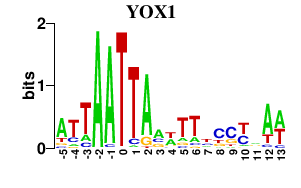
Results for YOX1
Z-value: 1.05
Transcription factors associated with YOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
YOX1
|
S000004489 | Homeobox transcriptional repressor |
Activity-expression correlation:
Activity profile of YOX1 motif
Sorted Z-values of YOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YJR094W-A | 6.71 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YOL086C | 6.52 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YFR056C | 6.17 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YFR055W | 5.86 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YHR180W-A | 4.57 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps dubious ORF YHR180C-B and long terminal repeat YHRCsigma3 |
||
| YPR119W | 3.79 |
CLB2
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YNR001W-A | 3.62 |
Dubious open reading frame unlikely to encode a functional protein; identified by homology |
||
| YHR181W | 3.27 |
SVP26
|
Integral membrane protein of the early Golgi apparatus and endoplasmic reticulum, involved in COP II vesicle transport; may also function to promote retention of proteins in the early Golgi compartment |
|
| YDR471W | 3.03 |
RPL27B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl27Ap and has similarity to rat L27 ribosomal protein |
|
| YLR333C | 3.00 |
RPS25B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps25Ap and has similarity to rat S25 ribosomal protein |
|
| YOR369C | 2.89 |
RPS12
|
Protein component of the small (40S) ribosomal subunit; has similarity to rat ribosomal protein S12 |
|
| YGR108W | 2.85 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YDR278C | 2.72 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOL085C | 2.72 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOL085W-A |
||
| YPR157W | 2.67 |
Putative protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YHR141C | 2.64 |
RPL42B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl42Ap and has similarity to rat L44; required for propagation of the killer toxin-encoding M1 double-stranded RNA satellite of the L-A double-stranded RNA virus |
|
| YOR096W | 2.46 |
RPS7A
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps7Bp; interacts with Kti11p; deletion causes hypersensitivity to zymocin; has similarity to rat S7 and Xenopus S8 ribosomal proteins |
|
| YDR279W | 2.30 |
RNH202
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YLR154C | 2.19 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YPL143W | 2.19 |
RPL33A
|
N-terminally acetylated ribosomal protein L37 of the large (60S) ribosomal subunit, nearly identical to Rpl33Bp and has similarity to rat L35a; rpl33a null mutant exhibits slow growth while rpl33a rpl33b double null mutant is inviable |
|
| YJR094C | 2.08 |
IME1
|
Master regulator of meiosis that is active only during meiotic events, activates transcription of early meiotic genes through interaction with Ume6p, degraded by the 26S proteasome following phosphorylation by Ime2p |
|
| YGR034W | 1.92 |
RPL26B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl26Ap and has similarity to E. coli L24 and rat L26 ribosomal proteins; binds to 5.8S rRNA |
|
| YER137C | 1.91 |
Putative protein of unknown function |
||
| YOR342C | 1.89 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and the nucleus |
||
| YOR137C | 1.86 |
SIA1
|
Protein of unassigned function involved in activation of the Pma1p plasma membrane H+-ATPase by glucose |
|
| YDR497C | 1.82 |
ITR1
|
Myo-inositol transporter with strong similarity to the minor myo-inositol transporter Itr2p, member of the sugar transporter superfamily; expression is repressed by inositol and choline via Opi1p and derepressed via Ino2p and Ino4p |
|
| YGL031C | 1.82 |
RPL24A
|
Ribosomal protein L30 of the large (60S) ribosomal subunit, nearly identical to Rpl24Bp and has similarity to rat L24 ribosomal protein; not essential for translation but may be required for normal translation rate |
|
| YMR032W | 1.79 |
HOF1
|
Bud neck-localized, SH3 domain-containing protein required for cytokinesis; regulates actomyosin ring dynamics and septin localization; interacts with the formins, Bni1p and Bnr1p, and with Cyk3p, Vrp1p, and Bni5p |
|
| YCR018C | 1.78 |
SRD1
|
Protein involved in the processing of pre-rRNA to mature rRNA; contains a C2/C2 zinc finger motif; srd1 mutation suppresses defects caused by the rrp1-1 mutation |
|
| YBR158W | 1.74 |
AMN1
|
Protein required for daughter cell separation, multiple mitotic checkpoints, and chromosome stability; contains 12 degenerate leucine-rich repeat motifs; expression is induced by the Mitotic Exit Network (MEN) |
|
| YGL039W | 1.73 |
Oxidoreductase, catalyzes NADPH-dependent reduction of the bicyclic diketone bicyclo[2.2.2]octane-2,6-dione (BCO2,6D) to the chiral ketoalcohol (1R,4S,6S)-6-hydroxybicyclo[2.2.2]octane-2-one (BCO2one6ol) |
||
| YHR216W | 1.73 |
IMD2
|
Inosine monophosphate dehydrogenase, catalyzes the first step of GMP biosynthesis, expression is induced by mycophenolic acid resulting in resistance to the drug, expression is repressed by nutrient limitation |
|
| YFR054C | 1.72 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YKL180W | 1.70 |
RPL17A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl17Bp and has similarity to E. coli L22 and rat L17 ribosomal proteins; copurifies with the Dam1 complex (aka DASH complex) |
|
| YBR084C-A | 1.68 |
RPL19A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl19Bp and has similarity to rat L19 ribosomal protein; rpl19a and rpl19b single null mutations result in slow growth, while the double null mutation is lethal |
|
| YIR021W | 1.67 |
MRS1
|
Protein required for the splicing of two mitochondrial group I introns (BI3 in COB and AI5beta in COX1); forms a splicing complex, containing four subunits of Mrs1p and two subunits of the BI3-encoded maturase, that binds to the BI3 RNA |
|
| YOR047C | 1.67 |
STD1
|
Protein involved in control of glucose-regulated gene expression; interacts with protein kinase Snf1p, glucose sensors Snf3p and Rgt2p, and TATA-binding protein Spt15p; acts as a regulator of the transcription factor Rgt1p |
|
| YPL090C | 1.66 |
RPS6A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps6Bp and has similarity to rat S6 ribosomal protein |
|
| YMR321C | 1.66 |
Putative protein of unknown function |
||
| YOR385W | 1.59 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YOR385W is not an essential gene |
||
| YNL043C | 1.57 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YIP3/YNL044W |
||
| YKL218C | 1.55 |
SRY1
|
3-hydroxyaspartate dehydratase, deaminates L-threo-3-hydroxyaspartate to form oxaloacetate and ammonia; required for survival in the presence of hydroxyaspartate |
|
| YHR094C | 1.51 |
HXT1
|
Low-affinity glucose transporter of the major facilitator superfamily, expression is induced by Hxk2p in the presence of glucose and repressed by Rgt1p when glucose is limiting |
|
| YMR246W | 1.47 |
FAA4
|
Long chain fatty acyl-CoA synthetase, regulates protein modification during growth in the presence of ethanol, functions to incorporate palmitic acid into phospholipids and neutral lipids |
|
| YLR167W | 1.43 |
RPS31
|
Fusion protein that is cleaved to yield a ribosomal protein of the small (40S) subunit and ubiquitin; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes; interacts genetically with translation factor eIF2B |
|
| YDR345C | 1.41 |
HXT3
|
Low affinity glucose transporter of the major facilitator superfamily, expression is induced in low or high glucose conditions |
|
| YDL241W | 1.39 |
Putative protein of unknown function; YDL241W is not an essential gene |
||
| YJL145W | 1.39 |
SFH5
|
Putative phosphatidylinositol transfer protein (PITP), exhibits phosphatidylinositol- but not phosphatidylcholine-transfer activity, localized to cytosol and microsomes, similar to Sec14p; may be PITP regulator rather than actual PITP |
|
| YDL082W | 1.38 |
RPL13A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl13Bp; not essential for viability; has similarity to rat L13 ribosomal protein |
|
| YNL337W | 1.35 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGL030W | 1.35 |
RPL30
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L30 ribosomal protein; involved in pre-rRNA processing in the nucleolus; autoregulates splicing of its transcript |
|
| YEL040W | 1.35 |
UTR2
|
Cell wall protein that functions in the transfer of chitin to beta(1-6)glucan; putative chitin transglycosidase; glycosylphosphatidylinositol (GPI)-anchored protein localized to the bud neck; has a role in cell wall maintenance |
|
| YPL007C | 1.34 |
TFC8
|
One of six subunits of RNA polymerase III transcription initiation factor complex (TFIIIC); part of TFIIIC TauB domain that binds BoxB promoter sites of tRNA and other genes; linker between TauB and TauA domains; human homolog is TFIIIC-90 |
|
| YGR229C | 1.27 |
SMI1
|
Protein involved in the regulation of cell wall synthesis; proposed to be involved in coordinating cell cycle progression with cell wall integrity |
|
| YGR151C | 1.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF RSR1/BUD1/YGR152C |
||
| YMR123W | 1.25 |
PKR1
|
V-ATPase assembly factor, functions with other V-ATPase assembly factors in the ER to efficiently assemble the V-ATPase membrane sector (V0); overproduction confers resistance to Pichia farinosa killer toxin |
|
| YDL083C | 1.25 |
RPS16B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps16Ap and has similarity to E. coli S9 and rat S16 ribosomal proteins |
|
| YBR085W | 1.22 |
AAC3
|
Mitochondrial inner membrane ADP/ATP translocator, exchanges cytosolic ADP for mitochondrially synthesized ATP; expressed under anaerobic conditions; similar to Pet9p and Aac1p; has roles in maintenance of viability and in respiration |
|
| YHL033C | 1.21 |
RPL8A
|
Ribosomal protein L4 of the large (60S) ribosomal subunit, nearly identical to Rpl8Bp and has similarity to rat L7a ribosomal protein; mutation results in decreased amounts of free 60S subunits |
|
| YAL003W | 1.20 |
EFB1
|
Translation elongation factor 1 beta; stimulates nucleotide exchange to regenerate EF-1 alpha-GTP for the next elongation cycle; part of the EF-1 complex, which facilitates binding of aminoacyl-tRNA to the ribosomal A site |
|
| YJL148W | 1.20 |
RPA34
|
RNA polymerase I subunit A34.5 |
|
| YGL179C | 1.19 |
TOS3
|
Protein kinase, related to and functionally redundant with Elm1p and Sak1p for the phosphorylation and activation of Snf1p; functionally orthologous to LKB1, a mammalian kinase associated with Peutz-Jeghers cancer-susceptibility syndrome |
|
| YAL067W-A | 1.18 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YAR018C | 1.15 |
KIN3
|
Nonessential protein kinase with unknown cellular role |
|
| YEL054C | 1.14 |
RPL12A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl12Bp; rpl12a rpl12b double mutant exhibits slow growth and slow translation; has similarity to E. coli L11 and rat L12 ribosomal proteins |
|
| YGL158W | 1.13 |
RCK1
|
Protein kinase involved in the response to oxidative stress; identified as suppressor of S. pombe cell cycle checkpoint mutations |
|
| YNL058C | 1.12 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YNL058C is not an essential gene |
||
| YHR201C | 1.12 |
PPX1
|
Exopolyphosphatase, hydrolyzes inorganic polyphosphate (poly P) into Pi residues; located in the cytosol, plasma membrane, and mitochondrial matrix |
|
| YDR098C | 1.11 |
GRX3
|
Hydroperoxide and superoxide-radical responsive glutathione-dependent oxidoreductase; monothiol glutaredoxin subfamily member along with Grx4p and Grx5p; protects cells from oxidative damage |
|
| YDR033W | 1.09 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YIL169C | 1.08 |
Putative protein of unknown function; serine/threonine rich and highly similar to YOL155C, a putative glucan alpha-1,4-glucosidase; transcript is induced in both high and low pH environments; YIL169C is a non-essential gene |
||
| YLR301W | 1.07 |
Protein of unknown function that interacts with Sec72p |
||
| YJR123W | 1.07 |
RPS5
|
Protein component of the small (40S) ribosomal subunit, the least basic of the non-acidic ribosomal proteins; phosphorylated in vivo; essential for viability; has similarity to E. coli S7 and rat S5 ribosomal proteins |
|
| YGR185C | 1.06 |
TYS1
|
Cytoplasmic tyrosyl-tRNA synthetase, class I aminoacyl-tRNA synthetase that aminoacylates tRNA(Tyr), required for cytoplasmic protein synthesis, interacts with positions 34 and 35 of the anticodon of tRNATyr |
|
| YPL183C | 1.05 |
RTT10
|
Cytoplasmic protein with a role in regulation of Ty1 transposition |
|
| YDL211C | 1.05 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YDR209C | 1.04 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps uncharacterized gene YDR210W. |
||
| YIL009W | 1.04 |
FAA3
|
Long chain fatty acyl-CoA synthetase, has a preference for C16 and C18 fatty acids; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery |
|
| YGL242C | 1.02 |
Putative protein of unknown function; deletion mutant is viable |
||
| YLR154W-C | 1.01 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YDR190C | 1.01 |
RVB1
|
Essential protein involved in transcription regulation; component of chromatin remodeling complexes; required for assembly and function of the INO80 complex; member of the RUVB-like protein family |
|
| YDR071C | 1.01 |
PAA1
|
Polyamine acetyltransferase; acetylates polyamines (e.g. putrescine, spermidine, spermine) and also aralkylamines (e.g. tryptamine, phenylethylamine); may be involved in transcription and/or DNA replication |
|
| YLR409C | 0.99 |
UTP21
|
Possible U3 snoRNP protein involved in maturation of pre-18S rRNA, based on computational analysis of large-scale protein-protein interaction data |
|
| YER056C-A | 0.98 |
RPL34A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl34Bp and has similarity to rat L34 ribosomal protein |
|
| YOR103C | 0.98 |
OST2
|
Epsilon subunit of the oligosaccharyltransferase complex of the ER lumen, which catalyzes asparagine-linked glycosylation of newly synthesized proteins |
|
| YBL023C | 0.97 |
MCM2
|
Protein involved in DNA replication; component of the Mcm2-7 hexameric complex that binds chromatin as a part of the pre-replicative complex |
|
| YOR104W | 0.96 |
PIN2
|
Protein that induces appearance of [PIN+] prion when overproduced |
|
| YKL154W | 0.96 |
SRP102
|
Signal recognition particle (SRP) receptor beta subunit; involved in SRP-dependent protein targeting; anchors Srp101p to the ER membrane |
|
| YFR051C | 0.96 |
RET2
|
Delta subunit of the coatomer complex (COPI), which coats Golgi-derived transport vesicles; involved in retrograde transport between Golgi and ER |
|
| YDR108W | 0.93 |
GSG1
|
Subunit of TRAPP (transport protein particle), a multi-subunit complex involved in targeting and/or fusion of ER-to-Golgi transport vesicles with their acceptor compartment; protein has late meiotic role, following DNA replication |
|
| YBR012C | 0.92 |
Dubious open reading frame, unlikely to encode a functional protein; expression induced by iron-regulated transcriptional activator Aft2p |
||
| YOR073W-A | 0.91 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially/completely overlaps the verified ORF CDC21/YOR074C; identified by RT-PCR |
||
| YNL079C | 0.91 |
TPM1
|
Major isoform of tropomyosin; binds to and stabilizes actin cables and filaments, which direct polarized cell growth and the distribution of several organelles; acetylated by the NatB complex and acetylated form binds actin most efficiently |
|
| YDR326C | 0.90 |
YSP2
|
Protein involved in programmed cell death; mutant shows resistance to cell death induced by amiodarone or intracellular acidification |
|
| YJL105W | 0.90 |
SET4
|
Protein of unknown function, contains a SET domain |
|
| YER131W | 0.87 |
RPS26B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps26Ap and has similarity to rat S26 ribosomal protein |
|
| YER135C | 0.86 |
Dubious open reading frame unlikely to encode a protein; YER135C is not an essential gene |
||
| YDR144C | 0.85 |
MKC7
|
GPI-anchored aspartyl protease (yapsin) involved in protein processing; shares functions with Yap3p and Kex2p |
|
| YBL108W | 0.81 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YNL078W | 0.81 |
NIS1
|
Protein localized in the bud neck at G2/M phase; physically interacts with septins; possibly involved in a mitotic signaling network |
|
| YNL042W-B | 0.80 |
Putative protein of unknown function |
||
| YER001W | 0.80 |
MNN1
|
Alpha-1,3-mannosyltransferase, integral membrane glycoprotein of the Golgi complex, required for addition of alpha1,3-mannose linkages to N-linked and O-linked oligosaccharides, one of five S. cerevisiae proteins of the MNN1 family |
|
| YKL073W | 0.79 |
LHS1
|
Molecular chaperone of the endoplasmic reticulum lumen, involved in polypeptide translocation and folding; member of the Hsp70 family; localizes to the lumen of the ER; regulated by the unfolded protein response pathway |
|
| YLR335W | 0.78 |
NUP2
|
Nucleoporin involved in nucleocytoplasmic transport, binds to either the nucleoplasmic or cytoplasmic faces of the nuclear pore complex depending on Ran-GTP levels; also has a role in chromatin organization |
|
| YLR154W-B | 0.78 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YKL223W | 0.76 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR089C | 0.76 |
GAR1
|
Protein component of the H/ACA snoRNP pseudouridylase complex, involved in the modification and cleavage of the 18S pre-rRNA |
|
| YGL123C-A | 0.75 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene RPS2/YGL123W |
||
| YDR021W | 0.75 |
FAL1
|
Nucleolar protein required for maturation of 18S rRNA, member of the eIF4A subfamily of DEAD-box ATP-dependent RNA helicases |
|
| YMR049C | 0.75 |
ERB1
|
Protein required for maturation of the 25S and 5.8S ribosomal RNAs; constituent of 66S pre-ribosomal particles; homologous to mammalian Bop1 |
|
| YLR274W | 0.74 |
MCM5
|
Component of the hexameric MCM complex, which is important for priming origins of DNA replication in G1 and becomes an active ATP-dependent helicase that promotes DNA melting and elongation when activated by Cdc7p-Dbf4p in S-phase |
|
| YEL034C-A | 0.74 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; completely overlaps HYP2/YEL034W, a verified gene that encodes eiF-5A |
||
| YNL069C | 0.73 |
RPL16B
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, binds to 5.8 S rRNA; has similarity to Rpl16Ap, E. coli L13 and rat L13a ribosomal proteins; transcriptionally regulated by Rap1p |
|
| YIL009C-A | 0.73 |
EST3
|
Component of the telomerase holoenzyme, involved in telomere replication |
|
| YGL085W | 0.73 |
Putative protein of unknown function, has homology to Staphylococcus aureus nuclease; green fluorescent protein (GFP)-fusion protein localizes to mitochondria and is induced in response to the DNA-damaging agent MMS |
||
| YGL123W | 0.72 |
RPS2
|
Protein component of the small (40S) subunit, essential for control of translational accuracy; has similarity to E. coli S5 and rat S2 ribosomal proteins |
|
| YEL029C | 0.72 |
BUD16
|
Putative pyridoxal kinase, key enzyme in vitamin B6 metabolism; involved in maintaining levels of pyridoxal 5'-phosphate, the active form of vitamin B6; required for genome integrity; homolog of E. coli PdxK; involved in bud-site selection |
|
| YKL153W | 0.71 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; transcription of both YLK153W and the overlapping essential gene GPM1 is reduced in the gcr1 null mutant |
||
| YDR415C | 0.71 |
Putative protein of unknown function |
||
| YNL057W | 0.71 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YBR011C | 0.69 |
IPP1
|
Cytoplasmic inorganic pyrophosphatase (PPase), catalyzes the rapid exchange of oxygens from Pi with water, highly expressed and essential for viability, active-site residues show identity to those from E. coli PPase |
|
| YGL157W | 0.69 |
Oxidoreductase, catalyzes NADPH-dependent reduction of the bicyclic diketone bicyclo[2.2.2]octane-2,6-dione (BCO2,6D) to the chiral ketoalcohol (1R,4S,6S)-6-hydroxybicyclo[2.2.2]octane-2-one (BCO2one6ol) |
||
| YBR248C | 0.69 |
HIS7
|
Imidazole glycerol phosphate synthase (glutamine amidotransferase:cyclase), catalyzes the fifth and sixth steps of histidine biosynthesis and also produces 5-aminoimidazole-4-carboxamide ribotide (AICAR), a purine precursor |
|
| YOR074C | 0.69 |
CDC21
|
Thymidylate synthase, required for de novo biosynthesis of pyrimidine deoxyribonucleotides; expression is induced at G1/S |
|
| YCR099C | 0.68 |
Putative protein of unknown function |
||
| YGR255C | 0.68 |
COQ6
|
Putative flavin-dependent monooxygenase, involved in ubiquinone (Coenzyme Q) biosynthesis; localizes to the matrix face of the mitochondrial inner membrane in a large complex with other ubiquinone biosynthetic enzymes |
|
| YMR108W | 0.68 |
ILV2
|
Acetolactate synthase, catalyses the first common step in isoleucine and valine biosynthesis and is the target of several classes of inhibitors, localizes to the mitochondria; expression of the gene is under general amino acid control |
|
| YER177W | 0.67 |
BMH1
|
14-3-3 protein, major isoform; controls proteome at post-transcriptional level, binds proteins and DNA, involved in regulation of many processes including exocytosis, vesicle transport, Ras/MAPK signaling, and rapamycin-sensitive signaling |
|
| YGR092W | 0.66 |
DBF2
|
Ser/Thr kinase involved in transcription and stress response; functions as part of a network of genes in exit from mitosis; localization is cell cycle regulated; activated by Cdc15p during the exit from mitosis |
|
| YLL024C | 0.65 |
SSA2
|
ATP binding protein involved in protein folding and vacuolar import of proteins; member of heat shock protein 70 (HSP70) family; associated with the chaperonin-containing T-complex; present in the cytoplasm, vacuolar membrane and cell wall |
|
| YPR010C | 0.65 |
RPA135
|
RNA polymerase I subunit A135 |
|
| YKL074C | 0.65 |
MUD2
|
Protein involved in early pre-mRNA splicing; component of the pre-mRNA-U1 snRNP complex, the commitment complex; interacts with Msl5p/BBP splicing factor and Sub2p; similar to metazoan splicing factor U2AF65 |
|
| YER134C | 0.65 |
Putative protein of unknown function; non-essential gene |
||
| YOR319W | 0.65 |
HSH49
|
U2-snRNP associated splicing factor with similarity to the mammalian splicing factor SAP49; proposed to function as a U2-snRNP assembly factor along with Hsh155p and binding partner Cus1p; contains two RNA recognition motifs (RRM) |
|
| YDR399W | 0.64 |
HPT1
|
Dimeric hypoxanthine-guanine phosphoribosyltransferase, catalyzes the formation of both inosine monophosphate and guanosine monophosphate; mutations in the human homolog HPRT1 can cause Lesch-Nyhan syndrome and Kelley-Seegmiller syndrome |
|
| YGL040C | 0.64 |
HEM2
|
Delta-aminolevulinate dehydratase, a homo-octameric enzyme, catalyzes the conversion of delta-aminolevulinic acid to porphobilinogen, the second step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus |
|
| YJL107C | 0.64 |
Putative protein of unknown function; expression is induced by activation of the HOG1 mitogen-activated signaling pathway and this induction is Hog1p/Pbs2p dependent; YJL107C and adjacent ORF, YJL108C are merged in related fungi |
||
| YJL126W | 0.64 |
NIT2
|
Nit protein, one of two proteins in S. cerevisiae with similarity to the Nit domain of NitFhit from fly and worm and to the mouse and human Nit protein which interacts with the Fhit tumor suppressor; nitrilase superfamily member |
|
| YKL128C | 0.64 |
PMU1
|
Putative phosphomutase, contains a region homologous to the active site of phosphomutases; overexpression suppresses the histidine auxotrophy of an ade3 ade16 ade17 triple mutant and the temperature sensitivity of a tps2 mutant |
|
| YDR445C | 0.63 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJR097W | 0.63 |
JJJ3
|
Protein of unknown function, contains a J-domain, which is a region with homology to the E. coli DnaJ protein |
|
| YJL169W | 0.63 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YJL168C/SET2 |
||
| YHR144C | 0.62 |
DCD1
|
Deoxycytidine monophosphate (dCMP) deaminase required for dCTP and dTTP synthesis; expression is NOT cell cycle regulated |
|
| YGL241W | 0.61 |
KAP114
|
Karyopherin, responsible for nuclear import of Spt15p, histones H2A and H2B, and Nap1p; amino terminus shows similarity to those of other importins, particularly Cse1p; localization is primarily nuclear |
|
| YNL016W | 0.61 |
PUB1
|
Poly(A)+ RNA-binding protein, abundant mRNP-component protein that binds mRNA and is required for stability of a number of mRNAs; not reported to associate with polyribosomes |
|
| YKL224C | 0.60 |
PAU16
|
Putative protein of unknown function |
|
| YLR196W | 0.60 |
PWP1
|
Protein with WD-40 repeats involved in rRNA processing; associates with trans-acting ribosome biogenesis factors; similar to beta-transducin superfamily |
|
| YGR139W | 0.59 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR082C | 0.59 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPL141C | 0.59 |
Putative protein kinase; similar to Kin4p; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YPL141C is not an essential gene |
||
| YMR106C | 0.59 |
YKU80
|
Subunit of the telomeric Ku complex (Yku70p-Yku80p), involved in telomere length maintenance, structure and telomere position effect; relocates to sites of double-strand cleavage to promote nonhomologous end joining during DSB repair |
|
| YOR004W | 0.59 |
UTP23
|
Essential nucleolar protein that is a component of the SSU (small subunit) processome involved in 40S ribosomal subunit biogenesis; has homology to PINc domain protein Fcf1p, although the PINc domain of Utp23p is not required for function |
|
| YGR040W | 0.58 |
KSS1
|
Mitogen-activated protein kinase (MAPK) involved in signal transduction pathways that control filamentous growth and pheromone response; the KSS1 gene is nonfunctional in S288C strains and functional in W303 strains |
|
| YHR018C | 0.58 |
ARG4
|
Argininosuccinate lyase, catalyzes the final step in the arginine biosynthesis pathway |
|
| YNL189W | 0.57 |
SRP1
|
Karyopherin alpha homolog, forms a dimer with karyopherin beta Kap95p to mediate import of nuclear proteins, binds the nuclear localization signal of the substrate during import; may also play a role in regulation of protein degradation |
|
| YBL097W | 0.57 |
BRN1
|
Essential protein required for chromosome condensation, likely to function as an intrinsic component of the condensation machinery, may influence multiple aspects of chromosome transmission and dynamics |
|
| YLR154W-A | 0.57 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YGR208W | 0.56 |
SER2
|
Phosphoserine phosphatase of the phosphoglycerate pathway, involved in serine and glycine biosynthesis, expression is regulated by the available nitrogen source |
|
| YGR138C | 0.56 |
TPO2
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; transcription of TPO2 is regulated by Haa1p; member of the major facilitator superfamily |
|
| YDR450W | 0.56 |
RPS18A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps18Bp and has similarity to E. coli S13 and rat S18 ribosomal proteins |
|
| YOR180C | 0.56 |
DCI1
|
Peroxisomal delta(3,5)-delta(2,4)-dienoyl-CoA isomerase, involved in fatty acid metabolism, contains peroxisome targeting signals at amino and carboxy termini |
|
| YDR037W | 0.55 |
KRS1
|
Lysyl-tRNA synthetase; also identified as a negative regulator of general control of amino acid biosynthesis |
|
| YJR071W | 0.55 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHR142W | 0.55 |
CHS7
|
Protein of unknown function, involved in chitin biosynthesis by regulating Chs3p export from the ER |
|
| YER136W | 0.55 |
GDI1
|
GDP dissociation inhibitor, regulates vesicle traffic in secretory pathways by regulating the dissociation of GDP from the Sec4/Ypt/rab family of GTP binding proteins |
|
| YCR090C | 0.55 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YCR090C is not an essential gene |
||
| YLR013W | 0.54 |
GAT3
|
Protein containing GATA family zinc finger motifs |
|
| YHR136C | 0.54 |
SPL2
|
Protein with similarity to cyclin-dependent kinase inhibitors, overproduction suppresses a plc1 null mutation; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YOR181W | 0.53 |
LAS17
|
Actin assembly factor, activates the Arp2/3 protein complex that nucleates branched actin filaments; localizes with the Arp2/3 complex to actin patches; homolog of the human Wiskott-Aldrich syndrome protein (WASP) |
|
| YCR098C | 0.53 |
GIT1
|
Plasma membrane permease, mediates uptake of glycerophosphoinositol and glycerophosphocholine as sources of the nutrients inositol and phosphate; expression and transport rate are regulated by phosphate and inositol availability |
|
| YER049W | 0.53 |
TPA1
|
Protein of unknown function; interacts with Sup45p (eRF1), Sup35p (eRF3) and Pab1p; has a role in translation termination efficiency, mRNA poly(A) tail length and mRNA stability |
|
| YPR060C | 0.53 |
ARO7
|
Chorismate mutase, catalyzes the conversion of chorismate to prephenate to initiate the tyrosine/phenylalanine-specific branch of aromatic amino acid biosynthesis |
|
| YPL256C | 0.53 |
CLN2
|
G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) |
|
| YML121W | 0.53 |
GTR1
|
Cytoplasmic GTP binding protein and negative regulator of the Ran/Tc4 GTPase cycle; component of GSE complex, which is required for sorting of Gap1p; involved in phosphate transport and telomeric silencing; similar to human RagA and RagB |
|
| YJR070C | 0.52 |
LIA1
|
Deoxyhypusine hydroxylase, a HEAT-repeat containing metalloenzyme that catalyses hypusine formation; binds to and is required for the modification of Hyp2p (eIF5A); complements S. pombe mmd1 mutants defective in mitochondrial positioning |
|
| YLL045C | 0.52 |
RPL8B
|
Ribosomal protein L4 of the large (60S) ribosomal subunit, nearly identical to Rpl8Ap and has similarity to rat L7a ribosomal protein; mutation results in decreased amounts of free 60S subunits |
|
| YPL239W | 0.52 |
YAR1
|
Cytoplasmic ankyrin-repeat containing protein of unknown function, proposed to link the processes of 40S ribosomal subunit biogenesis and adaptation to osmotic and oxidative stress; expression repressed by heat shock |
|
| YOR058C | 0.51 |
ASE1
|
Mitotic spindle midzone localized microtubule-associated protein (MAP) family member; required for spindle elongation and stabilization; undergoes cell cycle-regulated degradation by anaphase promoting complex; potential Cdc28p substrate |
|
| YMR119W | 0.50 |
ASI1
|
Putative integral membrane E3 ubiquitin ligase; acts with Asi2p and Asi3p to ensure the fidelity of SPS-sensor signalling by maintaining the dormant repressed state of gene expression in the absence of inducing signals |
|
| YDR416W | 0.50 |
SYF1
|
Component of the spliceosome complex involved in pre-mRNA splicing; involved in regulation of cell cycle progression; similar to Drosophila crooked neck protein |
|
| YGR156W | 0.50 |
PTI1
|
Pta1p Interacting protein |
|
| YOR356W | 0.50 |
Mitochondrial protein with similarity to flavoprotein-type oxidoreductases; found in a large supramolecular complex with other mitochondrial dehydrogenases |
||
| YEL027W | 0.50 |
CUP5
|
Proteolipid subunit of the vacuolar H(+)-ATPase V0 sector (subunit c; dicyclohexylcarbodiimide binding subunit); required for vacuolar acidification and important for copper and iron metal ion homeostasis |
|
| YDR060W | 0.49 |
MAK21
|
Constituent of 66S pre-ribosomal particles, required for large (60S) ribosomal subunit biogenesis; involved in nuclear export of pre-ribosomes; required for maintenance of dsRNA virus; homolog of human CAATT-binding protein |
|
| YGR107W | 0.49 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR133C | 0.48 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR134C |
||
| YBR104W | 0.48 |
YMC2
|
Mitochondrial protein, putative inner membrane transporter with a role in oleate metabolism and glutamate biosynthesis; member of the mitochondrial carrier (MCF) family; has similarity with Ymc1p |
|
| YGR140W | 0.48 |
CBF2
|
Essential kinetochore protein, component of the CBF3 multisubunit complex that binds to the CDEIII region of the centromere; Cbf2p also binds to the CDEII region possibly forming a different multimeric complex, ubiquitinated in vivo |
|
| YDR153C | 0.48 |
ENT5
|
Protein containing an N-terminal epsin-like domain involved in clathrin recruitment and traffic between the Golgi and endosomes; associates with the clathrin adaptor Gga2p, clathrin adaptor complex AP-1, and clathrin |
|
| YKL205W | 0.48 |
LOS1
|
Nuclear pore protein involved in nuclear export of pre-tRNA |
|
| YLR257W | 0.48 |
Putative protein of unknown function |
||
| YBR089W | 0.47 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; almost completely overlaps the verified gene POL30 |
||
| YGL054C | 0.47 |
ERV14
|
Protein localized to COPII-coated vesicles, involved in vesicle formation and incorporation of specific secretory cargo; required for the delivery of bud-site selection protein Axl2p to cell surface; related to Drosophila cornichon |
|
| YBR181C | 0.47 |
RPS6B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps6Ap and has similarity to rat S6 ribosomal protein |
|
| YJL208C | 0.47 |
NUC1
|
Major mitochondrial nuclease, has RNAse and DNA endo- and exonucleolytic activities; has roles in mitochondrial recombination, apoptosis and maintenance of polyploidy |
|
| YLR256W | 0.47 |
HAP1
|
Zinc finger transcription factor involved in the complex regulation of gene expression in response to levels of heme and oxygen; the S288C sequence differs from other strain backgrounds due to a Ty1 insertion in the carboxy terminus |
|
| YOR087W | 0.47 |
YVC1
|
Vacuolar cation channel, mediates release of Ca(2+) from the vacuole in response to hyperosmotic shock |
|
| YPR156C | 0.46 |
TPO3
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; member of the major facilitator superfamily |
|
| YIL117C | 0.46 |
PRM5
|
Pheromone-regulated protein, predicted to have 1 transmembrane segment; induced during cell integrity signaling |
|
| YPL112C | 0.46 |
PEX25
|
Peripheral peroxisomal membrane peroxin required for the regulation of peroxisome size and maintenance, recruits GTPase Rho1p to peroxisomes, induced by oleate, interacts with homologous protein Pex27p |
Network of associatons between targets according to the STRING database.
First level regulatory network of YOX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.9 | 6.3 | GO:0019655 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.9 | 6.2 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 0.7 | 3.0 | GO:0014070 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 0.7 | 4.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.6 | 1.7 | GO:0006183 | GTP biosynthetic process(GO:0006183) GTP metabolic process(GO:0046039) |
| 0.6 | 1.7 | GO:0070637 | nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) |
| 0.5 | 2.7 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.5 | 1.5 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.4 | 1.3 | GO:0048024 | regulation of RNA splicing(GO:0043484) regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.3 | 0.9 | GO:0006798 | polyphosphate catabolic process(GO:0006798) |
| 0.3 | 1.3 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 1.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.3 | 3.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.3 | 44.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.3 | 1.2 | GO:0000296 | spermine transport(GO:0000296) |
| 0.3 | 1.4 | GO:0017157 | regulation of exocytosis(GO:0017157) regulation of secretion(GO:0051046) regulation of secretion by cell(GO:1903530) |
| 0.3 | 0.8 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.3 | 2.0 | GO:0015791 | polyol transport(GO:0015791) |
| 0.2 | 0.7 | GO:0006343 | establishment of chromatin silencing(GO:0006343) |
| 0.2 | 0.7 | GO:0006407 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.2 | 1.0 | GO:0048313 | Golgi inheritance(GO:0048313) |
| 0.2 | 0.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.2 | 1.5 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.2 | 1.9 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.2 | 1.7 | GO:0000372 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.2 | 0.6 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.2 | 0.6 | GO:0000348 | mRNA branch site recognition(GO:0000348) |
| 0.2 | 1.2 | GO:0001009 | transcription from RNA polymerase III type 2 promoter(GO:0001009) transcription from a RNA polymerase III hybrid type promoter(GO:0001041) |
| 0.2 | 0.9 | GO:0007119 | budding cell isotropic bud growth(GO:0007119) |
| 0.2 | 2.6 | GO:0010383 | cell wall polysaccharide metabolic process(GO:0010383) |
| 0.2 | 0.5 | GO:1902223 | tyrosine biosynthetic process(GO:0006571) L-phenylalanine biosynthetic process(GO:0009094) erythrose 4-phosphate/phosphoenolpyruvate family amino acid biosynthetic process(GO:1902223) |
| 0.2 | 2.7 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.2 | 0.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.2 | 0.9 | GO:0007535 | donor selection(GO:0007535) |
| 0.2 | 0.7 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 0.8 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) maintenance of chromatin silencing at telomere(GO:0035392) |
| 0.2 | 0.3 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.2 | 0.2 | GO:0001015 | snoRNA transcription from an RNA polymerase II promoter(GO:0001015) |
| 0.2 | 1.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.6 | GO:0032231 | regulation of actin filament bundle assembly(GO:0032231) positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.3 | GO:1903313 | positive regulation of mRNA metabolic process(GO:1903313) |
| 0.1 | 2.2 | GO:0001100 | negative regulation of exit from mitosis(GO:0001100) |
| 0.1 | 1.4 | GO:0052803 | histidine biosynthetic process(GO:0000105) histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.1 | 0.6 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 0.1 | 0.3 | GO:0071825 | lipid tube assembly(GO:0060988) protein-lipid complex assembly(GO:0065005) protein-lipid complex subunit organization(GO:0071825) |
| 0.1 | 0.4 | GO:0019379 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.1 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.5 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 0.8 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.3 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 1.3 | GO:1903338 | regulation of cell wall organization or biogenesis(GO:1903338) |
| 0.1 | 0.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.5 | GO:0043097 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.1 | 0.4 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.4 | GO:0002252 | immune effector process(GO:0002252) immune system process(GO:0002376) response to virus(GO:0009615) response to external biotic stimulus(GO:0043207) defense response to virus(GO:0051607) response to other organism(GO:0051707) defense response to other organism(GO:0098542) |
| 0.1 | 0.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.3 | GO:0000917 | barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.1 | 0.6 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.1 | 0.5 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 0.2 | GO:0031384 | regulation of initiation of mating projection growth(GO:0031384) |
| 0.1 | 0.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 1.0 | GO:0042991 | transcription factor import into nucleus(GO:0042991) |
| 0.1 | 0.7 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.1 | 0.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.1 | GO:0097271 | protein localization to bud neck(GO:0097271) |
| 0.1 | 0.4 | GO:0060188 | regulation of protein desumoylation(GO:0060188) |
| 0.1 | 0.8 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) snRNA modification(GO:0040031) |
| 0.1 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.9 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 3.3 | GO:0051445 | regulation of meiotic nuclear division(GO:0040020) regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.9 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.1 | 0.2 | GO:0010452 | regulation of histone H3-K36 methylation(GO:0000414) histone H3-K36 methylation(GO:0010452) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.3 | GO:0060277 | obsolete negative regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0060277) |
| 0.1 | 0.7 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.1 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 0.2 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) |
| 0.1 | 0.4 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 0.2 | GO:0043419 | urea metabolic process(GO:0019627) urea catabolic process(GO:0043419) |
| 0.1 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of metal ion(GO:0051238) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.3 | GO:0009097 | isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.7 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.1 | 0.3 | GO:0034755 | intracellular copper ion transport(GO:0015680) iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.2 | GO:2000877 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 0.1 | 0.8 | GO:0042181 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) ketone biosynthetic process(GO:0042181) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.5 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 1.9 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.1 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 0.1 | GO:0009847 | spore germination(GO:0009847) |
| 0.1 | 1.4 | GO:0012501 | apoptotic process(GO:0006915) cell death(GO:0008219) programmed cell death(GO:0012501) |
| 0.1 | 0.3 | GO:0090158 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.8 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 1.6 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 0.1 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.1 | 0.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.5 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.1 | 0.2 | GO:0010674 | negative regulation of transcription from RNA polymerase II promoter involved in meiotic cell cycle(GO:0010674) |
| 0.1 | 0.5 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 0.3 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.2 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.3 | GO:0048017 | inositol lipid-mediated signaling(GO:0048017) |
| 0.1 | 0.5 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 0.9 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 0.9 | GO:0043094 | cellular metabolic compound salvage(GO:0043094) |
| 0.1 | 1.2 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.1 | 0.6 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.2 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 1.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.5 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) regulation of vacuole organization(GO:0044088) |
| 0.0 | 0.9 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 1.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 2.4 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.2 | GO:0051320 | mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.0 | 1.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:0036170 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.0 | 0.2 | GO:0034503 | protein localization to nucleolar rDNA repeats(GO:0034503) rDNA condensation(GO:0070550) |
| 0.0 | 0.4 | GO:0033673 | negative regulation of protein kinase activity(GO:0006469) negative regulation of kinase activity(GO:0033673) |
| 0.0 | 0.1 | GO:0000751 | mitotic cell cycle arrest in response to pheromone(GO:0000751) mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.3 | GO:0009263 | deoxyribonucleotide metabolic process(GO:0009262) deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.7 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.2 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.3 | GO:0090522 | vesicle tethering involved in exocytosis(GO:0090522) |
| 0.0 | 0.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0000743 | nuclear migration involved in conjugation with cellular fusion(GO:0000743) |
| 0.0 | 0.2 | GO:0006768 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.0 | 0.1 | GO:0030811 | regulation of glycolytic process(GO:0006110) regulation of nucleoside metabolic process(GO:0009118) regulation of nucleotide catabolic process(GO:0030811) regulation of ATP metabolic process(GO:1903578) |
| 0.0 | 0.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) |
| 0.0 | 0.3 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.3 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) copper ion homeostasis(GO:0055070) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0042173 | regulation of sporulation resulting in formation of a cellular spore(GO:0042173) regulation of sporulation(GO:0043937) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) regulation of phosphatidylglycerol biosynthetic process(GO:1901351) negative regulation of phosphatidylglycerol biosynthetic process(GO:1901352) |
| 0.0 | 0.2 | GO:0010526 | negative regulation of transposition, RNA-mediated(GO:0010526) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.2 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 0.2 | GO:0030473 | establishment of localization by movement along microtubule(GO:0010970) nuclear migration along microtubule(GO:0030473) organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.6 | GO:0051170 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) nuclear import(GO:0051170) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.9 | GO:0007088 | regulation of mitotic nuclear division(GO:0007088) |
| 0.0 | 0.0 | GO:0051093 | negative regulation of developmental process(GO:0051093) |
| 0.0 | 0.2 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.1 | GO:0001308 | progressive alteration of chromatin involved in cell aging(GO:0001301) progressive alteration of chromatin involved in replicative cell aging(GO:0001304) negative regulation of chromatin silencing involved in replicative cell aging(GO:0001308) heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.2 | GO:0044396 | actin cortical patch assembly(GO:0000147) actin cortical patch organization(GO:0044396) |
| 0.0 | 0.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.7 | GO:0051726 | regulation of cell cycle(GO:0051726) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0006797 | polyphosphate metabolic process(GO:0006797) |
| 0.0 | 0.0 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.3 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 1.0 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.0 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.3 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.3 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.4 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.3 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.1 | GO:0043489 | RNA stabilization(GO:0043489) mRNA stabilization(GO:0048255) |
| 0.0 | 0.1 | GO:0032845 | negative regulation of telomere maintenance(GO:0032205) negative regulation of homeostatic process(GO:0032845) |
| 0.0 | 0.1 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.0 | GO:0046021 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) |
| 0.0 | 0.2 | GO:0048311 | mitochondrion inheritance(GO:0000001) mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 1.8 | GO:0044697 | HICS complex(GO:0044697) |
| 0.4 | 31.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 17.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 1.0 | GO:0070209 | ASTRA complex(GO:0070209) R2TP complex(GO:0097255) |
| 0.3 | 1.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 1.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 3.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 1.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 0.8 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 2.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.7 | GO:0000142 | cellular bud neck contractile ring(GO:0000142) |
| 0.2 | 1.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 1.4 | GO:0000144 | cellular bud neck septin ring(GO:0000144) |
| 0.2 | 0.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 1.7 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 3.3 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 1.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.6 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.1 | 2.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.6 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.9 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.1 | 0.4 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) Sec62/Sec63 complex(GO:0031207) |
| 0.1 | 0.5 | GO:0034448 | EGO complex(GO:0034448) |
| 0.1 | 0.5 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.7 | GO:0031933 | nuclear heterochromatin(GO:0005720) nuclear telomeric heterochromatin(GO:0005724) telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.4 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.5 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.5 | GO:0008278 | nuclear cohesin complex(GO:0000798) cohesin complex(GO:0008278) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.6 | GO:0030867 | rough endoplasmic reticulum(GO:0005791) rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.2 | GO:0045298 | polar microtubule(GO:0005827) tubulin complex(GO:0045298) |
| 0.1 | 0.3 | GO:0071819 | DUBm complex(GO:0071819) |
| 0.1 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 4.6 | GO:0005816 | microtubule organizing center(GO:0005815) spindle pole body(GO:0005816) |
| 0.1 | 0.1 | GO:0031391 | Elg1 RFC-like complex(GO:0031391) |
| 0.1 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.3 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.1 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.2 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.1 | 0.5 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 2.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.0 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.3 | GO:0005940 | septin ring(GO:0005940) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0034456 | UTP-C complex(GO:0034456) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 2.0 | GO:0000131 | incipient cellular bud site(GO:0000131) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 3.6 | GO:0005933 | cellular bud(GO:0005933) |
| 0.0 | 1.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0070772 | PAS complex(GO:0070772) |
| 0.0 | 0.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0000500 | RNA polymerase I upstream activating factor complex(GO:0000500) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.7 | GO:0043332 | mating projection tip(GO:0043332) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0000799 | condensin complex(GO:0000796) nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) ERMES complex(GO:0032865) ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0034044 | exomer complex(GO:0034044) |
| 0.0 | 0.2 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.1 | 6.5 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 1.0 | 5.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.8 | 2.3 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.6 | 2.5 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.6 | 1.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 2.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.4 | 1.6 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.3 | 1.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 1.5 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 0.6 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.3 | 1.2 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.3 | 6.6 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.3 | 0.8 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.3 | 1.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 48.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 1.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.2 | 1.2 | GO:0001003 | RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) transcription factor activity, RNA polymerase III promoter sequence-specific binding, TFIIIB recruiting(GO:0001004) transcription factor activity, RNA polymerase III type 1 promoter sequence-specific binding, TFIIIB recruiting(GO:0001005) transcription factor activity, RNA polymerase III type 2 promoter sequence-specific binding, TFIIIB recruiting(GO:0001008) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.2 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 2.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 0.9 | GO:0043141 | 5'-3' DNA helicase activity(GO:0043139) ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 0.9 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 1.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 0.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.0 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.1 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 1.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.7 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.1 | GO:0015645 | fatty acid ligase activity(GO:0015645) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.6 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.2 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 0.5 | GO:0004338 | glucan exo-1,3-beta-glucosidase activity(GO:0004338) |
| 0.1 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.7 | GO:0043177 | carboxylic acid binding(GO:0031406) organic acid binding(GO:0043177) |
| 0.1 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.3 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.5 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 3.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 1.4 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.3 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.1 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.7 | GO:0004529 | exodeoxyribonuclease activity(GO:0004529) |
| 0.1 | 0.4 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.3 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 1.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.2 | GO:0042973 | glucan endo-1,3-beta-D-glucosidase activity(GO:0042973) |
| 0.1 | 0.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.3 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 0.2 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.1 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.1 | 0.4 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.1 | 3.5 | GO:0008092 | cytoskeletal protein binding(GO:0008092) |
| 0.1 | 0.6 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.7 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.5 | GO:0004526 | ribonuclease MRP activity(GO:0000171) ribonuclease P activity(GO:0004526) |
| 0.1 | 0.2 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.1 | 1.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.1 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.0 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.1 | GO:0001082 | transcription factor activity, RNA polymerase I transcription factor binding(GO:0001082) |
| 0.0 | 0.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.2 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 1.5 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.4 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.6 | GO:0015605 | organophosphate ester transmembrane transporter activity(GO:0015605) |
| 0.0 | 0.2 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.2 | GO:0004026 | alcohol O-acetyltransferase activity(GO:0004026) alcohol O-acyltransferase activity(GO:0034318) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.0 | 0.7 | GO:0004536 | endodeoxyribonuclease activity(GO:0004520) deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.3 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.7 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0001168 | RNA polymerase I transcription factor activity, sequence-specific DNA binding(GO:0001167) transcription factor activity, RNA polymerase I upstream control element sequence-specific binding(GO:0001168) |
| 0.0 | 0.8 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.5 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.3 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0016410 | N-acyltransferase activity(GO:0016410) |
| 0.0 | 1.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.3 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0042736 | NAD+ kinase activity(GO:0003951) NADH kinase activity(GO:0042736) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.4 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 0.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 0.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.0 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 1.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 0.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 2.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 0.2 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 0.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.1 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.4 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.0 | 0.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME M G1 TRANSITION | Genes involved in M/G1 Transition |
| 0.0 | 0.1 | REACTOME SIGNALLING TO ERKS | Genes involved in Signalling to ERKs |
| 0.0 | 0.1 | REACTOME APOPTOSIS | Genes involved in Apoptosis |
| 0.0 | 0.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.1 | REACTOME DEADENYLATION DEPENDENT MRNA DECAY | Genes involved in Deadenylation-dependent mRNA decay |
| 0.0 | 0.1 | REACTOME TRANSCRIPTION | Genes involved in Transcription |