG14
|
zgc:113944 |
| chr18_-_40913294 | 0.77 |
ENSDART00000059196
ENSDART00000098878 |
polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr9_+_42607138 | 0.77 |
ENSDART00000138133
ENSDART00000002027 |
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr10_+_41939963 | 0.77 |
ENSDART00000126248
|
tmem120b
|
transmembrane protein 120B |
| chr25_-_14087377 | 0.76 |
ENSDART00000124140
|
zgc:101566
|
zgc:101566 |
| chr24_+_5893134 | 0.76 |
ENSDART00000077941
|
mastl
|
microtubule associated serine/threonine kinase-like |
| chr10_+_43117661 | 0.76 |
ENSDART00000024644
ENSDART00000186932 |
xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr11_+_24348425 | 0.75 |
ENSDART00000089747
|
nfs1
|
NFS1 cysteine desulfurase |
| chr13_-_36761379 | 0.75 |
ENSDART00000131534
ENSDART00000029824 |
map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr9_+_23003208 | 0.74 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr11_+_1584747 | 0.74 |
ENSDART00000154583
|
si:dkey-40c23.2
|
si:dkey-40c23.2 |
| chr19_+_32807236 | 0.74 |
ENSDART00000008698
|
stk3
|
serine/threonine kinase 3 (STE20 homolog, yeast) |
| chr19_-_24135824 | 0.73 |
ENSDART00000189505
ENSDART00000104087 |
thap7
|
THAP domain containing 7 |
| chr8_+_23147609 | 0.72 |
ENSDART00000180284
|
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr3_-_37681824 | 0.72 |
ENSDART00000185858
|
gpatch8
|
G patch domain containing 8 |
| chr3_-_25492361 | 0.72 |
ENSDART00000147322
ENSDART00000055473 |
grb2b
|
growth factor receptor-bound protein 2b |
| chr7_-_50367326 | 0.72 |
ENSDART00000141926
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr19_+_4968947 | 0.72 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr18_-_14337450 | 0.72 |
ENSDART00000061435
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr22_+_29648854 | 0.72 |
ENSDART00000146280
|
bbip1
|
BBSome interacting protein 1 |
| chr15_+_1534644 | 0.72 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr17_-_32743044 | 0.71 |
ENSDART00000135517
|
cenpf
|
centromere protein F |
| chr5_+_44944778 | 0.71 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr17_+_32500387 | 0.71 |
ENSDART00000018423
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr3_+_53116172 | 0.71 |
ENSDART00000115117
|
brd4
|
bromodomain containing 4 |
| chr13_-_36844945 | 0.70 |
ENSDART00000129562
ENSDART00000150899 |
nin
|
ninein (GSK3B interacting protein) |
| chr14_+_8638353 | 0.70 |
ENSDART00000163240
|
si:dkeyp-115e12.6
|
si:dkeyp-115e12.6 |
| chr11_+_42422371 | 0.70 |
ENSDART00000163780
|
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chr4_-_20135919 | 0.70 |
ENSDART00000172230
|
cep83
|
centrosomal protein 83 |
| chr4_+_6833583 | 0.69 |
ENSDART00000165179
ENSDART00000186134 ENSDART00000174507 |
dock4b
|
dedicator of cytokinesis 4b |
| chr9_+_55455801 | 0.69 |
ENSDART00000144757
ENSDART00000186543 |
mxra5b
|
matrix-remodelling associated 5b |
| chr5_+_8196264 | 0.68 |
ENSDART00000174564
ENSDART00000161261 |
lmbrd2a
|
LMBR1 domain containing 2a |
| chr2_-_32384683 | 0.68 |
ENSDART00000182942
ENSDART00000141757 |
ubtfl
|
upstream binding transcription factor, like |
| chr23_-_912817 | 0.68 |
ENSDART00000192851
|
rhoac
|
ras homolog gene family, member Ac |
| chr13_+_37653851 | 0.68 |
ENSDART00000141988
ENSDART00000126902 ENSDART00000100352 |
phf3
|
PHD finger protein 3 |
| chr17_-_11417551 | 0.68 |
ENSDART00000128291
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr21_-_13856689 | 0.67 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr17_-_23709347 | 0.67 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr2_-_44777592 | 0.67 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr3_-_5228841 | 0.67 |
ENSDART00000092373
ENSDART00000182438 |
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr22_+_30112390 | 0.67 |
ENSDART00000191715
|
add3a
|
adducin 3 (gamma) a |
| chr1_+_51066671 | 0.67 |
ENSDART00000064007
|
srd5a2a
|
steroid-5-alpha-reductase, alpha polypeptide 2a |
| chr25_-_3647277 | 0.66 |
ENSDART00000166363
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr22_+_24603930 | 0.66 |
ENSDART00000180240
ENSDART00000164256 |
CU655842.1
|
|
| chr17_-_11417904 | 0.66 |
ENSDART00000103228
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr3_+_35510040 | 0.65 |
ENSDART00000162757
ENSDART00000182383 |
tnrc6a
|
trinucleotide repeat containing 6a |
| chr13_-_18011168 | 0.65 |
ENSDART00000144813
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr22_-_35063526 | 0.65 |
ENSDART00000162211
|
zgc:63733
|
zgc:63733 |
| chr2_+_2168547 | 0.65 |
ENSDART00000029347
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr23_-_25686894 | 0.65 |
ENSDART00000181420
ENSDART00000088208 |
lrp1ab
|
low density lipoprotein receptor-related protein 1Ab |
| chr8_+_16738282 | 0.65 |
ENSDART00000134265
ENSDART00000100698 |
ercc8
|
excision repair cross-complementation group 8 |
| chr7_+_26844261 | 0.64 |
ENSDART00000079165
|
ext2
|
exostosin glycosyltransferase 2 |
| chr23_-_21534738 | 0.63 |
ENSDART00000134587
|
rcc2
|
regulator of chromosome condensation 2 |
| chr4_+_16725960 | 0.63 |
ENSDART00000034441
|
tcp11l2
|
t-complex 11, testis-specific-like 2 |
| chr8_+_17167876 | 0.63 |
ENSDART00000134665
|
cenph
|
centromere protein H |
| chr2_+_44518636 | 0.63 |
ENSDART00000153733
|
pask
|
PAS domain containing serine/threonine kinase |
| chr10_+_17235370 | 0.63 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr22_-_3152357 | 0.63 |
ENSDART00000170983
|
lmnb2
|
lamin B2 |
| chr12_+_3912544 | 0.62 |
ENSDART00000013465
|
tbx6
|
T-box 6 |
| chr24_-_27256673 | 0.62 |
ENSDART00000181182
|
zmynd11
|
zinc finger, MYND-type containing 11 |
| chr18_+_20566817 | 0.62 |
ENSDART00000100716
|
bida
|
BH3 interacting domain death agonist |
| chr19_+_42047427 | 0.62 |
ENSDART00000180225
ENSDART00000145356 |
si:ch211-13c6.2
|
si:ch211-13c6.2 |
| chr18_+_13077800 | 0.61 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr4_-_20135406 | 0.61 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr20_-_45709990 | 0.61 |
ENSDART00000027482
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr1_+_53321878 | 0.61 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr19_-_47997424 | 0.61 |
ENSDART00000081675
|
ctnnb2
|
catenin, beta 2 |
| chr14_+_51056605 | 0.61 |
ENSDART00000159639
|
CABZ01078593.1
|
|
| chr7_+_7511914 | 0.61 |
ENSDART00000172848
|
clcn3
|
chloride channel 3 |
| chr10_+_17371356 | 0.61 |
ENSDART00000122663
|
sppl3
|
signal peptide peptidase 3 |
| chr5_-_42661012 | 0.61 |
ENSDART00000158339
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr24_-_23716097 | 0.60 |
ENSDART00000084954
ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr8_+_10862353 | 0.60 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr7_-_56766973 | 0.60 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr23_-_21534455 | 0.59 |
ENSDART00000139092
|
rcc2
|
regulator of chromosome condensation 2 |
| chr5_-_71722257 | 0.59 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr24_-_35707552 | 0.59 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr20_+_38671894 | 0.59 |
ENSDART00000146544
|
mpv17
|
MpV17 mitochondrial inner membrane protein |
| chr1_+_11107688 | 0.59 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr20_-_42378865 | 0.58 |
ENSDART00000139912
ENSDART00000015801 |
dcbld1
|
discoidin, CUB and LCCL domain containing 1 |
| chr4_+_6833735 | 0.58 |
ENSDART00000136355
|
dock4b
|
dedicator of cytokinesis 4b |
| chr5_-_19963537 | 0.57 |
ENSDART00000148146
|
si:dkey-234h16.7
|
si:dkey-234h16.7 |
| chr20_-_33487729 | 0.57 |
ENSDART00000061843
|
smek1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr19_-_9503473 | 0.57 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr18_-_15532016 | 0.56 |
ENSDART00000165279
|
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr17_+_45737992 | 0.56 |
ENSDART00000135073
ENSDART00000143525 ENSDART00000158165 ENSDART00000184167 ENSDART00000109525 |
aspg
|
asparaginase homolog (S. cerevisiae) |
| chr13_-_22961605 | 0.56 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr6_+_29791164 | 0.56 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr12_+_17436904 | 0.56 |
ENSDART00000079130
|
atad1b
|
ATPase family, AAA domain containing 1b |
| chr11_+_13024002 | 0.56 |
ENSDART00000104113
|
btf3l4
|
basic transcription factor 3-like 4 |
| chr7_+_25858380 | 0.56 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr7_+_21787507 | 0.55 |
ENSDART00000100936
|
tmem88b
|
transmembrane protein 88 b |
| chr19_-_10324182 | 0.55 |
ENSDART00000151352
ENSDART00000151162 ENSDART00000023571 |
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr23_+_22873415 | 0.55 |
ENSDART00000135130
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr6_-_34860574 | 0.55 |
ENSDART00000073957
|
slc35d1a
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1a |
| chr17_-_26719821 | 0.55 |
ENSDART00000186813
|
calm1a
|
calmodulin 1a |
| chr7_-_24995631 | 0.55 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr15_+_26941063 | 0.55 |
ENSDART00000149957
|
bcas3
|
breast carcinoma amplified sequence 3 |
| chr7_+_1550966 | 0.55 |
ENSDART00000177863
ENSDART00000126840 |
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr8_+_12930216 | 0.55 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr5_+_19314574 | 0.54 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr9_+_24088062 | 0.54 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr19_-_6134802 | 0.54 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr7_+_52712807 | 0.54 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr3_-_13546610 | 0.54 |
ENSDART00000159647
|
amdhd2
|
amidohydrolase domain containing 2 |
| chr9_-_28939796 | 0.54 |
ENSDART00000101269
|
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr10_+_16036573 | 0.54 |
ENSDART00000188757
|
lmnb1
|
lamin B1 |
| chr17_-_32743249 | 0.54 |
ENSDART00000176858
ENSDART00000189622 |
cenpf
|
centromere protein F |
| chr15_-_18432673 | 0.54 |
ENSDART00000146853
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr21_+_13327527 | 0.53 |
ENSDART00000114294
|
snrpd3l
|
small nuclear ribonucleoprotein D3 polypeptide, like |
| chr3_+_7771420 | 0.53 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr22_+_10232527 | 0.53 |
ENSDART00000139297
|
si:dkeyp-87e7.4
|
si:dkeyp-87e7.4 |
| chr10_+_16036246 | 0.52 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr21_-_36453594 | 0.52 |
ENSDART00000193176
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr20_-_53949798 | 0.52 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxf1+foxf2a+foxf2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.3 | 1.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 0.9 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.3 | 1.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 1.9 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.3 | 1.4 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.3 | 2.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 | 1.5 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 1.0 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.2 | 0.7 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.2 | 0.7 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 0.7 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 0.9 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 0.9 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.2 | 1.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 0.8 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 0.6 | GO:0000103 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 0.6 | GO:0046048 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.2 | 1.1 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.7 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 0.5 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 0.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 0.8 | GO:0006266 | DNA ligation(GO:0006266) immunoglobulin V(D)J recombination(GO:0033152) DNA ligation involved in DNA repair(GO:0051103) |
| 0.2 | 0.9 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.3 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.1 | 0.9 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.7 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 1.5 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.4 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 1.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 1.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.5 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 0.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 2.0 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 0.4 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.1 | 1.3 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 1.9 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.1 | 0.7 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.3 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 | 0.6 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 1.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.3 | GO:0034398 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.4 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.7 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.8 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.8 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 1.0 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.6 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.6 | GO:0009188 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.1 | 1.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 1.2 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 0.2 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.8 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 0.5 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 2.3 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.6 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.9 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.2 | GO:0061178 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.5 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.1 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.7 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.4 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.7 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.4 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.1 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.3 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 1.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.5 | GO:0061098 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of protein tyrosine kinase activity(GO:0061098) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.4 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.2 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 1.9 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.1 | 0.4 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.0 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.8 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.4 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.5 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 1.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 3.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.7 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.1 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.6 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.5 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.9 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 2.0 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.2 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.4 | GO:0008585 | female gonad development(GO:0008585) |
| 0.0 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 1.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.7 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 1.0 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.4 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.7 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.3 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.7 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 1.0 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.5 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.5 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.5 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.2 | GO:0090660 | regulation of microtubule polymerization(GO:0031113) cerebrospinal fluid circulation(GO:0090660) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.4 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.3 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.7 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 0.1 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.2 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.3 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:0070375 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.4 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.7 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 0.5 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.9 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.9 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.2 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.4 | GO:0003231 | cardiac ventricle development(GO:0003231) |
| 0.0 | 0.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.1 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.0 | 0.5 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.7 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 1.1 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.1 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.2 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.5 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.3 | 1.2 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.3 | 0.8 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 0.8 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.4 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 1.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.3 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 1.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.8 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 1.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.8 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.3 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 0.3 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.9 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.3 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 2.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 4.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 5.5 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 2.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) spindle midzone(GO:0051233) |
| 0.0 | 2.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 1.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 2.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.2 | GO:0098862 | stereocilium bundle(GO:0032421) cluster of actin-based cell projections(GO:0098862) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 4.4 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.4 | 2.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.9 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 1.4 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.2 | 0.6 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 1.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 0.7 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 1.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 0.6 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 0.6 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.6 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 2.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.0 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.6 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 0.6 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.5 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.8 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.3 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 1.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.5 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.3 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 1.0 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 1.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.3 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.1 | 0.8 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.0 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 1.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.4 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.4 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.6 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.1 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.2 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.1 | 0.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.6 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 2.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.9 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 1.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.8 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.5 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 1.5 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 1.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 2.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 1.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.9 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 3.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 2.0 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 2.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 1.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.4 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 1.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 0.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 2.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |