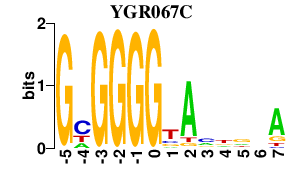
Results for YGR067C
Z-value: 0.87
Transcription factors associated with YGR067C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
|
S000003299 | Putative protein of unknown function |
Activity-expression correlation:
Activity profile of YGR067C motif
Sorted Z-values of YGR067C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YGR067C | 11.66 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YOR348C | 9.72 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YOR100C | 9.27 |
CRC1
|
Mitochondrial inner membrane carnitine transporter, required for carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria during fatty acid beta-oxidation |
|
| YDR536W | 6.45 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YHR217C | 5.82 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; located in the telomeric region TEL08R. |
||
| YDR342C | 5.77 |
HXT7
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels |
|
| YLR312C | 5.73 |
QNQ1
|
Protein of unknown function; null mutant quiescent and non-quiescent cells exhibit reduced reproductive capacity |
|
| YKL163W | 5.24 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YMR107W | 5.12 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YFL052W | 4.99 |
Putative zinc cluster protein that contains a DNA binding domain; null mutant sensitive to calcofluor white, low osmolarity and heat, suggesting a role for YFL052Wp in cell wall integrity |
||
| YIL057C | 4.66 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YDR096W | 4.64 |
GIS1
|
JmjC domain-containing histone demethylase; transcription factor involved in the expression of genes during nutrient limitation; also involved in the negative regulation of DPP1 and PHR1 |
|
| YAR053W | 4.61 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPL230W | 4.38 |
USV1
|
Putative transcription factor containing a C2H2 zinc finger; mutation affects transcriptional regulation of genes involved in protein folding, ATP binding, and cell wall biosynthesis |
|
| YFR053C | 4.38 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YOL052C-A | 4.26 |
DDR2
|
Multistress response protein, expression is activated by a variety of xenobiotic agents and environmental or physiological stresses |
|
| YDR277C | 4.23 |
MTH1
|
Negative regulator of the glucose-sensing signal transduction pathway, required for repression of transcription by Rgt1p; interacts with Rgt1p and the Snf3p and Rgt2p glucose sensors; phosphorylated by Yck1p, triggering Mth1p degradation |
|
| YNR073C | 4.16 |
Putative mannitol dehydrogenase |
||
| YGR243W | 4.04 |
FMP43
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YOR178C | 3.97 |
GAC1
|
Regulatory subunit for Glc7p type-1 protein phosphatase (PP1), tethers Glc7p to Gsy2p glycogen synthase, binds Hsf1p heat shock transcription factor, required for induction of some HSF-regulated genes under heat shock |
|
| YML042W | 3.94 |
CAT2
|
Carnitine acetyl-CoA transferase present in both mitochondria and peroxisomes, transfers activated acetyl groups to carnitine to form acetylcarnitine which can be shuttled across membranes |
|
| YLR023C | 3.91 |
IZH3
|
Membrane protein involved in zinc metabolism, member of the four-protein IZH family, expression induced by zinc deficiency; deletion reduces sensitivity to elevated zinc and shortens lag phase, overexpression reduces Zap1p activity |
|
| YNL036W | 3.86 |
NCE103
|
Carbonic anhydrase; poorly transcribed under aerobic conditions and at an undetectable level under anaerobic conditions; involved in non-classical protein export pathway |
|
| YML089C | 3.83 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression induced by calcium shortage |
||
| YMR194C-B | 3.70 |
Putative protein of unknown function |
||
| YOR383C | 3.69 |
FIT3
|
Mannoprotein that is incorporated into the cell wall via a glycosylphosphatidylinositol (GPI) anchor, involved in the retention of siderophore-iron in the cell wall |
|
| YFR017C | 3.53 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and is induced in response to the DNA-damaging agent MMS; YFR017C is not an essential gene |
||
| YOR382W | 3.46 |
FIT2
|
Mannoprotein that is incorporated into the cell wall via a glycosylphosphatidylinositol (GPI) anchor, involved in the retention of siderophore-iron in the cell wall |
|
| YFL011W | 3.42 |
HXT10
|
Putative hexose transporter, expressed at low levels and expression is repressed by glucose |
|
| YKL031W | 3.30 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species |
||
| YHR218W | 3.24 |
Helicase-like protein encoded within the telomeric Y' element |
||
| YHR211W | 3.16 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YAR060C | 3.15 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YMR191W | 3.14 |
SPG5
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YKL026C | 3.11 |
GPX1
|
Phospholipid hydroperoxide glutathione peroxidase induced by glucose starvation that protects cells from phospholipid hydroperoxides and nonphospholipid peroxides during oxidative stress |
|
| YKL217W | 3.06 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YPL061W | 3.03 |
ALD6
|
Cytosolic aldehyde dehydrogenase, activated by Mg2+ and utilizes NADP+ as the preferred coenzyme; required for conversion of acetaldehyde to acetate; constitutively expressed; locates to the mitochondrial outer surface upon oxidative stress |
|
| YJR146W | 3.02 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene HMS2 |
||
| YDR119W-A | 2.98 |
Putative protein of unknown function |
||
| YKL109W | 2.96 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YPL119C | 2.83 |
DBP1
|
Putative ATP-dependent RNA helicase of the DEAD-box protein family; mutants show reduced stability of the 40S ribosomal subunit scanning through 5' untranslated regions of mRNAs |
|
| YEL039C | 2.83 |
CYC7
|
Cytochrome c isoform 2, expressed under hypoxic conditions; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YHR212W-A | 2.83 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YHR212C | 2.72 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR023W | 2.63 |
MTL1
|
Protein with both structural and functional similarity to Mid2p, which is a plasma membrane sensor required for cell integrity signaling during pheromone-induced morphogenesis; suppresses rgd1 null mutations |
|
| YJR115W | 2.62 |
Putative protein of unknown function |
||
| YBR051W | 2.60 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partiallly overlaps the REG2/YBR050C regulatory subunit of the Glc7p type-1 protein phosphatase |
||
| YML090W | 2.56 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YML089C; exhibits growth defect on a non-fermentable (respiratory) carbon source |
||
| YPR030W | 2.54 |
CSR2
|
Nuclear protein with a potential regulatory role in utilization of galactose and nonfermentable carbon sources; overproduction suppresses the lethality at high temperature of a chs5 spa2 double null mutation; potential Cdc28p substrate |
|
| YGR022C | 2.52 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF MTL1/YGR023W |
||
| YMR195W | 2.50 |
ICY1
|
Protein of unknown function, required for viability in rich media of cells lacking mitochondrial DNA; mutants have an invasive growth defect with elongated morphology; induced by amino acid starvation |
|
| YFL063W | 2.49 |
Dubious open reading frame, based on available experimental and comparative sequence data |
||
| YEL070W | 2.48 |
DSF1
|
Deletion suppressor of mpt5 mutation |
|
| YLR004C | 2.47 |
THI73
|
Putative plasma membrane permease proposed to be involved in carboxylic acid uptake and repressed by thiamine; substrate of Dbf2p/Mob1p kinase; transcription is altered if mitochondrial dysfunction occurs |
|
| YMR081C | 2.44 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YGL146C | 2.41 |
Putative protein of unknown function, contains two putative transmembrane spans, shows no significant homology to any other known protein sequence, YGL146C is not an essential gene |
||
| YBR050C | 2.35 |
REG2
|
Regulatory subunit of the Glc7p type-1 protein phosphatase; involved with Reg1p, Glc7p, and Snf1p in regulation of glucose-repressible genes, also involved in glucose-induced proteolysis of maltose permease |
|
| YJL045W | 2.33 |
Minor succinate dehydrogenase isozyme; homologous to Sdh1p, the major isozyme reponsible for the oxidation of succinate and transfer of electrons to ubiquinone; induced during the diauxic shift in a Cat8p-dependent manner |
||
| YBR105C | 2.29 |
VID24
|
Peripheral membrane protein located at Vid (vacuole import and degradation) vesicles; regulates fructose-1,6-bisphosphatase (FBPase) targeting to the vacuole; involved in proteasome-dependent catabolite degradation of FBPase |
|
| YLR327C | 2.26 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YML091C | 2.25 |
RPM2
|
Protein subunit of mitochondrial RNase P, has roles in nuclear transcription, cytoplasmic and mitochondrial RNA processing, and mitochondrial translation; distributed to mitochondria, cytoplasmic processing bodies, and the nucleus |
|
| YOR235W | 2.24 |
IRC13
|
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YPR006C | 2.20 |
ICL2
|
2-methylisocitrate lyase of the mitochondrial matrix, functions in the methylcitrate cycle to catalyze the conversion of 2-methylisocitrate to succinate and pyruvate; ICL2 transcription is repressed by glucose and induced by ethanol |
|
| YOL051W | 2.16 |
GAL11
|
Subunit of the RNA polymerase II mediator complex; associates with core polymerase subunits to form the RNA polymerase II holoenzyme; affects transcription by acting as target of activators and repressors |
|
| YGR183C | 2.12 |
QCR9
|
Subunit 9 of the ubiquinol cytochrome-c reductase complex, which is a component of the mitochondrial inner membrane electron transport chain; required for electron transfer at the ubiquinol oxidase site of the complex |
|
| YIL055C | 2.11 |
Putative protein of unknown function |
||
| YEL011W | 2.06 |
GLC3
|
Glycogen branching enzyme, involved in glycogen accumulation; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YLR377C | 2.06 |
FBP1
|
Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway, required for glucose metabolism |
|
| YNL305C | 2.06 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YNL305C is not an essential gene |
||
| YFL064C | 2.04 |
Putative protein of unknown function |
||
| YOR343C | 2.02 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YDR343C | 2.01 |
HXT6
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt7p, expressed at high basal levels relative to other HXTs, repression of expression by high glucose requires SNF3 |
|
| YDL079C | 2.00 |
MRK1
|
Glycogen synthase kinase 3 (GSK-3) homolog; one of four GSK-3 homologs in S. cerevisiae that function to activate Msn2p-dependent transcription of stress responsive genes and that function in protein degradation |
|
| YPR184W | 1.96 |
GDB1
|
Glycogen debranching enzyme containing glucanotranferase and alpha-1,6-amyloglucosidase activities, required for glycogen degradation; phosphorylated in mitochondria |
|
| YPR026W | 1.96 |
ATH1
|
Acid trehalase required for utilization of extracellular trehalose |
|
| YOL084W | 1.94 |
PHM7
|
Protein of unknown function, expression is regulated by phosphate levels; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery and vacuole |
|
| YHR033W | 1.93 |
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm |
||
| YDL114W | 1.92 |
Putative protein of unknown function with similarity to acyl-carrier-protein reductases; YDL114W is not an essential gene |
||
| YJR151C | 1.89 |
DAN4
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YIL155C | 1.89 |
GUT2
|
Mitochondrial glycerol-3-phosphate dehydrogenase; expression is repressed by both glucose and cAMP and derepressed by non-fermentable carbon sources in a Snf1p, Rsf1p, Hap2/3/4/5 complex dependent manner |
|
| YMR280C | 1.87 |
CAT8
|
Zinc cluster transcriptional activator necessary for derepression of a variety of genes under non-fermentative growth conditions, active after diauxic shift, binds carbon source responsive elements |
|
| YKR097W | 1.83 |
PCK1
|
Phosphoenolpyruvate carboxykinase, key enzyme in gluconeogenesis, catalyzes early reaction in carbohydrate biosynthesis, glucose represses transcription and accelerates mRNA degradation, regulated by Mcm1p and Cat8p, located in the cytosol |
|
| YNL037C | 1.83 |
IDH1
|
Subunit of mitochondrial NAD(+)-dependent isocitrate dehydrogenase, which catalyzes the oxidation of isocitrate to alpha-ketoglutarate in the TCA cycle |
|
| YDL130W-A | 1.83 |
STF1
|
Protein involved in regulation of the mitochondrial F1F0-ATP synthase; Stf1p and Stf2p may act as stabilizing factors that enhance inhibitory action of the Inh1p protein |
|
| YCR068W | 1.79 |
ATG15
|
Lipase, required for intravacuolar lysis of autophagosomes; located in the endoplasmic reticulum membrane and targeted to intravacuolar vesicles during autophagy via the multivesicular body (MVB) pathway |
|
| YIL107C | 1.78 |
PFK26
|
6-phosphofructo-2-kinase, inhibited by phosphoenolpyruvate and sn-glycerol 3-phosphate, has negligible fructose-2,6-bisphosphatase activity, transcriptional regulation involves protein kinase A |
|
| YDR406W | 1.77 |
PDR15
|
Plasma membrane ATP binding cassette (ABC) transporter, multidrug transporter and general stress response factor implicated in cellular detoxification; regulated by Pdr1p, Pdr3p and Pdr8p; promoter contains a PDR responsive element |
|
| YEL012W | 1.70 |
UBC8
|
Ubiquitin-conjugating enzyme that negatively regulates gluconeogenesis by mediating the glucose-induced ubiquitination of fructose-1,6-bisphosphatase (FBPase); cytoplasmic enzyme that catalyzes the ubiquitination of histones in vitro |
|
| YOR376W | 1.69 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YOR376W is not an essential gene. |
||
| YPR150W | 1.68 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene SUE1/YPR151C |
||
| YLR332W | 1.67 |
MID2
|
O-glycosylated plasma membrane protein that acts as a sensor for cell wall integrity signaling and activates the pathway; interacts with Rom2p, a guanine nucleotide exchange factor for Rho1p, and with cell integrity pathway protein Zeo1p |
|
| YER187W | 1.67 |
Putative protein of unknown function; induced in respiratory-deficient cells |
||
| YMR105C | 1.60 |
PGM2
|
Phosphoglucomutase, catalyzes the conversion from glucose-1-phosphate to glucose-6-phosphate, which is a key step in hexose metabolism; functions as the acceptor for a Glc-phosphotransferase |
|
| YLR331C | 1.59 |
JIP3
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 98% of ORF overlaps the verified gene MID2 |
|
| YLR267W | 1.59 |
BOP2
|
Protein of unknown function, overproduction suppresses a pam1 slv3 double null mutation |
|
| YAL061W | 1.58 |
BDH2
|
Putative medium-chain alcohol dehydrogenase with similarity to BDH1; transcription induced by constitutively active PDR1 and PDR3; BDH2 is an essential gene |
|
| YKL044W | 1.58 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR111W | 1.57 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YMR175W | 1.57 |
SIP18
|
Protein of unknown function whose expression is induced by osmotic stress |
|
| YGL163C | 1.56 |
RAD54
|
DNA-dependent ATPase, stimulates strand exchange by modifying the topology of double-stranded DNA; involved in the recombinational repair of double-strand breaks in DNA during vegetative growth and meiosis; member of the SWI/SNF family |
|
| YCR007C | 1.56 |
Putative integral membrane protein, member of DUP240 gene family; YCR007C is not an essential gene |
||
| YER054C | 1.56 |
GIP2
|
Putative regulatory subunit of the protein phosphatase Glc7p, involved in glycogen metabolism; contains a conserved motif (GVNK motif) that is also found in Gac1p, Pig1p, and Pig2p |
|
| YNL339C | 1.55 |
YRF1-6
|
Helicase encoded by the Y' element of subtelomeric regions, highly expressed in the mutants lacking the telomerase component TLC1; potentially phosphorylated by Cdc28p |
|
| YOR394W | 1.52 |
PAU21
|
Hypothetical protein |
|
| YPL201C | 1.51 |
YIG1
|
Protein that interacts with glycerol 3-phosphatase and plays a role in anaerobic glycerol production; localizes to the nucleus and cytosol |
|
| YER188C-A | 1.49 |
Putative protein of unknown function |
||
| YLR438W | 1.47 |
CAR2
|
L-ornithine transaminase (OTAse), catalyzes the second step of arginine degradation, expression is dually-regulated by allophanate induction and a specific arginine induction process; not nitrogen catabolite repression sensitive |
|
| YKL141W | 1.42 |
SDH3
|
Cytochrome b subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
|
| YPL282C | 1.41 |
PAU22
|
Hypothetical protein |
|
| YDR231C | 1.40 |
COX20
|
Mitochondrial inner membrane protein, required for proteolytic processing of Cox2p and its assembly into cytochrome c oxidase |
|
| YDR259C | 1.39 |
YAP6
|
Putative basic leucine zipper (bZIP) transcription factor; overexpression increases sodium and lithium tolerance |
|
| YKL032C | 1.38 |
IXR1
|
Protein that binds DNA containing intrastrand cross-links formed by cisplatin, contains two HMG (high mobility group box) domains, which confer the ability to bend cisplatin-modified DNA; mediates aerobic transcriptional repression of COX5b |
|
| YIL060W | 1.38 |
Putative protein of unknown function; mutant accumulates less glycogen than does wild type; YIL060W is not an essential gene |
||
| YDR010C | 1.38 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YIL122W | 1.37 |
POG1
|
Putative transcriptional activator that promotes recovery from pheromone induced arrest; inhibits both alpha-factor induced G1 arrest and repression of CLN1 and CLN2 via SCB/MCB promoter elements; potential Cdc28p substrate; SBF regulated |
|
| YLL053C | 1.37 |
Putative protein; in the Sigma 1278B strain background YLL053C is contiguous with AQY2 which encodes an aquaporin |
||
| YBR021W | 1.36 |
FUR4
|
Uracil permease, localized to the plasma membrane; expression is tightly regulated by uracil levels and environmental cues |
|
| YOR113W | 1.36 |
AZF1
|
Zinc-finger transcription factor, involved in induction of CLN3 transcription in response to glucose; genetic and physical interactions indicate a possible role in mitochondrial transcription or genome maintenance |
|
| YLR176C | 1.35 |
RFX1
|
Major transcriptional repressor of DNA-damage-regulated genes, recruits repressors Tup1p and Cyc8p to their promoters; involved in DNA damage and replication checkpoint pathway; similar to a family of mammalian DNA binding RFX1-4 proteins |
|
| YKR102W | 1.34 |
FLO10
|
Lectin-like protein with similarity to Flo1p, thought to be involved in flocculation |
|
| YKR096W | 1.33 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the nucleus and cytoplasm; predicted to contain a PINc (PilT N terminus) domain |
||
| YDR216W | 1.33 |
ADR1
|
Carbon source-responsive zinc-finger transcription factor, required for transcription of the glucose-repressed gene ADH2, of peroxisomal protein genes, and of genes required for ethanol, glycerol, and fatty acid utilization |
|
| YPR151C | 1.33 |
SUE1
|
Mitochondrial protein required for degradation of unstable forms of cytochrome c |
|
| YGL117W | 1.32 |
Putative protein of unknown function |
||
| YLR122C | 1.31 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR123C |
||
| YOL047C | 1.29 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
||
| YLR437C-A | 1.25 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF CAR2/YLR438W |
||
| YMR272C | 1.22 |
SCS7
|
Sphingolipid alpha-hydroxylase, functions in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids, has both cytochrome b5-like and hydroxylase/desaturase domains, not essential for growth |
|
| YLL052C | 1.22 |
AQY2
|
Water channel that mediates the transport of water across cell membranes, only expressed in proliferating cells, controlled by osmotic signals, may be involved in freeze tolerance; disrupted by a stop codon in many S. cerevisiae strains |
|
| YIL045W | 1.22 |
PIG2
|
Putative type-1 protein phosphatase targeting subunit that tethers Glc7p type-1 protein phosphatase to Gsy2p glycogen synthase |
|
| YIL112W | 1.20 |
HOS4
|
Subunit of the Set3 complex, which is a meiotic-specific repressor of sporulation specific genes that contains deacetylase activity; potential Cdc28p substrate |
|
| YMR213W | 1.19 |
CEF1
|
Essential splicing factor; associated with Prp19p and the spliceosome, contains an N-terminal c-Myb DNA binding motif necessary for cell viability but not for Prp19p association, evolutionarily conserved and homologous to S. pombe Cdc5p |
|
| YBR072W | 1.19 |
HSP26
|
Small heat shock protein (sHSP) with chaperone activity; forms hollow, sphere-shaped oligomers that suppress unfolded proteins aggregation; oligomer activation requires a heat-induced conformational change; not expressed in unstressed cells |
|
| YJL103C | 1.19 |
GSM1
|
Putative zinc cluster protein of unknown function; proposed to be involved in the regulation of energy metabolism, based on patterns of expression and sequence analysis |
|
| YPR013C | 1.18 |
Putative zinc finger protein; YPR013C is not an essential gene |
||
| YER189W | 1.18 |
Putative protein of unknown function |
||
| YOR378W | 1.17 |
Putative protein of unknown function; belongs to the DHA2 family of drug:H+ antiporters; YOR378W is not an essential gene |
||
| YLR124W | 1.17 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR015C | 1.16 |
Putative protein of unknown function |
||
| YGR236C | 1.15 |
SPG1
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YNL337W | 1.13 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR123C | 1.12 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR122C; contains characteristic aminoacyl-tRNA motif |
||
| YHR210C | 1.10 |
Putative protein of unknown function; non-essential gene; highly expressed under anaeorbic conditions; sequence similarity to aldose 1-epimerases such as GAL10 |
||
| YDL113C | 1.10 |
ATG20
|
Sorting nexin family member required for the cytoplasm-to-vacuole targeting (Cvt) pathway and for endosomal sorting; has a Phox homology domain that binds phosphatidylinositol-3-phosphate; interacts with Snx4p; potential Cdc28p substrate |
|
| YNL018C | 1.10 |
Putative protein of unknown function |
||
| YHR145C | 1.09 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPR010C-A | 1.09 |
Putative protein of unknown function; conserved among Saccharomyces sensu stricto species |
||
| YPL057C | 1.08 |
SUR1
|
Probable catalytic subunit of a mannosylinositol phosphorylceramide (MIPC) synthase, forms a complex with probable regulatory subunit Csg2p; function in sphingolipid biosynthesis is overlapping with that of Csh1p |
|
| YHR205W | 1.08 |
SCH9
|
Protein kinase involved in transcriptional activation of osmostress-responsive genes; regulates G1 progression, cAPK activity, nitrogen activation of the FGM pathway; involved in life span regulation; homologous to mammalian Akt/PKB |
|
| YCL001W-B | 1.07 |
Putative protein of unknown function; identified by homology |
||
| YER142C | 1.07 |
MAG1
|
3-methyl-adenine DNA glycosylase involved in protecting DNA against alkylating agents; initiates base excision repair by removing damaged bases to create abasic sites that are subsequently repaired |
|
| YER158C | 1.04 |
Protein of unknown function, has similarity to Afr1p; potentially phosphorylated by Cdc28p |
||
| YJR160C | 1.03 |
MPH3
|
Alpha-glucoside permease, transports maltose, maltotriose, alpha-methylglucoside, and turanose; identical to Mph2p; encoded in a subtelomeric position in a region likely to have undergone duplication |
|
| YPR005C | 1.02 |
HAL1
|
Cytoplasmic protein involved in halotolerance; decreases intracellular Na+ (via Ena1p) and increases intracellular K+ by decreasing efflux; expression repressed by Ssn6p-Tup1p and Sko1p and induced by NaCl, KCl, and sorbitol through Gcn4p |
|
| YBR132C | 1.02 |
AGP2
|
High affinity polyamine permease, preferentially uses spermidine over putrescine; expression is down-regulated by osmotic stress; plasma membrane carnitine transporter, also functions as a low-affinity amino acid permease |
|
| YKL043W | 1.02 |
PHD1
|
Transcriptional activator that enhances pseudohyphal growth; regulates expression of FLO11, an adhesin required for pseudohyphal filament formation; similar to StuA, an A. nidulans developmental regulator; potential Cdc28p substrate |
|
| YKL085W | 1.01 |
MDH1
|
Mitochondrial malate dehydrogenase, catalyzes interconversion of malate and oxaloacetate; involved in the tricarboxylic acid (TCA) cycle; phosphorylated |
|
| YEL044W | 0.99 |
IES6
|
Protein that associates with the INO80 chromatin remodeling complex under low-salt conditions |
|
| YJR120W | 0.98 |
Protein of unknown function; essential for growth under anaerobic conditions; mutation causes decreased expression of ATP2, impaired respiration, defective sterol uptake, and altered levels/localization of ABC transporters Aus1p and Pdr11p |
||
| YJR159W | 0.97 |
SOR1
|
Sorbitol dehydrogenase; expression is induced in the presence of sorbitol |
|
| YLR223C | 0.97 |
IFH1
|
Essential protein with a highly acidic N-terminal domain; IFH1 exhibits genetic interactions with FHL1, overexpression interferes with silencing at telomeres and HM loci; potential Cdc28p substrate |
|
| YOL060C | 0.96 |
MAM3
|
Protein required for normal mitochondrial morphology, has similarity to hemolysins |
|
| YBR033W | 0.95 |
EDS1
|
Putative zinc cluster protein; YBR033W is not an essential gene |
|
| YAR035W | 0.95 |
YAT1
|
Outer mitochondrial carnitine acetyltransferase, minor ethanol-inducible enzyme involved in transport of activated acyl groups from the cytoplasm into the mitochondrial matrix; phosphorylated |
|
| YER111C | 0.94 |
SWI4
|
DNA binding component of the SBF complex (Swi4p-Swi6p), a transcriptional activator that in concert with MBF (Mbp1-Swi6p) regulates late G1-specific transcription of targets including cyclins and genes required for DNA synthesis and repair |
|
| YMR190C | 0.94 |
SGS1
|
Nucleolar DNA helicase of the RecQ family involved in maintenance of genome integrity, regulates chromosome synapsis and meiotic crossing over; has similarity to human BLM and WRN helicases implicated in Bloom and Werner syndromes |
|
| YKR095W | 0.93 |
MLP1
|
Myosin-like protein associated with the nuclear envelope, connects the nuclear pore complex with the nuclear interior; involved with Tel1p in telomere length control; involved with Pml1p and Pml39p in nuclear retention of unspliced mRNAs |
|
| YDR389W | 0.93 |
SAC7
|
GTPase activating protein (GAP) for Rho1p, involved in signaling to the actin cytoskeleton, null mutations suppress tor2 mutations and temperature sensitive mutations in actin; potential Cdc28p substrate |
|
| YIL136W | 0.93 |
OM45
|
Protein of unknown function, major constituent of the mitochondrial outer membrane; located on the outer (cytosolic) face of the outer membrane |
|
| YPR169W-A | 0.93 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps two other dubious ORFs: YPR170C and YPR170W-B |
||
| YIL125W | 0.92 |
KGD1
|
Component of the mitochondrial alpha-ketoglutarate dehydrogenase complex, which catalyzes a key step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation of alpha-ketoglutarate to form succinyl-CoA |
|
| YOL085W-A | 0.91 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YOL085C |
||
| YJL102W | 0.90 |
MEF2
|
Mitochondrial elongation factor involved in translational elongation |
|
| YBR001C | 0.90 |
NTH2
|
Putative neutral trehalase, required for thermotolerance and may mediate resistance to other cellular stresses |
|
| YCR010C | 0.90 |
ADY2
|
Acetate transporter required for normal sporulation; phosphorylated in mitochondria |
|
| YLR334C | 0.89 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps a stand-alone long terminal repeat sequence whose presence indicates a retrotransposition event occurred here |
||
| YGR258C | 0.88 |
RAD2
|
Single-stranded DNA endonuclease, cleaves single-stranded DNA during nucleotide excision repair to excise damaged DNA; subunit of Nucleotide Excision Repair Factor 3 (NEF3); homolog of human XPG protein |
|
| YGL033W | 0.88 |
HOP2
|
Meiosis-specific protein that localizes to chromosomes, preventing synapsis between nonhomologous chromosomes and ensuring synapsis between homologs; complexes with Mnd1p to promote homolog pairing and meiotic double-strand break repair |
|
| YHR202W | 0.88 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole, while HA-tagged protein is found in the soluble fraction, suggesting cytoplasmic localization |
||
| YBR218C | 0.88 |
PYC2
|
Pyruvate carboxylase isoform, cytoplasmic enzyme that converts pyruvate to oxaloacetate; highly similar to isoform Pyc1p but differentially regulated; mutations in the human homolog are associated with lactic acidosis |
|
| YPR154W | 0.88 |
PIN3
|
Protein that induces appearance of [PIN+] prion when overproduced |
|
| YDL244W | 0.86 |
THI13
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YER116C | 0.86 |
SLX8
|
Subunit of the Slx5-Slx8 substrate-specific ubiquitin ligase complex; stimulated by prior attachment of SUMO to the substrate |
|
| YAL062W | 0.85 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YFL033C | 0.85 |
RIM15
|
Glucose-repressible protein kinase involved in signal transduction during cell proliferation in response to nutrients, specifically the establishment of stationary phase; identified as a regulator of IME2; substrate of Pho80p-Pho85p kinase |
|
| YMR135C | 0.84 |
GID8
|
Protein of unknown function, involved in proteasome-dependent catabolite inactivation of fructose-1,6-bisphosphatase; contains LisH and CTLH domains, like Vid30p; dosage-dependent regulator of START |
|
| YOR120W | 0.83 |
GCY1
|
Putative NADP(+) coupled glycerol dehydrogenase, proposed to be involved in an alternative pathway for glycerol catabolism; member of the aldo-keto reductase (AKR) family |
|
| YDR186C | 0.83 |
Putative protein of unknown function; may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YLR174W | 0.83 |
IDP2
|
Cytosolic NADP-specific isocitrate dehydrogenase, catalyzes oxidation of isocitrate to alpha-ketoglutarate; levels are elevated during growth on non-fermentable carbon sources and reduced during growth on glucose |
|
| YLR149C | 0.82 |
Putative protein of unknown function; YLR149C is not an essential gene |
||
| YJL199C | 0.81 |
MBB1
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; protein detected in large-scale protein-protein interaction studies |
|
| YCL025C | 0.81 |
AGP1
|
Low-affinity amino acid permease with broad substrate range, involved in uptake of asparagine, glutamine, and other amino acids; expression is regulated by the SPS plasma membrane amino acid sensor system (Ssy1p-Ptr3p-Ssy5p) |
|
| YPL250C | 0.80 |
ICY2
|
Protein of unknown function; mobilized into polysomes upon a shift from a fermentable to nonfermentable carbon source; potential Cdc28p substrate |
|
| YOR023C | 0.79 |
AHC1
|
Subunit of the Ada histone acetyltransferase complex, required for structural integrity of the complex |
|
| YMR291W | 0.79 |
Putative kinase of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YMR291W is not an essential gene |
||
| YOR236W | 0.78 |
DFR1
|
Dihydrofolate reductase, part of the dTTP biosynthetic pathway, involved in folate metabolism, possibly required for mitochondrial function |
|
| YJR147W | 0.78 |
HMS2
|
Protein with similarity to heat shock transcription factors; overexpression suppresses the pseudohyphal filamentation defect of a diploid mep1 mep2 homozygous null mutant |
|
| YGR121C | 0.78 |
MEP1
|
Ammonium permease; belongs to a ubiquitous family of cytoplasmic membrane proteins that transport only ammonium (NH4+); expression is under the nitrogen catabolite repression regulation |
|
| YOR186C-A | 0.78 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YDR232W | 0.78 |
HEM1
|
5-aminolevulinate synthase, catalyzes the first step in the heme biosynthetic pathway; an N-terminal signal sequence is required for localization to the mitochondrial matrix; expression is regulated by Hap2p-Hap3p |
Network of associatons between targets according to the STRING database.
First level regulatory network of YGR067C
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 9.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 1.6 | 4.9 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 1.5 | 4.4 | GO:0015755 | fructose transport(GO:0015755) |
| 1.4 | 5.6 | GO:0006848 | pyruvate transport(GO:0006848) |
| 1.3 | 5.1 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 1.2 | 6.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 1.0 | 6.0 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 1.0 | 3.9 | GO:0015976 | carbon utilization(GO:0015976) |
| 1.0 | 2.9 | GO:0006068 | ethanol catabolic process(GO:0006068) primary alcohol catabolic process(GO:0034310) |
| 1.0 | 2.9 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 0.8 | 2.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.8 | 3.3 | GO:0019563 | alditol catabolic process(GO:0019405) glycerol catabolic process(GO:0019563) |
| 0.8 | 4.0 | GO:0061416 | regulation of transcription from RNA polymerase II promoter in response to salt stress(GO:0061416) |
| 0.8 | 6.1 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.8 | 2.3 | GO:0051469 | vesicle fusion with vacuole(GO:0051469) |
| 0.7 | 0.7 | GO:0019413 | acetate biosynthetic process(GO:0019413) |
| 0.7 | 1.5 | GO:0071462 | response to water(GO:0009415) cellular response to water stimulus(GO:0071462) |
| 0.7 | 2.2 | GO:0019629 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 0.7 | 0.7 | GO:0043902 | positive regulation of multi-organism process(GO:0043902) |
| 0.6 | 13.5 | GO:0008645 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.6 | 1.9 | GO:0035948 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 0.6 | 1.8 | GO:0006624 | vacuolar protein processing(GO:0006624) multivesicular body organization(GO:0036257) |
| 0.6 | 5.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.6 | 11.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.5 | 2.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.5 | 1.9 | GO:0001324 | age-dependent general metabolic decline involved in chronological cell aging(GO:0001323) age-dependent response to oxidative stress involved in chronological cell aging(GO:0001324) |
| 0.5 | 1.4 | GO:0052547 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 0.5 | 4.1 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.5 | 2.3 | GO:0032071 | regulation of deoxyribonuclease activity(GO:0032070) regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.4 | 0.4 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.4 | 1.7 | GO:0030969 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) IRE1-mediated unfolded protein response(GO:0036498) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) |
| 0.4 | 1.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.4 | 1.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.4 | 2.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.4 | 1.5 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) |
| 0.4 | 1.1 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.3 | 1.4 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.3 | 2.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 3.0 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.3 | 0.6 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.3 | 0.9 | GO:0031292 | gene conversion at mating-type locus, DNA double-strand break processing(GO:0031292) |
| 0.3 | 0.9 | GO:0090527 | actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.3 | 0.9 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.3 | 2.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.3 | 1.4 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.3 | 1.1 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.3 | 0.8 | GO:0042843 | D-xylose catabolic process(GO:0042843) |
| 0.3 | 1.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.3 | 1.0 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.7 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.2 | 0.7 | GO:0042743 | hydrogen peroxide metabolic process(GO:0042743) hydrogen peroxide catabolic process(GO:0042744) |
| 0.2 | 0.9 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.2 | 0.7 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.2 | 0.9 | GO:1901925 | cellular response to biotic stimulus(GO:0071216) response to cell cycle checkpoint signaling(GO:0072396) response to mitotic cell cycle checkpoint signaling(GO:0072414) response to spindle checkpoint signaling(GO:0072417) response to mitotic spindle checkpoint signaling(GO:0072476) response to mitotic cell cycle spindle assembly checkpoint signaling(GO:0072479) response to spindle assembly checkpoint signaling(GO:0072485) negative regulation of protein import into nucleus during spindle assembly checkpoint(GO:1901925) |
| 0.2 | 1.4 | GO:0071322 | cellular response to carbohydrate stimulus(GO:0071322) |
| 0.2 | 0.7 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.2 | 2.4 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.2 | 0.9 | GO:0051126 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.2 | 1.0 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.2 | 0.6 | GO:0034090 | maintenance of meiotic sister chromatid cohesion(GO:0034090) |
| 0.2 | 0.6 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.2 | 0.9 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.2 | 0.7 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.2 | 0.6 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.2 | 0.8 | GO:0090294 | nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) nitrogen catabolite activation of transcription(GO:0090294) |
| 0.1 | 0.4 | GO:0043419 | urea metabolic process(GO:0019627) urea catabolic process(GO:0043419) |
| 0.1 | 7.2 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.1 | 0.5 | GO:2000284 | positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 0.6 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.5 | GO:0051101 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.1 | 1.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.4 | GO:0032443 | regulation of steroid metabolic process(GO:0019218) regulation of ergosterol biosynthetic process(GO:0032443) regulation of steroid biosynthetic process(GO:0050810) |
| 0.1 | 0.5 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.5 | GO:0051303 | meiotic telomere clustering(GO:0045141) establishment of chromosome localization(GO:0051303) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.1 | 0.8 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 1.9 | GO:0009228 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.1 | 1.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 0.9 | GO:0000321 | re-entry into mitotic cell cycle after pheromone arrest(GO:0000321) |
| 0.1 | 0.2 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.1 | 1.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.7 | GO:0070058 | tRNA gene clustering(GO:0070058) |
| 0.1 | 0.6 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 0.2 | GO:0051098 | regulation of binding(GO:0051098) |
| 0.1 | 0.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 2.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.7 | GO:0097034 | mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.2 | GO:0034033 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 1.7 | GO:0042493 | response to drug(GO:0042493) |
| 0.1 | 0.4 | GO:0031135 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.1 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.1 | 0.5 | GO:0046352 | disaccharide catabolic process(GO:0046352) |
| 0.1 | 0.1 | GO:0033241 | regulation of cellular amine catabolic process(GO:0033241) |
| 0.1 | 0.4 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 0.3 | GO:0071902 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) positive regulation of protein serine/threonine kinase activity(GO:0071902) |
| 0.1 | 0.3 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.1 | 0.2 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.1 | 0.5 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.1 | 0.2 | GO:0016320 | endoplasmic reticulum membrane fusion(GO:0016320) |
| 0.1 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.2 | GO:0055078 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.1 | 1.1 | GO:0007129 | synapsis(GO:0007129) |
| 0.1 | 0.1 | GO:0006827 | high-affinity iron ion transmembrane transport(GO:0006827) |
| 0.1 | 0.3 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.1 | 0.1 | GO:0045980 | negative regulation of nucleotide metabolic process(GO:0045980) |
| 0.1 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.3 | GO:0046461 | triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.1 | 1.0 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 0.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 1.2 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.1 | 0.1 | GO:0072350 | tricarboxylic acid metabolic process(GO:0072350) |
| 0.1 | 2.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.2 | GO:0060237 | regulation of fungal-type cell wall organization(GO:0060237) |
| 0.1 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.5 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.1 | GO:0001178 | regulation of transcriptional start site selection at RNA polymerase II promoter(GO:0001178) |
| 0.1 | 1.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 3.2 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.1 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.1 | GO:0009743 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.2 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) cyclic-nucleotide-mediated signaling(GO:0019935) |
| 0.0 | 1.8 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.2 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.9 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.0 | 0.1 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.3 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.2 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.7 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0031032 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin structure organization(GO:0031032) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 1.1 | GO:0030258 | lipid modification(GO:0030258) |
| 0.0 | 0.4 | GO:0001101 | response to acid chemical(GO:0001101) |
| 0.0 | 0.1 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.2 | GO:0070192 | chromosome organization involved in meiotic cell cycle(GO:0070192) |
| 0.0 | 0.2 | GO:0033674 | positive regulation of kinase activity(GO:0033674) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0000751 | mitotic cell cycle arrest in response to pheromone(GO:0000751) mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.1 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.2 | GO:0042168 | porphyrin-containing compound metabolic process(GO:0006778) tetrapyrrole metabolic process(GO:0033013) heme metabolic process(GO:0042168) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.9 | GO:0042176 | regulation of protein catabolic process(GO:0042176) |
| 0.0 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.2 | GO:0034087 | establishment of sister chromatid cohesion(GO:0034085) establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.0 | 0.4 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0019932 | second-messenger-mediated signaling(GO:0019932) |
| 0.0 | 0.4 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.0 | 0.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.0 | 0.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.8 | 3.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.7 | 3.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.6 | 1.8 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.6 | 2.9 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.5 | 2.0 | GO:0030287 | cell wall-bounded periplasmic space(GO:0030287) |
| 0.5 | 3.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.5 | 1.4 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.4 | 1.5 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.3 | 3.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.3 | 0.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.3 | 1.4 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) mitochondrial inner membrane protein insertion complex(GO:0042721) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 0.9 | GO:0033309 | SBF transcription complex(GO:0033309) |
| 0.2 | 0.6 | GO:0045265 | mitochondrial proton-transporting ATP synthase, stator stalk(GO:0000274) proton-transporting ATP synthase, stator stalk(GO:0045265) |
| 0.2 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 1.2 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.2 | 1.3 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 0.2 | 8.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 0.7 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.2 | 0.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 1.9 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.2 | 3.9 | GO:0098852 | fungal-type vacuole membrane(GO:0000329) lytic vacuole membrane(GO:0098852) |
| 0.2 | 2.8 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 1.5 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.1 | 7.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.8 | GO:0070772 | PAS complex(GO:0070772) |
| 0.1 | 0.7 | GO:0032545 | CURI complex(GO:0032545) |
| 0.1 | 1.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.7 | GO:0005619 | ascospore wall(GO:0005619) |
| 0.1 | 3.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 0.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 2.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 16.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 0.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 1.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.3 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.0 | 0.8 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.3 | GO:0000133 | polarisome(GO:0000133) |
| 0.0 | 0.2 | GO:0000817 | COMA complex(GO:0000817) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.8 | GO:0000323 | storage vacuole(GO:0000322) lytic vacuole(GO:0000323) fungal-type vacuole(GO:0000324) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0032221 | Rpd3S complex(GO:0032221) |
| 0.0 | 0.1 | GO:0030907 | MBF transcription complex(GO:0030907) |
| 0.0 | 0.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 1.2 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0000799 | condensin complex(GO:0000796) nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.3 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.1 | GO:0005825 | half bridge of spindle pole body(GO:0005825) |
| 0.0 | 1.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.1 | GO:0045334 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle(GO:0030139) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 9.1 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.0 | GO:0072380 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.7 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 1.8 | 7.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 1.6 | 4.9 | GO:0016406 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 1.6 | 4.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 1.3 | 5.2 | GO:0005537 | mannose binding(GO:0005537) |
| 1.0 | 4.0 | GO:0004396 | hexokinase activity(GO:0004396) |
| 1.0 | 3.0 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 1.0 | 2.9 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.9 | 2.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.6 | 8.7 | GO:0015578 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.6 | 7.4 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.6 | 3.0 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.5 | 2.2 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.5 | 2.6 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.5 | 1.0 | GO:0042947 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.5 | 1.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.5 | 2.8 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.5 | 1.4 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 0.4 | 2.1 | GO:0050253 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.4 | 1.7 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.4 | 2.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.4 | 0.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.3 | 1.4 | GO:0015294 | solute:cation symporter activity(GO:0015294) |
| 0.3 | 1.0 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.3 | 5.7 | GO:0046906 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.3 | 6.0 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.3 | 2.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.3 | 2.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 1.0 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.2 | 0.7 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.2 | 0.9 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.2 | 1.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 1.1 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.2 | 2.6 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.2 | 0.9 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.2 | 0.8 | GO:0016405 | CoA-ligase activity(GO:0016405) acid-thiol ligase activity(GO:0016878) |
| 0.2 | 0.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 3.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 2.3 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.2 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 0.9 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.2 | 0.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.2 | 0.5 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.2 | 1.7 | GO:0099600 | transmembrane signaling receptor activity(GO:0004888) signaling receptor activity(GO:0038023) transmembrane receptor activity(GO:0099600) |
| 0.2 | 1.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.4 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 2.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.4 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.9 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.7 | GO:0015038 | peptide disulfide oxidoreductase activity(GO:0015037) glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 1.3 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.1 | 2.3 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.1 | 0.1 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.1 | 1.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 4.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.2 | GO:0001026 | transcription factor activity, core RNA polymerase III binding(GO:0000995) TFIIIB-type transcription factor activity(GO:0001026) |
| 0.1 | 0.3 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.1 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.5 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.9 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.1 | 0.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.2 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 1.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.8 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 1.1 | GO:0016769 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.5 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 0.2 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 0.5 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.1 | 0.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 1.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 0.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.2 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.2 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.0 | 0.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 2.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 2.4 | GO:0032403 | protein complex binding(GO:0032403) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.4 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.6 | GO:0004871 | signal transducer activity(GO:0004871) |
| 0.0 | 0.2 | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen(GO:0016705) |
| 0.0 | 1.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.0 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.4 | GO:0004843 | obsolete ubiquitin thiolesterase activity(GO:0004221) thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.0 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.0 | 0.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 0.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 38.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.3 | 1.1 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.2 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 0.5 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.2 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME CHROMOSOME MAINTENANCE | Genes involved in Chromosome Maintenance |
| 0.0 | 0.1 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 35.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME TRANSCRIPTION | Genes involved in Transcription |