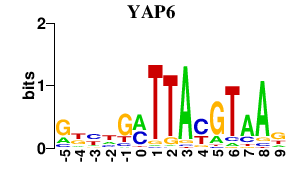
Results for YAP6
Z-value: 0.59
Transcription factors associated with YAP6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
YAP6
|
S000002667 | Basic leucine zipper (bZIP) transcription factor |
Activity-expression correlation:
Activity profile of YAP6 motif
Sorted Z-values of YAP6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YKL217W | 1.82 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YDL210W | 1.28 |
UGA4
|
Permease that serves as a gamma-aminobutyrate (GABA) transport protein involved in the utilization of GABA as a nitrogen source; catalyzes the transport of putrescine and delta-aminolevulinic acid (ALA); localized to the vacuolar membrane |
|
| YNR034W-A | 1.26 |
Putative protein of unknown function; expression is regulated by Msn2p/Msn4p |
||
| YML131W | 1.25 |
Putative protein of unknown function with similarity to oxidoreductases; HOG1 and SKO1-dependent mRNA expression is induced after osmotic shock; GFP-fusion protein localizes to the cytoplasm and is induced by the DNA-damaging agent MMS |
||
| YAL062W | 1.22 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YGL258W-A | 1.15 |
Putative protein of unknown function |
||
| YKL044W | 1.10 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR342C | 1.07 |
HXT7
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels |
|
| YMR107W | 1.06 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YLR327C | 1.00 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YCL025C | 1.00 |
AGP1
|
Low-affinity amino acid permease with broad substrate range, involved in uptake of asparagine, glutamine, and other amino acids; expression is regulated by the SPS plasma membrane amino acid sensor system (Ssy1p-Ptr3p-Ssy5p) |
|
| YKL043W | 0.87 |
PHD1
|
Transcriptional activator that enhances pseudohyphal growth; regulates expression of FLO11, an adhesin required for pseudohyphal filament formation; similar to StuA, an A. nidulans developmental regulator; potential Cdc28p substrate |
|
| YHR217C | 0.85 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; located in the telomeric region TEL08R. |
||
| YOL128C | 0.84 |
YGK3
|
Protein kinase related to mammalian glycogen synthase kinases of the GSK-3 family; GSK-3 homologs (Mck1p, Rim11p, Mrk1p, Ygk3p) are involved in control of Msn2p-dependent transcription of stress responsive genes and in protein degradation |
|
| YDR277C | 0.80 |
MTH1
|
Negative regulator of the glucose-sensing signal transduction pathway, required for repression of transcription by Rgt1p; interacts with Rgt1p and the Snf3p and Rgt2p glucose sensors; phosphorylated by Yck1p, triggering Mth1p degradation |
|
| YLR377C | 0.77 |
FBP1
|
Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway, required for glucose metabolism |
|
| YFL051C | 0.77 |
Putative protein of unknown function; YFL051C is not an essential gene |
||
| YFR053C | 0.77 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YMR081C | 0.75 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YDR540C | 0.74 |
IRC4
|
Putative protein of unknown function; null mutant displays increased levels of spontaneous Rad52p foci; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
|
| YFL052W | 0.73 |
Putative zinc cluster protein that contains a DNA binding domain; null mutant sensitive to calcofluor white, low osmolarity and heat, suggesting a role for YFL052Wp in cell wall integrity |
||
| YFR026C | 0.71 |
Putative protein of unknown function |
||
| YLR258W | 0.70 |
GSY2
|
Glycogen synthase, similar to Gsy1p; expression induced by glucose limitation, nitrogen starvation, heat shock, and stationary phase; activity regulated by cAMP-dependent, Snf1p and Pho85p kinases as well as by the Gac1p-Glc7p phosphatase |
|
| YPL189C-A | 0.69 |
COA2
|
Putative cytochrome oxidase assembly factor; identified by homology to Ashbya gossypii; null mutation results in respiratory deficiency with specific loss of cytochrome oxidase activity |
|
| YAR060C | 0.69 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLL041C | 0.67 |
SDH2
|
Iron-sulfur protein subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
|
| YAR053W | 0.67 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR178C | 0.65 |
GAC1
|
Regulatory subunit for Glc7p type-1 protein phosphatase (PP1), tethers Glc7p to Gsy2p glycogen synthase, binds Hsf1p heat shock transcription factor, required for induction of some HSF-regulated genes under heat shock |
|
| YJR061W | 0.64 |
Putative protein of unknown function; non-essential gene with similarity to Mnn4, a putative membrane protein involved in glycosylation; transcription repressed by Rm101p |
||
| YIL162W | 0.62 |
SUC2
|
Invertase, sucrose hydrolyzing enzyme; a secreted, glycosylated form is regulated by glucose repression, and an intracellular, nonglycosylated enzyme is produced constitutively |
|
| YKL067W | 0.61 |
YNK1
|
Nucleoside diphosphate kinase, catalyzes the transfer of gamma phosphates from nucleoside triphosphates, usually ATP, to nucleoside diphosphates by a mechanism that involves formation of an autophosphorylated enzyme intermediate |
|
| YPR010C-A | 0.60 |
Putative protein of unknown function; conserved among Saccharomyces sensu stricto species |
||
| YLR122C | 0.60 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR123C |
||
| YHR212W-A | 0.59 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YJL089W | 0.59 |
SIP4
|
C6 zinc cluster transcriptional activator that binds to the carbon source-responsive element (CSRE) of gluconeogenic genes; involved in the positive regulation of gluconeogenesis; regulated by Snf1p protein kinase; localized to the nucleus |
|
| YLR259C | 0.59 |
HSP60
|
Tetradecameric mitochondrial chaperonin required for ATP-dependent folding of precursor polypeptides and complex assembly; prevents aggregation and mediates protein refolding after heat shock; role in mtDNA transmission; phosphorylated |
|
| YPR184W | 0.59 |
GDB1
|
Glycogen debranching enzyme containing glucanotranferase and alpha-1,6-amyloglucosidase activities, required for glycogen degradation; phosphorylated in mitochondria |
|
| YOR343C | 0.58 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YBL015W | 0.57 |
ACH1
|
Acetyl-coA hydrolase, primarily localized to mitochondria; phosphorylated; required for acetate utilization and for diploid pseudohyphal growth |
|
| YEL009C-A | 0.56 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER150W | 0.55 |
SPI1
|
GPI-anchored cell wall protein involved in weak acid resistance; basal expression requires Msn2p/Msn4p; expression is induced under conditions of stress and during the diauxic shift; similar to Sed1p |
|
| YDR455C | 0.55 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YDR456W |
||
| YER067W | 0.55 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YER067W is not an essential gene |
||
| YDR456W | 0.55 |
NHX1
|
Endosomal Na+/H+ exchanger, required for intracellular sequestration of Na+; required for osmotolerance to acute hypertonic shock |
|
| YEL010W | 0.54 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHR078W | 0.54 |
High osmolarity-regulated gene of unknown function |
||
| YOL154W | 0.53 |
ZPS1
|
Putative GPI-anchored protein; transcription is induced under low-zinc conditions, as mediated by the Zap1p transcription factor, and at alkaline pH |
|
| YEL070W | 0.53 |
DSF1
|
Deletion suppressor of mpt5 mutation |
|
| YOL157C | 0.52 |
Putative protein of unknown function |
||
| YOR387C | 0.52 |
Putative protein of unknown function; regulated by the metal-responsive Aft1p transcription factor; highly inducible in zinc-depleted conditions; localizes to the soluble fraction |
||
| YER133W-A | 0.51 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps uncharacterized gene YER134C. |
||
| YHR218W | 0.51 |
Helicase-like protein encoded within the telomeric Y' element |
||
| YGL187C | 0.51 |
COX4
|
Subunit IV of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; N-terminal 25 residues of precursor are cleaved during mitochondrial import; phosphorylated |
|
| YDR241W | 0.51 |
BUD26
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 1% of ORF overlaps the verified gene SNU56; diploid mutant displays a weak budding pattern phenotype in a systematic assay |
|
| YGL255W | 0.51 |
ZRT1
|
High-affinity zinc transporter of the plasma membrane, responsible for the majority of zinc uptake; transcription is induced under low-zinc conditions by the Zap1p transcription factor |
|
| YLL054C | 0.51 |
Putative protein of unknown function with similarity to Pip2p, an oleate-specific transcriptional activator of peroxisome proliferation; YLL054C is not an essential gene |
||
| YHR212C | 0.51 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR123C | 0.50 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR122C; contains characteristic aminoacyl-tRNA motif |
||
| YPL250C | 0.50 |
ICY2
|
Protein of unknown function; mobilized into polysomes upon a shift from a fermentable to nonfermentable carbon source; potential Cdc28p substrate |
|
| YPL171C | 0.50 |
OYE3
|
Widely conserved NADPH oxidoreductase containing flavin mononucleotide (FMN), homologous to Oye2p with slight differences in ligand binding and catalytic properties; may be involved in sterol metabolism |
|
| YEL009C | 0.49 |
GCN4
|
Transcriptional activator of amino acid biosynthetic genes in response to amino acid starvation; expression is tightly regulated at both the transcriptional and translational levels |
|
| YOR381W | 0.48 |
FRE3
|
Ferric reductase, reduces siderophore-bound iron prior to uptake by transporters; expression induced by low iron levels |
|
| YDR536W | 0.47 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YLL053C | 0.46 |
Putative protein; in the Sigma 1278B strain background YLL053C is contiguous with AQY2 which encodes an aquaporin |
||
| YLR124W | 0.46 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR346W | 0.45 |
REV1
|
Deoxycytidyl transferase, forms a complex with the subunits of DNA polymerase zeta, Rev3p and Rev7p; involved in repair of abasic sites in damaged DNA |
|
| YPR036W-A | 0.45 |
Protein of unknown function; transcription is regulated by Pdr1p |
||
| YOL052C-A | 0.45 |
DDR2
|
Multistress response protein, expression is activated by a variety of xenobiotic agents and environmental or physiological stresses |
|
| YPR030W | 0.45 |
CSR2
|
Nuclear protein with a potential regulatory role in utilization of galactose and nonfermentable carbon sources; overproduction suppresses the lethality at high temperature of a chs5 spa2 double null mutation; potential Cdc28p substrate |
|
| YGR182C | 0.44 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF TIM13/YGR181W |
||
| YOR348C | 0.44 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YOR010C | 0.43 |
TIR2
|
Putative cell wall mannoprotein of the Srp1p/Tip1p family of serine-alanine-rich proteins; transcription is induced by cold shock and anaerobiosis |
|
| YEL008W | 0.43 |
Hypothetical protein predicted to be involved in metabolism |
||
| YER038W-A | 0.43 |
FMP49
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 99% of ORF overlaps the verified gene HVG1; protein product detected in mitochondria |
|
| YPR160W | 0.43 |
GPH1
|
Non-essential glycogen phosphorylase required for the mobilization of glycogen, activity is regulated by cyclic AMP-mediated phosphorylation, expression is regulated by stress-response elements and by the HOG MAP kinase pathway |
|
| YPR016W-A | 0.43 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR393W | 0.42 |
ERR1
|
Protein of unknown function, has similarity to enolases |
|
| YDR542W | 0.42 |
PAU10
|
Hypothetical protein |
|
| YEL028W | 0.41 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YAL063C | 0.40 |
FLO9
|
Lectin-like protein with similarity to Flo1p, thought to be expressed and involved in flocculation |
|
| YHL046W-A | 0.39 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLL052C | 0.39 |
AQY2
|
Water channel that mediates the transport of water across cell membranes, only expressed in proliferating cells, controlled by osmotic signals, may be involved in freeze tolerance; disrupted by a stop codon in many S. cerevisiae strains |
|
| YOL001W | 0.38 |
PHO80
|
Cyclin, negatively regulates phosphate metabolism; Pho80p-Pho85p (cyclin-CDK complex) phosphorylates Pho4p and Swi5p; deletion of PHO80 leads to aminoglycoside supersensitivity; truncated form of PHO80 affects vacuole inheritance |
|
| YER015W | 0.38 |
FAA2
|
Long chain fatty acyl-CoA synthetase; accepts a wider range of acyl chain lengths than Faa1p, preferring C9:0-C13:0; involved in the activation of endogenous pools of fatty acids |
|
| YAR047C | 0.38 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YJR146W | 0.38 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene HMS2 |
||
| YPL018W | 0.37 |
CTF19
|
Outer kinetochore protein, required for accurate mitotic chromosome segregation; component of the kinetochore sub-complex COMA (Ctf19p, Okp1p, Mcm21p, Ame1p) that functions as a platform for kinetochore assembly |
|
| YDR338C | 0.37 |
Hypothetical protein |
||
| YMR027W | 0.37 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus and cytoplasm; YMR027W is not an essential gene |
||
| YOL060C | 0.36 |
MAM3
|
Protein required for normal mitochondrial morphology, has similarity to hemolysins |
|
| YNL194C | 0.36 |
Integral membrane protein localized to eisosomes; sporulation and plasma membrane sphingolipid content are altered in mutants; has homologs SUR7 and FMP45; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YOR394W | 0.36 |
PAU21
|
Hypothetical protein |
|
| YDR541C | 0.36 |
Putative dihydrokaempferol 4-reductase |
||
| YEL052W | 0.36 |
AFG1
|
Conserved protein that may act as a chaperone in the degradation of misfolded or unassembled cytochrome c oxidase subunits; localized to matrix face of the mitochondrial inner membrane; member of the AAA family but lacks a protease domain |
|
| YCR005C | 0.36 |
CIT2
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate, peroxisomal isozyme involved in glyoxylate cycle; expression is controlled by Rtg1p and Rtg2p transcription factors |
|
| YLR312C | 0.36 |
QNQ1
|
Protein of unknown function; null mutant quiescent and non-quiescent cells exhibit reduced reproductive capacity |
|
| YCR073W-A | 0.34 |
SOL2
|
Protein with a possible role in tRNA export; shows similarity to 6-phosphogluconolactonase non-catalytic domains but does not exhibit this enzymatic activity; homologous to Sol1p, Sol3p, and Sol4p |
|
| YGR190C | 0.34 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene HIP1/YGR191W |
||
| YOR377W | 0.34 |
ATF1
|
Alcohol acetyltransferase with potential roles in lipid and sterol metabolism; responsible for the major part of volatile acetate ester production during fermentation |
|
| YJR149W | 0.34 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YNL144C | 0.34 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YNL144C is not an essential gene |
||
| YMR250W | 0.33 |
GAD1
|
Glutamate decarboxylase, converts glutamate into gamma-aminobutyric acid (GABA) during glutamate catabolism; involved in response to oxidative stress |
|
| YBR076C-A | 0.33 |
Dubious open reading frame unlikely to encode a protein; partially overlaps verified gene ECM8; identified by fungal homology and RT-PCR |
||
| YGR191W | 0.33 |
HIP1
|
High-affinity histidine permease, also involved in the transport of manganese ions |
|
| YPL156C | 0.33 |
PRM4
|
Pheromone-regulated protein, predicted to have 1 transmembrane segment; transcriptionally regulated by Ste12p during mating and by Cat8p during the diauxic shift |
|
| YDR119W-A | 0.33 |
Putative protein of unknown function |
||
| YCR096C | 0.33 |
HMRA2
|
Silenced copy of a2 at HMR; similarity to Alpha2p; required along with a1p for inhibiting expression of the HO endonuclease in a/alpha HO/HO diploid cells with an active mating-type interconversion system |
|
| YHR211W | 0.33 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YHR048W | 0.32 |
YHK8
|
Presumed antiporter of the DHA1 family of multidrug resistance transporters; contains 12 predicted transmembrane spans; expression of gene is up-regulated in cells exhibiting reduced susceptibility to azoles |
|
| YIR039C | 0.31 |
YPS6
|
Putative GPI-anchored aspartic protease |
|
| YBR117C | 0.31 |
TKL2
|
Transketolase, similar to Tkl1p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YOR152C | 0.31 |
Putative protein of unknown function; has no similarity to any known protein; YOR152C is not an essential gene |
||
| YMR271C | 0.31 |
URA10
|
Minor orotate phosphoribosyltransferase (OPRTase) isozyme that catalyzes the fifth enzymatic step in the de novo biosynthesis of pyrimidines, converting orotate into orotidine-5'-phosphate; major OPRTase encoded by URA5 |
|
| YDR537C | 0.29 |
Dubious open reading frame unlikely to encode a protein, almost completely overlaps verified ORF PAD1/YDR538W |
||
| YDR102C | 0.29 |
Dubious open reading frame; homozygous diploid deletion strain exhibits high budding index |
||
| YLR149C | 0.29 |
Putative protein of unknown function; YLR149C is not an essential gene |
||
| YMR090W | 0.29 |
Putative protein of unknown function with similarity to DTDP-glucose 4,6-dehydratases; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YMR090W is not an essential gene |
||
| YEL006W | 0.29 |
YEA6
|
Putative mitochondrial NAD+ transporter, member of the mitochondrial carrier subfamily (see also YIA6); has putative human ortholog |
|
| YBR054W | 0.29 |
YRO2
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in a phosphorylated state in highly purified mitochondria in high-throughput studies; transcriptionally regulated by Haa1p |
|
| YER076W-A | 0.28 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YER076C |
||
| YER186C | 0.28 |
Putative protein of unknown function |
||
| YDR368W | 0.28 |
YPR1
|
2-methylbutyraldehyde reductase, may be involved in isoleucine catabolism |
|
| YPR027C | 0.28 |
Putative protein of unknown function |
||
| YAR050W | 0.28 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YKL163W | 0.28 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YKR052C | 0.28 |
MRS4
|
Mitochondrial iron transporter of the mitochondrial carrier family (MCF), very similar to and functionally redundant with Mrs3p; functions under low-iron conditions; may transport other cations in addition to iron |
|
| YNR039C | 0.28 |
ZRG17
|
Endoplasmic reticulum protein of unknown function, transcription is induced under conditions of zinc deficiency; mutant phenotype suggests a role in uptake of zinc |
|
| YFR007W | 0.28 |
YFH7
|
Putative kinase with similarity to the phosphoribulokinase/uridine kinase/bacterial pantothenate kinase (PRK/URK/PANK) subfamily of P-loop kinases; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) |
|
| YLR334C | 0.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps a stand-alone long terminal repeat sequence whose presence indicates a retrotransposition event occurred here |
||
| YOR345C | 0.27 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene REV1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YIL059C | 0.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YIL060W |
||
| YJL133C-A | 0.27 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YIL045W | 0.27 |
PIG2
|
Putative type-1 protein phosphatase targeting subunit that tethers Glc7p type-1 protein phosphatase to Gsy2p glycogen synthase |
|
| YBR182C | 0.27 |
SMP1
|
Putative transcription factor involved in regulating the response to osmotic stress; member of the MADS-box family of transcription factors |
|
| YOR391C | 0.27 |
HSP33
|
Possible chaperone and cysteine protease with similarity to E. coli Hsp31 and S. cerevisiae Hsp31p, Hsp32p, and Sno4p; member of the DJ-1/ThiJ/PfpI superfamily, which includes human DJ-1 involved in Parkinson's disease |
|
| YDR014W-A | 0.26 |
HED1
|
Meiosis-specific protein that down-regulates Rad51p-mediated mitotic recombination when the meiotic recombination machinery is impaired; early meiotic gene, transcribed specifically during meiotic prophase |
|
| YEL024W | 0.26 |
RIP1
|
Ubiquinol-cytochrome-c reductase, a Rieske iron-sulfur protein of the mitochondrial cytochrome bc1 complex; transfers electrons from ubiquinol to cytochrome c1 during respiration |
|
| YFL062W | 0.26 |
COS4
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YGR288W | 0.26 |
MAL13
|
MAL-activator protein, part of complex locus MAL1; nonfunctional in genomic reference strain S288C |
|
| YER101C | 0.26 |
AST2
|
Protein that may have a role in targeting of plasma membrane [H+]ATPase (Pma1p) to the plasma membrane, as suggested by analysis of genetic interactions |
|
| YNL237W | 0.26 |
YTP1
|
Probable type-III integral membrane protein of unknown function, has regions of similarity to mitochondrial electron transport proteins |
|
| YLR294C | 0.26 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene ATP14 |
||
| YPL258C | 0.26 |
THI21
|
Hydroxymethylpyrimidine phosphate kinase, involved in the last steps in thiamine biosynthesis; member of a gene family with THI20 and THI22; functionally redundant with Thi20p |
|
| YBR021W | 0.26 |
FUR4
|
Uracil permease, localized to the plasma membrane; expression is tightly regulated by uracil levels and environmental cues |
|
| YLR356W | 0.26 |
Putative protein of unknown function with similarity to SCM4; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR356W is not an essential gene |
||
| YER163C | 0.25 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YKL178C | 0.25 |
STE3
|
Receptor for a factor receptor, transcribed in alpha cells and required for mating by alpha cells, couples to MAP kinase cascade to mediate pheromone response; ligand bound receptors are endocytosed and recycled to the plasma membrane; GPCR |
|
| YNL100W | 0.25 |
AIM37
|
Putative protein of unknown function; non-tagged protein is detected in purified mitochondria; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) and severe growth defect in minimal glycerol media |
|
| YHL040C | 0.25 |
ARN1
|
Transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; responsible for uptake of iron bound to ferrirubin, ferrirhodin, and related siderophores |
|
| YDR034C | 0.25 |
LYS14
|
Transcriptional activator involved in regulation of genes of the lysine biosynthesis pathway; requires 2-aminoadipate semialdehyde as co-inducer |
|
| YEL012W | 0.25 |
UBC8
|
Ubiquitin-conjugating enzyme that negatively regulates gluconeogenesis by mediating the glucose-induced ubiquitination of fructose-1,6-bisphosphatase (FBPase); cytoplasmic enzyme that catalyzes the ubiquitination of histones in vitro |
|
| YPL017C | 0.25 |
IRC15
|
Putative S-adenosylmethionine-dependent methyltransferase of the seven beta-strand family, required for accurate meiotic chromosome segregation; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YLR271W | 0.25 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and the nucleus and is induced in response to the DNA-damaging agent MMS |
||
| YJL221C | 0.25 |
FSP2
|
Protein of unknown function, expression is induced during nitrogen limitation |
|
| YNL142W | 0.25 |
MEP2
|
Ammonium permease involved in regulation of pseudohyphal growth; belongs to a ubiquitous family of cytoplasmic membrane proteins that transport only ammonium (NH4+); expression is under the nitrogen catabolite repression regulation |
|
| YCR097W | 0.25 |
HMRA1
|
Silenced copy of a1 at HMR; homeobox corepressor that interacts with Alpha2p to repress haploid-specific gene transcription in diploid cells |
|
| YML132W | 0.24 |
COS3
|
Protein involved in salt resistance; interacts with sodium:hydrogen antiporter Nha1p; member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YFL011W | 0.24 |
HXT10
|
Putative hexose transporter, expressed at low levels and expression is repressed by glucose |
|
| YGL192W | 0.24 |
IME4
|
Probable mRNA N6-adenosine methyltransferase that is required for IME1 transcript accumulation and for sporulation; expression is induced in starved MATa/MAT alpha diploid cells |
|
| YPR196W | 0.24 |
Putative maltose activator |
||
| YJL220W | 0.24 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YJL221C/FSP2 |
||
| YHL047C | 0.24 |
ARN2
|
Transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; responsible for uptake of iron bound to the siderophore triacetylfusarinine C |
|
| YDR132C | 0.24 |
Putative protein of unknown function |
||
| YBR085C-A | 0.24 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and to the nucleus |
||
| YNR040W | 0.23 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YMR026C | 0.23 |
PEX12
|
C3HC4-type RING-finger peroxisomal membrane peroxin required for peroxisome biogenesis and peroxisomal matrix protein import; forms translocation subcomplex with Pex2p and Pex10p; mutations in human homolog cause peroxisomal disorders |
|
| YKL038W | 0.23 |
RGT1
|
Glucose-responsive transcription factor that regulates expression of several glucose transporter (HXT) genes in response to glucose; binds to promoters and acts both as a transcriptional activator and repressor |
|
| YOL151W | 0.23 |
GRE2
|
3-methylbutanal reductase and NADPH-dependent methylglyoxal reductase (D-lactaldehyde dehydrogenase); stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YAR035W | 0.23 |
YAT1
|
Outer mitochondrial carnitine acetyltransferase, minor ethanol-inducible enzyme involved in transport of activated acyl groups from the cytoplasm into the mitochondrial matrix; phosphorylated |
|
| YMR082C | 0.23 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR152C | 0.23 |
Putative protein of unknown function; YLR152C is not an essential gene |
||
| YKL177W | 0.23 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene STE3 |
||
| YKR011C | 0.23 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus |
||
| YGL193C | 0.23 |
Haploid-specific gene repressed by a1-alpha2, turned off in sir3 null strains, absence enhances the sensitivity of rad52-327 cells to campothecin almost 100-fold |
||
| YNL241C | 0.23 |
ZWF1
|
Glucose-6-phosphate dehydrogenase (G6PD), catalyzes the first step of the pentose phosphate pathway; involved in adapting to oxidatve stress; homolog of the human G6PD which is deficient in patients with hemolytic anemia |
|
| YOR389W | 0.23 |
Putative protein of unknown function; expression regulated by copper levels |
||
| YER076C | 0.22 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; analysis of HA-tagged protein suggests a membrane localization |
||
| YDR036C | 0.22 |
EHD3
|
3-hydroxyisobutyryl-CoA hydrolase, member of a family of enoyl-CoA hydratase/isomerases; non-tagged protein is detected in highly purified mitochondria in high-throughput studies; phosphorylated; mutation affects fluid-phase endocytosis |
|
| YPR202W | 0.22 |
Putative protein of unknown function with similarity to telomere-encoded helicases; YPR202W is not an essential gene; transcript is predicted to be spliced but there is no evidence that it is spliced in vivo |
||
| YDL079C | 0.22 |
MRK1
|
Glycogen synthase kinase 3 (GSK-3) homolog; one of four GSK-3 homologs in S. cerevisiae that function to activate Msn2p-dependent transcription of stress responsive genes and that function in protein degradation |
|
| YLR001C | 0.22 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YLR001C is not an essential gene |
||
| YDR340W | 0.22 |
Putative protein of unknown function |
||
| YMR034C | 0.22 |
Putative transporter, member of the SLC10 carrier family; identified in a transposon mutagenesis screen as a gene involved in azole resistance; YMR034C is not an essential gene |
||
| YDR124W | 0.22 |
Putative protein of unknown function; non-essential gene; expression is strongly induced by alpha factor |
||
| YBR272C | 0.22 |
HSM3
|
Protein of unknown function, involved in DNA mismatch repair during slow growth; has weak similarity to Msh1p |
|
| YPL189W | 0.22 |
GUP2
|
Probable membrane protein with a possible role in proton symport of glycerol; member of the MBOAT family of putative membrane-bound O-acyltransferases; Gup1p homolog |
|
| YLR125W | 0.21 |
Putative protein of unknown function; mutant has decreased Ty3 transposition; YLR125W is not an essential gene |
||
| YFL063W | 0.21 |
Dubious open reading frame, based on available experimental and comparative sequence data |
||
| YMR041C | 0.21 |
ARA2
|
NAD-dependent arabinose dehydrogenase, involved in biosynthesis of erythroascorbic acid; similar to plant L-galactose dehydrogenase |
|
| YNL336W | 0.21 |
COS1
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YCR021C | 0.21 |
HSP30
|
Hydrophobic plasma membrane localized, stress-responsive protein that negatively regulates the H(+)-ATPase Pma1p; induced by heat shock, ethanol treatment, weak organic acid, glucose limitation, and entry into stationary phase |
|
| YNL179C | 0.21 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YKL109W | 0.21 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YNL015W | 0.21 |
PBI2
|
Cytosolic inhibitor of vacuolar proteinase B, required for efficient vacuole inheritance; with thioredoxin forms protein complex LMA1, which assists in priming SNARE molecules and promotes vacuole fusion |
|
| YGL041C-B | 0.21 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YOR049C | 0.21 |
RSB1
|
Suppressor of sphingoid long chain base (LCB) sensitivity of an LCB-lyase mutation; putative integral membrane transporter or flippase that may transport LCBs from the cytoplasmic side toward the extracytoplasmic side of the membrane |
|
| YML089C | 0.20 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression induced by calcium shortage |
||
| YNR071C | 0.20 |
Putative protein of unknown function |
||
| YDR043C | 0.20 |
NRG1
|
Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters; mediates glucose repression and negatively regulates a variety of processes including filamentous growth and alkaline pH response |
|
| YLR228C | 0.20 |
ECM22
|
Sterol regulatory element binding protein, regulates transcription of the sterol biosynthetic genes ERG2 and ERG3; member of the fungus-specific Zn[2]-Cys[6] binuclear cluster family of transcription factors; homologous to Upc2p |
Network of associatons between targets according to the STRING database.
First level regulatory network of YAP6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.3 | 1.3 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.3 | 0.8 | GO:0015755 | fructose transport(GO:0015755) |
| 0.2 | 0.7 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.2 | 1.2 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.2 | 0.6 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 0.2 | 0.2 | GO:0044247 | glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.9 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.1 | 1.1 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.1 | 0.7 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.7 | GO:0032102 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.1 | 0.6 | GO:0009209 | CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) |
| 0.1 | 0.5 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.1 | 0.2 | GO:0046323 | glucose import(GO:0046323) |
| 0.1 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.7 | GO:0015758 | glucose transport(GO:0015758) |
| 0.1 | 1.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.1 | 0.3 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.1 | 0.4 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) |
| 0.1 | 0.1 | GO:0051222 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 0.1 | 0.3 | GO:0042843 | D-xylose catabolic process(GO:0042843) |
| 0.1 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.1 | 0.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.3 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.2 | GO:0045980 | negative regulation of nucleotide metabolic process(GO:0045980) |
| 0.1 | 0.5 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 0.6 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.6 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 1.1 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.5 | GO:0045117 | azole transport(GO:0045117) |
| 0.1 | 0.2 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.1 | 0.6 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.1 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.1 | GO:0051606 | detection of chemical stimulus(GO:0009593) detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) detection of stimulus(GO:0051606) |
| 0.1 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 0.1 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.1 | 0.3 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) thiamine diphosphate metabolic process(GO:0042357) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.2 | GO:0009106 | lipoate metabolic process(GO:0009106) lipoate biosynthetic process(GO:0009107) |
| 0.1 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.2 | GO:0043065 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.1 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 0.2 | GO:0061393 | positive regulation of transcription from RNA polymerase II promoter in response to osmotic stress(GO:0061393) |
| 0.1 | 0.4 | GO:0034762 | regulation of transmembrane transport(GO:0034762) |
| 0.1 | 0.5 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.2 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) regulation of membrane lipid metabolic process(GO:1905038) |
| 0.1 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.4 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.3 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.1 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 | 0.5 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.0 | GO:0001315 | age-dependent response to reactive oxygen species(GO:0001315) cellular age-dependent response to reactive oxygen species(GO:0072353) |
| 0.0 | 0.2 | GO:0051101 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.0 | 0.1 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.0 | 0.2 | GO:2000284 | positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 0.1 | GO:0043200 | response to amino acid(GO:0043200) |
| 0.0 | 0.1 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.0 | 0.0 | GO:0035786 | protein complex oligomerization(GO:0035786) |
| 0.0 | 0.7 | GO:0072593 | reactive oxygen species metabolic process(GO:0072593) |
| 0.0 | 0.2 | GO:0051865 | cellular bud neck septin ring organization(GO:0032186) protein autoubiquitination(GO:0051865) protein localization to bud neck(GO:0097271) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.4 | GO:0045912 | negative regulation of cellular carbohydrate metabolic process(GO:0010677) negative regulation of gluconeogenesis(GO:0045721) negative regulation of carbohydrate metabolic process(GO:0045912) |
| 0.0 | 0.1 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.2 | GO:0007532 | regulation of mating-type specific transcription, DNA-templated(GO:0007532) |
| 0.0 | 0.7 | GO:0001101 | response to acid chemical(GO:0001101) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.5 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 0.1 | GO:0070304 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.0 | 0.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.3 | GO:0043687 | C-terminal protein amino acid modification(GO:0018410) post-translational protein modification(GO:0043687) |
| 0.0 | 0.5 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.6 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.5 | GO:0045991 | carbon catabolite activation of transcription(GO:0045991) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.4 | GO:0015749 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.0 | 0.1 | GO:0006641 | neutral lipid metabolic process(GO:0006638) acylglycerol metabolic process(GO:0006639) triglyceride metabolic process(GO:0006641) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:0035459 | cargo loading into vesicle(GO:0035459) cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.4 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.3 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.0 | 0.7 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.2 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:0032120 | ascospore-type prospore membrane assembly(GO:0032120) membrane biogenesis(GO:0044091) membrane assembly(GO:0071709) |
| 0.0 | 0.1 | GO:0098743 | aggregation of unicellular organisms(GO:0098630) cell aggregation(GO:0098743) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.0 | GO:0015848 | spermidine transport(GO:0015848) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.0 | GO:0015867 | ATP transport(GO:0015867) adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.1 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0031138 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.0 | 0.9 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0000161 | MAPK cascade involved in osmosensory signaling pathway(GO:0000161) |
| 0.0 | 0.4 | GO:1902532 | negative regulation of intracellular signal transduction(GO:1902532) |
| 0.0 | 0.1 | GO:0019933 | cAMP-mediated signaling(GO:0019933) cyclic-nucleotide-mediated signaling(GO:0019935) |
| 0.0 | 0.2 | GO:0000755 | cytogamy(GO:0000755) |
| 0.0 | 0.1 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.1 | GO:0001009 | transcription from RNA polymerase III type 2 promoter(GO:0001009) transcription from a RNA polymerase III hybrid type promoter(GO:0001041) |
| 0.0 | 0.3 | GO:0042724 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0034395 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.0 | 0.0 | GO:0015691 | cadmium ion transport(GO:0015691) |
| 0.0 | 0.1 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.2 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 | 0.1 | GO:0006771 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.0 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0048255 | RNA stabilization(GO:0043489) mRNA stabilization(GO:0048255) |
| 0.0 | 0.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0070096 | mitochondrial outer membrane translocase complex assembly(GO:0070096) |
| 0.0 | 0.1 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.0 | 0.2 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.0 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.0 | GO:0031111 | negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.1 | GO:0035753 | maintenance of DNA trinucleotide repeats(GO:0035753) |
| 0.0 | 0.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0000736 | double-strand break repair via single-strand annealing, removal of nonhomologous ends(GO:0000736) |
| 0.0 | 0.1 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.0 | GO:0071454 | response to anoxia(GO:0034059) cellular response to anoxia(GO:0071454) |
| 0.0 | 0.1 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.0 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.1 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.2 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.0 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.2 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0016559 | mitochondrial fission(GO:0000266) peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) |
| 0.0 | 0.1 | GO:0000321 | re-entry into mitotic cell cycle after pheromone arrest(GO:0000321) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) pantothenate biosynthetic process(GO:0015940) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.7 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.4 | GO:0000817 | COMA complex(GO:0000817) |
| 0.1 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.3 | GO:0044284 | mitochondrial crista junction(GO:0044284) |
| 0.1 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.7 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.2 | GO:0032176 | split septin rings(GO:0032176) cellular bud neck split septin rings(GO:0032177) |
| 0.1 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 2.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0005756 | mitochondrial proton-transporting ATP synthase, central stalk(GO:0005756) proton-transporting ATP synthase, central stalk(GO:0045269) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0030287 | cell wall-bounded periplasmic space(GO:0030287) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.0 | 0.6 | GO:0031226 | integral component of plasma membrane(GO:0005887) intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 4.0 | GO:0000324 | storage vacuole(GO:0000322) lytic vacuole(GO:0000323) fungal-type vacuole(GO:0000324) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 2.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0048188 | histone methyltransferase complex(GO:0035097) Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.0 | 0.1 | GO:0030666 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle(GO:0030139) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.0 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0031307 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.0 | 0.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0044455 | mitochondrial membrane part(GO:0044455) |
| 0.0 | 0.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.6 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.3 | 1.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 0.8 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.2 | 0.9 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 0.2 | 0.6 | GO:0004564 | beta-fructofuranosidase activity(GO:0004564) sucrose alpha-glucosidase activity(GO:0004575) |
| 0.2 | 0.8 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.2 | 0.6 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.2 | 0.6 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.2 | 0.7 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 0.7 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.2 | 0.8 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.7 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.5 | GO:0015603 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.1 | 0.4 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.1 | 0.1 | GO:0015288 | porin activity(GO:0015288) wide pore channel activity(GO:0022829) |
| 0.1 | 0.3 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 0.9 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 0.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.4 | GO:0034318 | alcohol O-acetyltransferase activity(GO:0004026) alcohol O-acyltransferase activity(GO:0034318) |
| 0.1 | 0.3 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 0.5 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.2 | GO:0016406 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.1 | 0.7 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.2 | GO:0008902 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.1 | 0.2 | GO:0030414 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 0.1 | 0.8 | GO:0022838 | substrate-specific channel activity(GO:0022838) |
| 0.1 | 0.6 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.2 | GO:0035591 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.1 | 0.2 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.4 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.7 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 0.5 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 0.4 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.1 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.0 | 0.5 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 0.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.0 | 0.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.3 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.0 | GO:0016722 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions(GO:0016722) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.0 | 0.3 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.3 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.1 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.1 | GO:0097372 | NAD-dependent histone deacetylase activity (H3-K18 specific)(GO:0097372) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.0 | GO:0019902 | phosphatase binding(GO:0019902) protein phosphatase binding(GO:0019903) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0001031 | RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) transcription factor activity, RNA polymerase III promoter sequence-specific binding, TFIIIB recruiting(GO:0001004) transcription factor activity, RNA polymerase III type 1 promoter sequence-specific binding, TFIIIB recruiting(GO:0001005) transcription factor activity, RNA polymerase III type 2 promoter sequence-specific binding, TFIIIB recruiting(GO:0001008) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.0 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.0 | GO:0016435 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.2 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.2 | GO:0019205 | nucleobase-containing compound kinase activity(GO:0019205) |
| 0.0 | 0.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.0 | GO:0004691 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.0 | GO:0016885 | ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.0 | 0.1 | GO:0048256 | 5'-flap endonuclease activity(GO:0017108) flap endonuclease activity(GO:0048256) |
| 0.0 | 0.0 | GO:0015186 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.0 | 0.1 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 0.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 22.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME ACTIVATED TLR4 SIGNALLING | Genes involved in Activated TLR4 signalling |
| 0.1 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 22.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |