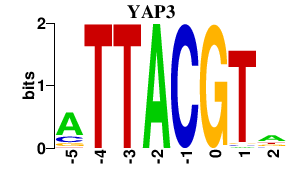
Results for YAP3
Z-value: 0.69
Transcription factors associated with YAP3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
YAP3
|
S000001001 | Basic leucine zipper (bZIP) transcription factor |
Activity-expression correlation:
Activity profile of YAP3 motif
Sorted Z-values of YAP3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YAR053W | 3.08 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YAR060C | 2.54 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR212W-A | 2.53 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YPL223C | 2.03 |
GRE1
|
Hydrophilin of unknown function; stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YLR307C-A | 2.02 |
Putative protein of unknown function |
||
| YHR212C | 2.01 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHR095W | 1.92 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YAL062W | 1.90 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YMR175W | 1.59 |
SIP18
|
Protein of unknown function whose expression is induced by osmotic stress |
|
| YKR039W | 1.42 |
GAP1
|
General amino acid permease; localization to the plasma membrane is regulated by nitrogen source |
|
| YDR043C | 1.38 |
NRG1
|
Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters; mediates glucose repression and negatively regulates a variety of processes including filamentous growth and alkaline pH response |
|
| YMR107W | 1.29 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YLR327C | 1.25 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YOR178C | 1.23 |
GAC1
|
Regulatory subunit for Glc7p type-1 protein phosphatase (PP1), tethers Glc7p to Gsy2p glycogen synthase, binds Hsf1p heat shock transcription factor, required for induction of some HSF-regulated genes under heat shock |
|
| YNL274C | 1.08 |
GOR1
|
Glyoxylate reductase; null mutation results in increased biomass after diauxic shift; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YAR050W | 1.07 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YML128C | 0.96 |
MSC1
|
Protein of unknown function; mutant is defective in directing meiotic recombination events to homologous chromatids; the authentic, non-tagged protein is detected in highly purified mitochondria and is phosphorylated |
|
| YAL063C | 0.94 |
FLO9
|
Lectin-like protein with similarity to Flo1p, thought to be expressed and involved in flocculation |
|
| YAL067W-A | 0.94 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YFR057W | 0.94 |
Putative protein of unknown function |
||
| YPL119C | 0.93 |
DBP1
|
Putative ATP-dependent RNA helicase of the DEAD-box protein family; mutants show reduced stability of the 40S ribosomal subunit scanning through 5' untranslated regions of mRNAs |
|
| YMR206W | 0.93 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YER101C | 0.93 |
AST2
|
Protein that may have a role in targeting of plasma membrane [H+]ATPase (Pma1p) to the plasma membrane, as suggested by analysis of genetic interactions |
|
| YAR047C | 0.90 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YAR035W | 0.90 |
YAT1
|
Outer mitochondrial carnitine acetyltransferase, minor ethanol-inducible enzyme involved in transport of activated acyl groups from the cytoplasm into the mitochondrial matrix; phosphorylated |
|
| YKR040C | 0.88 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YKR041W |
||
| YIL057C | 0.85 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YNL036W | 0.85 |
NCE103
|
Carbonic anhydrase; poorly transcribed under aerobic conditions and at an undetectable level under anaerobic conditions; involved in non-classical protein export pathway |
|
| YMR090W | 0.84 |
Putative protein of unknown function with similarity to DTDP-glucose 4,6-dehydratases; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YMR090W is not an essential gene |
||
| YMR164C | 0.82 |
MSS11
|
Transcription factor involved in regulation of invasive growth and starch degradation; controls the activation of MUC1 and STA2 in response to nutritional signals |
|
| YDL022W | 0.81 |
GPD1
|
NAD-dependent glycerol-3-phosphate dehydrogenase, key enzyme of glycerol synthesis, essential for growth under osmotic stress; expression regulated by high-osmolarity glycerol response pathway; homolog of Gpd2p |
|
| YDL023C | 0.79 |
Dubious open reading frame, unlikely to encode a protein; not conserved in other Saccharomyces species; overlaps the verified gene GPD1; deletion confers sensitivity to GSAO; deletion in cyr1 mutant results in loss of stress resistance |
||
| YGR087C | 0.77 |
PDC6
|
Minor isoform of pyruvate decarboxylase, decarboxylates pyruvate to acetaldehyde, involved in amino acid catabolism; transcription is glucose- and ethanol-dependent, and is strongly induced during sulfur limitation |
|
| YJL116C | 0.77 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YLR366W | 0.74 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR364C-A |
||
| YKL044W | 0.73 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHL040C | 0.72 |
ARN1
|
Transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; responsible for uptake of iron bound to ferrirubin, ferrirhodin, and related siderophores |
|
| YLR270W | 0.72 |
DCS1
|
Non-essential hydrolase involved in mRNA decapping, may function in a feedback mechanism to regulate deadenylation, contains pyrophosphatase activity and a HIT (histidine triad) motif; interacts with neutral trehalase Nth1p |
|
| YDR374C | 0.72 |
Putative protein of unknown function |
||
| YMR013C | 0.71 |
SEC59
|
Dolichol kinase, catalyzes the terminal step in dolichyl monophosphate (Dol-P) biosynthesis; required for viability and for normal rates of lipid intermediate synthesis and protein N-glycosylation |
|
| YNL194C | 0.70 |
Integral membrane protein localized to eisosomes; sporulation and plasma membrane sphingolipid content are altered in mutants; has homologs SUR7 and FMP45; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YBR071W | 0.69 |
Putative protein of unknown function; (GFP)-fusion and epitope-tagged proteins localize to the cytoplasm; mRNA expression may be regulated by the cell cycle and/or cell wall stress |
||
| YMR174C | 0.68 |
PAI3
|
Cytoplasmic proteinase A (Pep4p) inhibitor, dependent on Pbs2p and Hog1p protein kinases for osmotic induction; intrinsically unstructured, N-terminal half becomes ordered in the active site of proteinase A upon contact |
|
| YJL057C | 0.67 |
IKS1
|
Putative serine/threonine kinase; expression is induced during mild heat stress; deletion mutants are hypersensitive to copper sulphate and resistant to sorbate; interacts with an N-terminal fragment of Sst2p |
|
| YJR137C | 0.67 |
ECM17
|
Sulfite reductase beta subunit, involved in amino acid biosynthesis, transcription repressed by methionine |
|
| YOL151W | 0.67 |
GRE2
|
3-methylbutanal reductase and NADPH-dependent methylglyoxal reductase (D-lactaldehyde dehydrogenase); stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YPR005C | 0.66 |
HAL1
|
Cytoplasmic protein involved in halotolerance; decreases intracellular Na+ (via Ena1p) and increases intracellular K+ by decreasing efflux; expression repressed by Ssn6p-Tup1p and Sko1p and induced by NaCl, KCl, and sorbitol through Gcn4p |
|
| YPR007C | 0.66 |
REC8
|
Meiosis-specific component of sister chromatid cohesion complex; maintains cohesion between sister chromatids during meiosis I; maintains cohesion between centromeres of sister chromatids until meiosis II; homolog of S. pombe Rec8p |
|
| YNL142W | 0.65 |
MEP2
|
Ammonium permease involved in regulation of pseudohyphal growth; belongs to a ubiquitous family of cytoplasmic membrane proteins that transport only ammonium (NH4+); expression is under the nitrogen catabolite repression regulation |
|
| YOL126C | 0.65 |
MDH2
|
Cytoplasmic malate dehydrogenase, one of three isozymes that catalyze interconversion of malate and oxaloacetate; involved in the glyoxylate cycle and gluconeogenesis during growth on two-carbon compounds; interacts with Pck1p and Fbp1 |
|
| YEL007W | 0.64 |
Putative protein with sequence similarity to S. pombe gti1+ (gluconate transport inducer 1) |
||
| YLR365W | 0.64 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps dubious gene YLR364C-A; YLR365W is not an essential gene |
||
| YOR192C-C | 0.63 |
Putative protein of unknown function; identified by expression profiling and mass spectrometry |
||
| YJR138W | 0.62 |
IML1
|
Protein of unknown function, green fluorescent protein (GFP)-fusion protein localizes to the vacuolar membrane |
|
| YBR182C | 0.61 |
SMP1
|
Putative transcription factor involved in regulating the response to osmotic stress; member of the MADS-box family of transcription factors |
|
| YAR019W-A | 0.61 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YMR081C | 0.61 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YKL223W | 0.60 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPL222W | 0.59 |
FMP40
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YLR004C | 0.59 |
THI73
|
Putative plasma membrane permease proposed to be involved in carboxylic acid uptake and repressed by thiamine; substrate of Dbf2p/Mob1p kinase; transcription is altered if mitochondrial dysfunction occurs |
|
| YNR002C | 0.58 |
ATO2
|
Putative transmembrane protein involved in export of ammonia, a starvation signal that promotes cell death in aging colonies; phosphorylated in mitochondria; member of the TC 9.B.33 YaaH family; homolog of Ady2p and Y. lipolytica Gpr1p |
|
| YOR394W | 0.57 |
PAU21
|
Hypothetical protein |
|
| YBR076C-A | 0.57 |
Dubious open reading frame unlikely to encode a protein; partially overlaps verified gene ECM8; identified by fungal homology and RT-PCR |
||
| YHR075C | 0.56 |
PPE1
|
Protein with carboxyl methyl esterase activity that may have a role in demethylation of the phosphoprotein phosphatase catalytic subunit; also identified as a small subunit mitochondrial ribosomal protein |
|
| YMR273C | 0.56 |
ZDS1
|
Protein that interacts with silencing proteins at the telomere, involved in transcriptional silencing; has a role in localization of Bcy1p, a regulatory subunit of protein kinase A; implicated in mRNA nuclear export |
|
| YKL224C | 0.56 |
PAU16
|
Putative protein of unknown function |
|
| YLL023C | 0.55 |
Protein of unknown function; highly conserved across species and orthologous to human TMEM33; may interact with ribosomes, based on co-purification experiments; GFP-fusion protein localizes to the endoplasmic reticulum |
||
| YHL041W | 0.55 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YFR053C | 0.55 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YNL143C | 0.54 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR268C | 0.53 |
Putative protein of unknown function; sporulation is abnormal in homozygous diploid; YOR268C is not an essential gene |
||
| YOR062C | 0.53 |
Protein of unknown function; similar to YKR075Cp and Reg1p; expression regulated by glucose and Rgt1p; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YGR144W | 0.53 |
THI4
|
Thiazole synthase, catalyzes formation of the thiazole moiety of thiamin pyrophosphate; required for thiamine biosynthesis and for mitochondrial genome stability |
|
| YDR069C | 0.52 |
DOA4
|
Ubiquitin isopeptidase, required for recycling ubiquitin from proteasome-bound ubiquitinated intermediates, acts at the late endosome/prevacuolar compartment to recover ubiquitin from ubiquitinated membrane proteins en route to the vacuole |
|
| YMR180C | 0.52 |
CTL1
|
RNA 5'-triphosphatase, localizes to both the nucleus and cytoplasm |
|
| YKL016C | 0.52 |
ATP7
|
Subunit d of the stator stalk of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YER181C | 0.52 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative data; extensively overlaps a Ty1 LTR; protein product is detected in highly purified mitochondria in high-throughput studies |
||
| YLR122C | 0.52 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR123C |
||
| YAL061W | 0.51 |
BDH2
|
Putative medium-chain alcohol dehydrogenase with similarity to BDH1; transcription induced by constitutively active PDR1 and PDR3; BDH2 is an essential gene |
|
| YAR023C | 0.51 |
Putative integral membrane protein, member of DUP240 gene family |
||
| YIR041W | 0.50 |
PAU15
|
Hypothetical protein |
|
| YKL161C | 0.50 |
Protein kinase implicated in the Slt2p mitogen-activated (MAP) kinase signaling pathway; associates with Rlm1p |
||
| YPL062W | 0.50 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YPL062W is not an essential gene; homozygous diploid mutant shows a decrease in glycogen accumulation |
||
| YMR105C | 0.49 |
PGM2
|
Phosphoglucomutase, catalyzes the conversion from glucose-1-phosphate to glucose-6-phosphate, which is a key step in hexose metabolism; functions as the acceptor for a Glc-phosphotransferase |
|
| YCR068W | 0.49 |
ATG15
|
Lipase, required for intravacuolar lysis of autophagosomes; located in the endoplasmic reticulum membrane and targeted to intravacuolar vesicles during autophagy via the multivesicular body (MVB) pathway |
|
| YLR251W | 0.49 |
SYM1
|
Protein required for ethanol metabolism; induced by heat shock and localized to the inner mitochondrial membrane; homologous to mammalian peroxisomal membrane protein Mpv17 |
|
| YNL144C | 0.49 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YNL144C is not an essential gene |
||
| YNR034W-A | 0.49 |
Putative protein of unknown function; expression is regulated by Msn2p/Msn4p |
||
| YFR026C | 0.48 |
Putative protein of unknown function |
||
| YOR161C | 0.47 |
PNS1
|
Protein of unknown function; has similarity to Torpedo californica tCTL1p, which is postulated to be a choline transporter, neither null mutation nor overexpression affects choline transport |
|
| YAL054C | 0.47 |
ACS1
|
Acetyl-coA synthetase isoform which, along with Acs2p, is the nuclear source of acetyl-coA for histone acetlyation; expressed during growth on nonfermentable carbon sources and under aerobic conditions |
|
| YDL169C | 0.47 |
UGX2
|
Protein of unknown function, transcript accumulates in response to any combination of stress conditions |
|
| YLR356W | 0.46 |
Putative protein of unknown function with similarity to SCM4; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR356W is not an essential gene |
||
| YER167W | 0.46 |
BCK2
|
Protein rich in serine and threonine residues involved in protein kinase C signaling pathway, which controls cell integrity; overproduction suppresses pkc1 mutations |
|
| YML012C-A | 0.46 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene SEL1 |
||
| YER188W | 0.45 |
Hypothetical protein |
||
| YPR192W | 0.45 |
AQY1
|
Spore-specific water channel that mediates the transport of water across cell membranes, developmentally controlled; may play a role in spore maturation, probably by allowing water outflow, may be involved in freeze tolerance |
|
| YEL060C | 0.45 |
PRB1
|
Vacuolar proteinase B (yscB), a serine protease of the subtilisin family; involved in protein degradation in the vacuole and required for full protein degradation during sporulation |
|
| YPR015C | 0.45 |
Putative protein of unknown function |
||
| YIL108W | 0.45 |
Putative metalloprotease |
||
| YEL008W | 0.45 |
Hypothetical protein predicted to be involved in metabolism |
||
| YLL030C | 0.45 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR211W | 0.44 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YLR123C | 0.44 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR122C; contains characteristic aminoacyl-tRNA motif |
||
| YPR065W | 0.44 |
ROX1
|
Heme-dependent repressor of hypoxic genes; contains an HMG domain that is responsible for DNA bending activity |
|
| YEL059W | 0.44 |
Dubious open reading frame unlikely to encode a functional protein |
||
| YGL262W | 0.44 |
Putative protein of unknown function; null mutant displays elevated sensitivity to expression of a mutant huntingtin fragment or of alpha-synuclein; YGL262W is not an essential gene |
||
| YBR169C | 0.43 |
SSE2
|
Member of the heat shock protein 70 (HSP70) family; may be involved in protein folding; localized to the cytoplasm; highly homologous to the heat shock protein Sse1p |
|
| YBR147W | 0.43 |
RTC2
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays fluconazole resistance and suppresses cdc13-1 temperature sensitivity |
|
| YFR027W | 0.43 |
ECO1
|
Acetyltransferase required for the establishment of sister chromatid cohesion during DNA replication and in response to double-strand breaks; also required for postreplicative double-strand break repair |
|
| YMR068W | 0.43 |
AVO2
|
Component of a complex containing the Tor2p kinase and other proteins, which may have a role in regulation of cell growth |
|
| YMR169C | 0.43 |
ALD3
|
Cytoplasmic aldehyde dehydrogenase, involved in beta-alanine synthesis; uses NAD+ as the preferred coenzyme; very similar to Ald2p; expression is induced by stress and repressed by glucose |
|
| YPL080C | 0.43 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YBL078C | 0.42 |
ATG8
|
Conserved protein that is a component of autophagosomes and Cvt vesicles; undergoes C-terminal conjugation to phosphatidylethanolamine (PE), Atg8p-PE is anchored to membranes and may mediate membrane fusion during autophagosome formation |
|
| YFR016C | 0.41 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and bud; interacts with Spa2p; YFL016C is not an essential gene |
||
| YJR159W | 0.41 |
SOR1
|
Sorbitol dehydrogenase; expression is induced in the presence of sorbitol |
|
| YAR019C | 0.41 |
CDC15
|
Protein kinase of the Mitotic Exit Network that is localized to the spindle pole bodies at late anaphase; promotes mitotic exit by directly switching on the kinase activity of Dbf2p |
|
| YBL049W | 0.41 |
MOH1
|
Protein of unknown function, has homology to kinase Snf7p; not required for growth on nonfermentable carbon sources; essential for viability in stationary phase |
|
| YBL064C | 0.41 |
PRX1
|
Mitochondrial peroxiredoxin (1-Cys Prx) with thioredoxin peroxidase activity, has a role in reduction of hydroperoxides; induced during respiratory growth and under conditions of oxidative stress; phosphorylated |
|
| YFL054C | 0.40 |
Putative channel-like protein; similar to Fps1p; mediates passive diffusion of glycerol in the presence of ethanol |
||
| YOL164W-A | 0.40 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YGR149W | 0.40 |
Putative protein of unknown function; predicted to be an integal membrane protein |
||
| YIL119C | 0.40 |
RPI1
|
Putative transcriptional regulator; overexpression suppresses the heat shock sensitivity of wild-type RAS2 overexpression and also suppresses the cell lysis defect of an mpk1 mutation |
|
| YBR292C | 0.39 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR292C is not an essential gene |
||
| YER103W | 0.39 |
SSA4
|
Heat shock protein that is highly induced upon stress; plays a role in SRP-dependent cotranslational protein-membrane targeting and translocation; member of the HSP70 family; cytoplasmic protein that concentrates in nuclei upon starvation |
|
| YML131W | 0.39 |
Putative protein of unknown function with similarity to oxidoreductases; HOG1 and SKO1-dependent mRNA expression is induced after osmotic shock; GFP-fusion protein localizes to the cytoplasm and is induced by the DNA-damaging agent MMS |
||
| YDL222C | 0.39 |
FMP45
|
Integral membrane protein localized to mitochondria (untagged protein) and eisosomes, immobile patches at the cortex associated with endocytosis; sporulation and sphingolipid content are altered in mutants; has homologs SUR7 and YNL194C |
|
| YJR095W | 0.39 |
SFC1
|
Mitochondrial succinate-fumarate transporter, transports succinate into and fumarate out of the mitochondrion; required for ethanol and acetate utilization |
|
| YLR352W | 0.38 |
Putative protein of unknown function with similarity to F-box proteins; interacts with Skp1p and Cdc53p; YLR352W is not an essential gene |
||
| YDL214C | 0.38 |
PRR2
|
Serine/threonine protein kinase that inhibits pheromone induced signalling downstream of MAPK, possibly at the level of the Ste12p transcription factor |
|
| YLR273C | 0.38 |
PIG1
|
Putative targeting subunit for the type-1 protein phosphatase Glc7p that tethers it to the Gsy2p glycogen synthase |
|
| YHR125W | 0.37 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPR036W-A | 0.37 |
Protein of unknown function; transcription is regulated by Pdr1p |
||
| YJL089W | 0.37 |
SIP4
|
C6 zinc cluster transcriptional activator that binds to the carbon source-responsive element (CSRE) of gluconeogenic genes; involved in the positive regulation of gluconeogenesis; regulated by Snf1p protein kinase; localized to the nucleus |
|
| YJL005W | 0.36 |
CYR1
|
Adenylate cyclase, required for cAMP production and cAMP-dependent protein kinase signaling; the cAMP pathway controls a variety of cellular processes, including metabolism, cell cycle, stress response, stationary phase, and sporulation |
|
| YER150W | 0.36 |
SPI1
|
GPI-anchored cell wall protein involved in weak acid resistance; basal expression requires Msn2p/Msn4p; expression is induced under conditions of stress and during the diauxic shift; similar to Sed1p |
|
| YDL085W | 0.36 |
NDE2
|
Mitochondrial external NADH dehydrogenase, catalyzes the oxidation of cytosolic NADH; Nde1p and Nde2p are involved in providing the cytosolic NADH to the mitochondrial respiratory chain |
|
| YAL066W | 0.35 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHL027W | 0.35 |
RIM101
|
Transcriptional repressor involved in response to pH and in cell wall construction; required for alkaline pH-stimulated haploid invasive growth and sporulation; activated by proteolytic processing; similar to A. nidulans PacC |
|
| YHR033W | 0.35 |
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm |
||
| YGR287C | 0.35 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; has similarity to alpha-D-glucosidase (maltase); authentic, non-tagged protein detected in purified mitochondria in high-throughput studies |
||
| YLR124W | 0.34 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YAL067C | 0.34 |
SEO1
|
Putative permease, member of the allantoate transporter subfamily of the major facilitator superfamily; mutation confers resistance to ethionine sulfoxide |
|
| YLR296W | 0.34 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YNL025C | 0.34 |
SSN8
|
Cyclin-like component of the RNA polymerase II holoenzyme, involved in phosphorylation of the RNA polymerase II C-terminal domain; involved in glucose repression and telomere maintenance |
|
| YIL045W | 0.34 |
PIG2
|
Putative type-1 protein phosphatase targeting subunit that tethers Glc7p type-1 protein phosphatase to Gsy2p glycogen synthase |
|
| YDR461C-A | 0.34 |
Putative protein of unknown function |
||
| YIR040C | 0.34 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YKL107W | 0.34 |
Putative protein of unknown function |
||
| YKL124W | 0.34 |
SSH4
|
Specificity factor required for Rsp5p-dependent ubiquitination and sorting of cargo proteins at the multivesicular body; identified as a high-copy suppressor of a SHR3 deletion, increasing steady-state levels of amino acid permeases |
|
| YOR376W | 0.34 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YOR376W is not an essential gene. |
||
| YDR540C | 0.33 |
IRC4
|
Putative protein of unknown function; null mutant displays increased levels of spontaneous Rad52p foci; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
|
| YGR289C | 0.33 |
MAL11
|
Inducible high-affinity maltose transporter (alpha-glucoside transporter); encoded in the MAL1 complex locus; member of the 12 transmembrane domain superfamily of sugar transporters; broad substrate specificity that includes maltotriose |
|
| YDR077W | 0.33 |
SED1
|
Major stress-induced structural GPI-cell wall glycoprotein in stationary-phase cells, associates with translating ribosomes, possible role in mitochondrial genome maintenance; ORF contains two distinct variable minisatellites |
|
| YFL053W | 0.33 |
DAK2
|
Dihydroxyacetone kinase, required for detoxification of dihydroxyacetone (DHA); involved in stress adaptation |
|
| YJL130C | 0.33 |
URA2
|
Bifunctional carbamoylphosphate synthetase (CPSase)-aspartate transcarbamylase (ATCase), catalyzes the first two enzymatic steps in the de novo biosynthesis of pyrimidines; both activities are subject to feedback inhibition by UTP |
|
| YHR210C | 0.33 |
Putative protein of unknown function; non-essential gene; highly expressed under anaeorbic conditions; sequence similarity to aldose 1-epimerases such as GAL10 |
||
| YLR121C | 0.33 |
YPS3
|
Aspartic protease, attached to the plasma membrane via a glycosylphosphatidylinositol (GPI) anchor |
|
| YDR141C | 0.33 |
DOP1
|
Protein of unknown function, essential for viability, involved in establishing cellular polarity and morphogenesis; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YPL258C | 0.33 |
THI21
|
Hydroxymethylpyrimidine phosphate kinase, involved in the last steps in thiamine biosynthesis; member of a gene family with THI20 and THI22; functionally redundant with Thi20p |
|
| YLR425W | 0.32 |
TUS1
|
Guanine nucleotide exchange factor (GEF) that functions to modulate Rho1p activity as part of the cell integrity signaling pathway; multicopy suppressor of tor2 mutation and ypk1 ypk2 double mutation; potential Cdc28p substrate |
|
| YGR292W | 0.32 |
MAL12
|
Maltase (alpha-D-glucosidase), inducible protein involved in maltose catabolism; encoded in the MAL1 complex locus |
|
| YDR171W | 0.32 |
HSP42
|
Small heat shock protein (sHSP) with chaperone activity; forms barrel-shaped oligomers that suppress unfolded protein aggregation; involved in cytoskeleton reorganization after heat shock |
|
| YBL067C | 0.32 |
UBP13
|
Putative ubiquitin carboxyl-terminal hydrolase, ubiquitin-specific protease that cleaves ubiquitin-protein fusions |
|
| YFL033C | 0.32 |
RIM15
|
Glucose-repressible protein kinase involved in signal transduction during cell proliferation in response to nutrients, specifically the establishment of stationary phase; identified as a regulator of IME2; substrate of Pho80p-Pho85p kinase |
|
| YCR091W | 0.32 |
KIN82
|
Putative serine/threonine protein kinase, most similar to cyclic nucleotide-dependent protein kinase subfamily and the protein kinase C subfamily |
|
| YNL037C | 0.32 |
IDH1
|
Subunit of mitochondrial NAD(+)-dependent isocitrate dehydrogenase, which catalyzes the oxidation of isocitrate to alpha-ketoglutarate in the TCA cycle |
|
| YCR021C | 0.32 |
HSP30
|
Hydrophobic plasma membrane localized, stress-responsive protein that negatively regulates the H(+)-ATPase Pma1p; induced by heat shock, ethanol treatment, weak organic acid, glucose limitation, and entry into stationary phase |
|
| YKL216W | 0.31 |
URA1
|
Dihydroorotate dehydrogenase, catalyzes the fourth enzymatic step in the de novo biosynthesis of pyrimidines, converting dihydroorotic acid into orotic acid |
|
| YML089C | 0.31 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression induced by calcium shortage |
||
| YPL278C | 0.31 |
Putative protein of unknown function; gene expression regulated by copper levels |
||
| YGR039W | 0.31 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the Autonomously Replicating Sequence ARS722 |
||
| YKL028W | 0.31 |
TFA1
|
TFIIE large subunit, involved in recruitment of RNA polymerase II to the promoter, activation of TFIIH, and promoter opening |
|
| YOR177C | 0.31 |
MPC54
|
Component of the meiotic outer plaque, a membrane-organizing center which is assembled on the cytoplasmic face of the spindle pole body during meiosis II and triggers the formation of the prospore membrane; potential Cdc28p substrate |
|
| YEL069C | 0.30 |
HXT13
|
Hexose transporter, induced in the presence of non-fermentable carbon sources, induced by low levels of glucose, repressed by high levels of glucose |
|
| YPR202W | 0.30 |
Putative protein of unknown function with similarity to telomere-encoded helicases; YPR202W is not an essential gene; transcript is predicted to be spliced but there is no evidence that it is spliced in vivo |
||
| YER187W | 0.30 |
Putative protein of unknown function; induced in respiratory-deficient cells |
||
| YGR290W | 0.30 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; putative HLH protein; partially overlaps the verified ORF MAL11/YGR289C (a high-affinity maltose transporter) |
||
| YCR106W | 0.30 |
RDS1
|
Zinc cluster transcription factor involved in conferring resistance to cycloheximide |
|
| YOR192C | 0.30 |
THI72
|
Transporter of thiamine or related compound; shares sequence similarity with Thi7p |
|
| YPR010C-A | 0.29 |
Putative protein of unknown function; conserved among Saccharomyces sensu stricto species |
||
| YOR152C | 0.29 |
Putative protein of unknown function; has no similarity to any known protein; YOR152C is not an essential gene |
||
| YLR269C | 0.29 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR386W | 0.29 |
PHR1
|
DNA photolyase involved in photoreactivation, repairs pyrimidine dimers in the presence of visible light; induced by DNA damage; regulated by transcriptional repressor Rph1p |
|
| YDR222W | 0.29 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
||
| YGL071W | 0.29 |
AFT1
|
Transcription factor involved in iron utilization and homeostasis; binds the consensus site PyPuCACCCPu and activates the expression of target genes in response to changes in iron availability |
|
| YGL163C | 0.29 |
RAD54
|
DNA-dependent ATPase, stimulates strand exchange by modifying the topology of double-stranded DNA; involved in the recombinational repair of double-strand breaks in DNA during vegetative growth and meiosis; member of the SWI/SNF family |
|
| YCR089W | 0.28 |
FIG2
|
Cell wall adhesin, expressed specifically during mating; may be involved in maintenance of cell wall integrity during mating |
|
| YKL115C | 0.28 |
Dubious open reading frame, unlikely to encode a protein; partially overlaps the verified gene PRR1 |
||
| YPR064W | 0.28 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YLR456W | 0.28 |
Putative protein of unknown function; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YPR145W | 0.28 |
ASN1
|
Asparagine synthetase, isozyme of Asn2p; catalyzes the synthesis of L-asparagine from L-aspartate in the asparagine biosynthetic pathway |
|
| YHL045W | 0.28 |
Putative protein of unknown function; not an essential gene |
||
| YDR374W-A | 0.28 |
Putative protein of unknown function |
||
| YOR199W | 0.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YNL273W | 0.27 |
TOF1
|
Subunit of a replication-pausing checkpoint complex (Tof1p-Mrc1p-Csm3p) that acts at the stalled replication fork to promote sister chromatid cohesion after DNA damage, facilitating gap repair of damaged DNA; interacts with the MCM helicase |
|
| YCR066W | 0.27 |
RAD18
|
Protein involved in postreplication repair; binds single-stranded DNA and has single-stranded DNA dependent ATPase activity; forms heterodimer with Rad6p; contains RING-finger motif |
|
| YOL089C | 0.27 |
HAL9
|
Putative transcription factor containing a zinc finger; overexpression increases salt tolerance through increased expression of the ENA1 (Na+/Li+ extrusion pump) gene while gene disruption decreases both salt tolerance and ENA1 expression |
|
| YJL150W | 0.27 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YCR007C | 0.27 |
Putative integral membrane protein, member of DUP240 gene family; YCR007C is not an essential gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of YAP3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.5 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.5 | 1.4 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.4 | 1.3 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.4 | 1.5 | GO:0009415 | response to water(GO:0009415) cellular response to water stimulus(GO:0071462) |
| 0.3 | 0.9 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.2 | 0.7 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.2 | 0.9 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.2 | 0.7 | GO:0052547 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 0.2 | 1.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.2 | 0.7 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.2 | 0.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 0.8 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.2 | 1.2 | GO:0032069 | regulation of nuclease activity(GO:0032069) |
| 0.2 | 0.8 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.2 | 0.5 | GO:0015755 | fructose transport(GO:0015755) |
| 0.2 | 0.5 | GO:0036257 | multivesicular body organization(GO:0036257) |
| 0.2 | 0.3 | GO:0010446 | response to alkaline pH(GO:0010446) cellular response to alkaline pH(GO:0071469) |
| 0.2 | 3.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.1 | 0.3 | GO:0043270 | positive regulation of ion transport(GO:0043270) |
| 0.1 | 0.5 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) |
| 0.1 | 0.9 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.4 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.1 | 0.4 | GO:0006067 | ethanol metabolic process(GO:0006067) |
| 0.1 | 0.6 | GO:0061393 | positive regulation of transcription from RNA polymerase II promoter in response to osmotic stress(GO:0061393) |
| 0.1 | 0.4 | GO:0061414 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 0.1 | 0.4 | GO:0019405 | alditol catabolic process(GO:0019405) glycerol catabolic process(GO:0019563) |
| 0.1 | 1.3 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.1 | 0.6 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.4 | GO:0001324 | age-dependent general metabolic decline involved in chronological cell aging(GO:0001323) age-dependent response to oxidative stress involved in chronological cell aging(GO:0001324) |
| 0.1 | 0.4 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.1 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.4 | GO:0031135 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.1 | 0.4 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.1 | 0.1 | GO:1900544 | positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.1 | 0.3 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.6 | GO:0090294 | nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) nitrogen catabolite activation of transcription(GO:0090294) |
| 0.1 | 0.4 | GO:0006171 | cAMP biosynthetic process(GO:0006171) cyclic nucleotide biosynthetic process(GO:0009190) cyclic purine nucleotide metabolic process(GO:0052652) |
| 0.1 | 0.3 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.3 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.1 | 0.4 | GO:0044746 | amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 0.1 | 0.7 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 0.3 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.1 | 0.3 | GO:0042357 | thiamine diphosphate biosynthetic process(GO:0009229) thiamine diphosphate metabolic process(GO:0042357) |
| 0.1 | 0.2 | GO:0010039 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.1 | 0.2 | GO:0042744 | hydrogen peroxide metabolic process(GO:0042743) hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.2 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.1 | 0.4 | GO:1902707 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.1 | 0.9 | GO:0009228 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.1 | 0.6 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.1 | 0.2 | GO:0032874 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.1 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.2 | GO:0043388 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.1 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.0 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.1 | GO:0044247 | glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.3 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.1 | 0.4 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.2 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 0.4 | GO:0070058 | tRNA gene clustering(GO:0070058) |
| 0.1 | 0.3 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.5 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 0.2 | GO:0019320 | hexose catabolic process(GO:0019320) |
| 0.1 | 0.2 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 0.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) peptidyl-serine modification(GO:0018209) |
| 0.1 | 0.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.8 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.1 | 0.2 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 0.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0006078 | (1->6)-beta-D-glucan biosynthetic process(GO:0006078) beta-glucan biosynthetic process(GO:0051274) |
| 0.0 | 0.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.3 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) positive regulation of fatty acid beta-oxidation(GO:0032000) regulation of fatty acid oxidation(GO:0046320) positive regulation of fatty acid oxidation(GO:0046321) |
| 0.0 | 0.2 | GO:0070542 | response to lipid(GO:0033993) response to oleic acid(GO:0034201) response to fatty acid(GO:0070542) cellular response to lipid(GO:0071396) cellular response to fatty acid(GO:0071398) cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.3 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.4 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.0 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.0 | 0.3 | GO:0045764 | positive regulation of cellular amino acid metabolic process(GO:0045764) |
| 0.0 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:0045996 | negative regulation of transcription by pheromones(GO:0045996) negative regulation of transcription from RNA polymerase II promoter by pheromones(GO:0046020) |
| 0.0 | 0.2 | GO:0000411 | regulation of transcription by galactose(GO:0000409) positive regulation of transcription by galactose(GO:0000411) |
| 0.0 | 0.4 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.3 | GO:0006641 | neutral lipid metabolic process(GO:0006638) acylglycerol metabolic process(GO:0006639) triglyceride metabolic process(GO:0006641) |
| 0.0 | 0.2 | GO:0060237 | regulation of fungal-type cell wall organization(GO:0060237) |
| 0.0 | 0.2 | GO:0070814 | hydrogen sulfide metabolic process(GO:0070813) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:0060195 | antisense RNA transcription(GO:0009300) regulation of antisense RNA transcription(GO:0060194) negative regulation of antisense RNA transcription(GO:0060195) |
| 0.0 | 0.8 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.2 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.0 | 0.2 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0006183 | GTP biosynthetic process(GO:0006183) GTP metabolic process(GO:0046039) |
| 0.0 | 0.1 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.0 | 0.3 | GO:0022610 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 0.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.3 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.0 | 0.3 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.0 | 0.2 | GO:0032443 | regulation of steroid metabolic process(GO:0019218) regulation of ergosterol biosynthetic process(GO:0032443) regulation of steroid biosynthetic process(GO:0050810) |
| 0.0 | 0.6 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.2 | GO:0048209 | regulation of COPII vesicle coating(GO:0003400) regulation of vesicle targeting, to, from or within Golgi(GO:0048209) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.1 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.6 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 0.1 | GO:0061412 | positive regulation of transcription from RNA polymerase II promoter in response to amino acid starvation(GO:0061412) |
| 0.0 | 0.2 | GO:0034965 | intronic snoRNA processing(GO:0031070) intronic box C/D snoRNA processing(GO:0034965) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 1.3 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 0.0 | 0.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.6 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0030048 | actin filament-based movement(GO:0030048) actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0000916 | actomyosin contractile ring contraction(GO:0000916) contractile ring contraction(GO:0036213) |
| 0.0 | 0.0 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.1 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.0 | 0.1 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.3 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 0.1 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0042868 | antisense RNA metabolic process(GO:0042868) antisense RNA transcript catabolic process(GO:0071041) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.0 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0000114 | obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of cellular carbohydrate metabolic process(GO:0010677) negative regulation of gluconeogenesis(GO:0045721) negative regulation of carbohydrate metabolic process(GO:0045912) |
| 0.0 | 0.0 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0035753 | maintenance of DNA trinucleotide repeats(GO:0035753) |
| 0.0 | 0.3 | GO:0030474 | spindle pole body duplication(GO:0030474) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0051259 | protein oligomerization(GO:0051259) |
| 0.0 | 0.1 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.0 | GO:0051222 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 0.0 | 0.0 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0006031 | aminoglycan biosynthetic process(GO:0006023) chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.0 | 0.7 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.2 | GO:0046854 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0032258 | CVT pathway(GO:0032258) |
| 0.0 | 0.0 | GO:0042173 | regulation of sporulation resulting in formation of a cellular spore(GO:0042173) |
| 0.0 | 0.7 | GO:0009272 | fungal-type cell wall biogenesis(GO:0009272) |
| 0.0 | 0.2 | GO:0001101 | response to acid chemical(GO:0001101) |
| 0.0 | 0.0 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.0 | GO:0043207 | immune effector process(GO:0002252) immune system process(GO:0002376) response to virus(GO:0009615) response to external biotic stimulus(GO:0043207) defense response to virus(GO:0051607) response to other organism(GO:0051707) defense response to other organism(GO:0098542) |
| 0.0 | 0.1 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0006276 | plasmid maintenance(GO:0006276) |
| 0.0 | 0.1 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.0 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.0 | GO:0042773 | respiratory electron transport chain(GO:0022904) ATP synthesis coupled electron transport(GO:0042773) mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.1 | GO:0009743 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.1 | GO:0043487 | regulation of RNA stability(GO:0043487) regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.4 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.0 | 0.0 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.0 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.0 | GO:0042182 | ketone catabolic process(GO:0042182) L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.0 | 0.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.3 | 0.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 0.5 | GO:0000274 | mitochondrial proton-transporting ATP synthase, stator stalk(GO:0000274) proton-transporting ATP synthase, stator stalk(GO:0045265) |
| 0.2 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 0.6 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.1 | 0.4 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.1 | 0.5 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.3 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.1 | 0.2 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.1 | 0.2 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.1 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.5 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.4 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.0 | 0.4 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.1 | GO:0044426 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0033309 | SBF transcription complex(GO:0033309) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0032221 | Rpd3S complex(GO:0032221) |
| 0.0 | 0.3 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.0 | 0.2 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.0 | GO:0001400 | mating projection base(GO:0001400) |
| 0.0 | 0.2 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0032806 | carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 0.1 | GO:0035361 | Cul8-RING ubiquitin ligase complex(GO:0035361) |
| 0.0 | 0.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.0 | 0.2 | GO:0000142 | cellular bud neck contractile ring(GO:0000142) |
| 0.0 | 0.1 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 0.0 | 0.4 | GO:0042764 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 1.6 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.0 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.0 | 0.6 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.1 | GO:0005823 | central plaque of spindle pole body(GO:0005823) |
| 0.0 | 0.0 | GO:0031391 | Elg1 RFC-like complex(GO:0031391) |
| 0.0 | 0.1 | GO:0098552 | side of membrane(GO:0098552) cytoplasmic side of membrane(GO:0098562) |
| 0.0 | 0.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.7 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.5 | GO:0000131 | incipient cellular bud site(GO:0000131) |
| 0.0 | 1.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.0 | GO:0051233 | spindle midzone(GO:0051233) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.4 | 1.6 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.3 | 0.9 | GO:0016406 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.2 | 0.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 0.2 | 0.8 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 0.2 | 0.5 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.2 | 0.7 | GO:0015603 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.2 | 1.4 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.2 | 0.5 | GO:0016880 | AMP binding(GO:0016208) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.4 | GO:0042947 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.1 | 0.7 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.6 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.1 | 0.7 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 0.4 | GO:0008972 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.1 | 1.4 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.4 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.6 | GO:0050253 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 1.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.3 | GO:0004564 | beta-fructofuranosidase activity(GO:0004564) sucrose alpha-glucosidase activity(GO:0004575) |
| 0.1 | 0.3 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.3 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.1 | 0.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.4 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.1 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.3 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.1 | 0.3 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.1 | 0.2 | GO:0004805 | trehalose-phosphatase activity(GO:0004805) |
| 0.1 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.5 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.3 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.1 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.2 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.1 | 1.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 0.4 | GO:0015294 | solute:cation symporter activity(GO:0015294) |
| 0.1 | 1.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 0.2 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.3 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.1 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.5 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 1.7 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.3 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 0.3 | GO:0015085 | calcium ion transmembrane transporter activity(GO:0015085) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.3 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.2 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.0 | 0.1 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.0 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.0 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0019212 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.1 | GO:0042736 | NAD+ kinase activity(GO:0003951) NADH kinase activity(GO:0042736) |
| 0.0 | 0.1 | GO:0008897 | holo-[acyl-carrier-protein] synthase activity(GO:0008897) |
| 0.0 | 0.2 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0016684 | peroxidase activity(GO:0004601) oxidoreductase activity, acting on peroxide as acceptor(GO:0016684) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 1.7 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 0.5 | GO:0050662 | coenzyme binding(GO:0050662) |
| 0.0 | 0.1 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.0 | 0.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.2 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 1.2 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0015079 | potassium ion transmembrane transporter activity(GO:0015079) |
| 0.0 | 0.1 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.6 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.0 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0019787 | ubiquitin-like protein transferase activity(GO:0019787) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 0.2 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.0 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 34.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.1 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 1.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.2 | 0.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 0.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.7 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.2 | REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | Genes involved in E2F mediated regulation of DNA replication |
| 0.0 | 0.0 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 34.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.1 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |