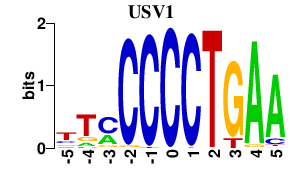
Results for USV1
Z-value: 0.73
Transcription factors associated with USV1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
USV1
|
S000006151 | Putative transcription factor containing a C2H2 zinc finger |
Activity-expression correlation:
Activity profile of USV1 motif
Sorted Z-values of USV1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YPR160W | 3.34 |
GPH1
|
Non-essential glycogen phosphorylase required for the mobilization of glycogen, activity is regulated by cyclic AMP-mediated phosphorylation, expression is regulated by stress-response elements and by the HOG MAP kinase pathway |
|
| YIL162W | 2.73 |
SUC2
|
Invertase, sucrose hydrolyzing enzyme; a secreted, glycosylated form is regulated by glucose repression, and an intracellular, nonglycosylated enzyme is produced constitutively |
|
| YFR053C | 2.72 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YHL021C | 2.41 |
AIM17
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) |
|
| YFR015C | 2.25 |
GSY1
|
Glycogen synthase with similarity to Gsy2p, the more highly expressed yeast homolog; expression induced by glucose limitation, nitrogen starvation, environmental stress, and entry into stationary phase |
|
| YER067W | 1.87 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YER067W is not an essential gene |
||
| YEL039C | 1.69 |
CYC7
|
Cytochrome c isoform 2, expressed under hypoxic conditions; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YDR516C | 1.64 |
EMI2
|
Non-essential protein of unknown function required for transcriptional induction of the early meiotic-specific transcription factor IME1; required for sporulation; expression is regulated by glucose-repression transcription factors Mig1/2p |
|
| YOL154W | 1.63 |
ZPS1
|
Putative GPI-anchored protein; transcription is induced under low-zinc conditions, as mediated by the Zap1p transcription factor, and at alkaline pH |
|
| YBR072W | 1.57 |
HSP26
|
Small heat shock protein (sHSP) with chaperone activity; forms hollow, sphere-shaped oligomers that suppress unfolded proteins aggregation; oligomer activation requires a heat-induced conformational change; not expressed in unstressed cells |
|
| YLR258W | 1.54 |
GSY2
|
Glycogen synthase, similar to Gsy1p; expression induced by glucose limitation, nitrogen starvation, heat shock, and stationary phase; activity regulated by cAMP-dependent, Snf1p and Pho85p kinases as well as by the Gac1p-Glc7p phosphatase |
|
| YCR021C | 1.50 |
HSP30
|
Hydrophobic plasma membrane localized, stress-responsive protein that negatively regulates the H(+)-ATPase Pma1p; induced by heat shock, ethanol treatment, weak organic acid, glucose limitation, and entry into stationary phase |
|
| YNL045W | 1.38 |
Leucyl aminopeptidase (leukotriene A4 hydrolase) with epoxide hydrolase activity, metalloenzyme containing one zinc atom; role in vivo is not defined; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YFL054C | 1.22 |
Putative channel-like protein; similar to Fps1p; mediates passive diffusion of glycerol in the presence of ethanol |
||
| YGR254W | 1.19 |
ENO1
|
Enolase I, a phosphopyruvate hydratase that catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate during glycolysis and the reverse reaction during gluconeogenesis; expression is repressed in response to glucose |
|
| YDR342C | 1.18 |
HXT7
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels |
|
| YPR184W | 1.18 |
GDB1
|
Glycogen debranching enzyme containing glucanotranferase and alpha-1,6-amyloglucosidase activities, required for glycogen degradation; phosphorylated in mitochondria |
|
| YER066C-A | 1.09 |
Dubious open reading frame unlikely to encode a protein, partially overlaps uncharacterized ORF YER067W |
||
| YOR343C | 1.08 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YFL052W | 1.02 |
Putative zinc cluster protein that contains a DNA binding domain; null mutant sensitive to calcofluor white, low osmolarity and heat, suggesting a role for YFL052Wp in cell wall integrity |
||
| YOL081W | 0.97 |
IRA2
|
GTPase-activating protein that negatively regulates RAS by converting it from the GTP- to the GDP-bound inactive form, required for reducing cAMP levels under nutrient limiting conditions, has similarity to Ira1p and human neurofibromin |
|
| YEL011W | 0.96 |
GLC3
|
Glycogen branching enzyme, involved in glycogen accumulation; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YBR183W | 0.96 |
YPC1
|
Alkaline ceramidase that also has reverse (CoA-independent) ceramide synthase activity, catalyzes both breakdown and synthesis of phytoceramide; overexpression confers fumonisin B1 resistance |
|
| YKR066C | 0.94 |
CCP1
|
Mitochondrial cytochrome-c peroxidase; degrades reactive oxygen species in mitochondria, involved in the response to oxidative stress |
|
| YMR169C | 0.94 |
ALD3
|
Cytoplasmic aldehyde dehydrogenase, involved in beta-alanine synthesis; uses NAD+ as the preferred coenzyme; very similar to Ald2p; expression is induced by stress and repressed by glucose |
|
| YBR132C | 0.92 |
AGP2
|
High affinity polyamine permease, preferentially uses spermidine over putrescine; expression is down-regulated by osmotic stress; plasma membrane carnitine transporter, also functions as a low-affinity amino acid permease |
|
| YML120C | 0.92 |
NDI1
|
NADH:ubiquinone oxidoreductase, transfers electrons from NADH to ubiquinone in the respiratory chain but does not pump protons, in contrast to the higher eukaryotic multisubunit respiratory complex I; phosphorylated; homolog of human AMID |
|
| YFR017C | 0.88 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and is induced in response to the DNA-damaging agent MMS; YFR017C is not an essential gene |
||
| YER103W | 0.87 |
SSA4
|
Heat shock protein that is highly induced upon stress; plays a role in SRP-dependent cotranslational protein-membrane targeting and translocation; member of the HSP70 family; cytoplasmic protein that concentrates in nuclei upon starvation |
|
| YPL274W | 0.86 |
SAM3
|
High-affinity S-adenosylmethionine permease, required for utilization of S-adenosylmethionine as a sulfur source; has similarity to S-methylmethionine permease Mmp1p |
|
| YNR014W | 0.84 |
Putative protein of unknown function; expression is cell-cycle regulated, Azf1p-dependent, and heat-inducible |
||
| YER066W | 0.84 |
Putative protein of unknown function; YER066W is not an essential gene |
||
| YHR087W | 0.82 |
RTC3
|
Protein of unknown function involved in RNA metabolism; has structural similarity to SBDS, the human protein mutated in Shwachman-Diamond Syndrome (the yeast SBDS ortholog = SDO1); null mutation suppresses cdc13-1 temperature sensitivity |
|
| YJR146W | 0.82 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene HMS2 |
||
| YDR133C | 0.82 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR134C |
||
| YBL045C | 0.80 |
COR1
|
Core subunit of the ubiquinol-cytochrome c reductase complex (bc1 complex), which is a component of the mitochondrial inner membrane electron transport chain |
|
| YBR298C | 0.79 |
MAL31
|
Maltose permease, high-affinity maltose transporter (alpha-glucoside transporter); encoded in the MAL3 complex locus; member of the 12 transmembrane domain superfamily of sugar transporters; functional in genomic reference strain S288C |
|
| YNR034W-A | 0.78 |
Putative protein of unknown function; expression is regulated by Msn2p/Msn4p |
||
| YLR377C | 0.76 |
FBP1
|
Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway, required for glucose metabolism |
|
| YGL229C | 0.75 |
SAP4
|
Protein required for function of the Sit4p protein phosphatase, member of a family of similar proteins that form complexes with Sit4p, including Sap155p, Sap185p, and Sap190p |
|
| YJR120W | 0.74 |
Protein of unknown function; essential for growth under anaerobic conditions; mutation causes decreased expression of ATP2, impaired respiration, defective sterol uptake, and altered levels/localization of ABC transporters Aus1p and Pdr11p |
||
| YFL053W | 0.73 |
DAK2
|
Dihydroxyacetone kinase, required for detoxification of dihydroxyacetone (DHA); involved in stress adaptation |
|
| YGR127W | 0.72 |
Putative protein of unknown function; expression is regulated by Msn2p/Msn4p, indicating a possible role in stress response |
||
| YKL093W | 0.72 |
MBR1
|
Protein involved in mitochondrial functions and stress response; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants |
|
| YER088C | 0.71 |
DOT6
|
Protein of unknown function, involved in telomeric gene silencing and filamentation |
|
| YER088C-A | 0.71 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR541C | 0.68 |
Putative dihydrokaempferol 4-reductase |
||
| YNL300W | 0.68 |
Glycosylphosphatidylinositol-dependent cell wall protein, expression is periodic and decreases in respone to ergosterol perturbation or upon entry into stationary phase; depletion increases resistance to lactic acid |
||
| YDR536W | 0.66 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YPR028W | 0.65 |
YOP1
|
Membrane protein that interacts with Yip1p to mediate membrane traffic; overexpression results in cell death and accumulation of internal cell membranes; regulates vesicular traffic in stressed cells |
|
| YOR317W | 0.63 |
FAA1
|
Long chain fatty acyl-CoA synthetase with a preference for C12:0-C16:0 fatty acids; involved in the activation of imported fatty acids; localized to both lipid particles and mitochondrial outer membrane; essential for stationary phase |
|
| YLR342W | 0.63 |
FKS1
|
Catalytic subunit of 1,3-beta-D-glucan synthase, functionally redundant with alternate catalytic subunit Gsc2p; binds to regulatory subunit Rho1p; involved in cell wall synthesis and maintenance; localizes to sites of cell wall remodeling |
|
| YER065C | 0.62 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YBR085C-A | 0.61 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and to the nucleus |
||
| YDR178W | 0.60 |
SDH4
|
Membrane anchor subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
|
| YMR105C | 0.60 |
PGM2
|
Phosphoglucomutase, catalyzes the conversion from glucose-1-phosphate to glucose-6-phosphate, which is a key step in hexose metabolism; functions as the acceptor for a Glc-phosphotransferase |
|
| YPR027C | 0.60 |
Putative protein of unknown function |
||
| YLR130C | 0.59 |
ZRT2
|
Low-affinity zinc transporter of the plasma membrane; transcription is induced under low-zinc conditions by the Zap1p transcription factor |
|
| YGR249W | 0.59 |
MGA1
|
Protein similar to heat shock transcription factor; multicopy suppressor of pseudohyphal growth defects of ammonium permease mutants |
|
| YAL054C | 0.56 |
ACS1
|
Acetyl-coA synthetase isoform which, along with Acs2p, is the nuclear source of acetyl-coA for histone acetlyation; expressed during growth on nonfermentable carbon sources and under aerobic conditions |
|
| YKL067W | 0.56 |
YNK1
|
Nucleoside diphosphate kinase, catalyzes the transfer of gamma phosphates from nucleoside triphosphates, usually ATP, to nucleoside diphosphates by a mechanism that involves formation of an autophosphorylated enzyme intermediate |
|
| YGR191W | 0.56 |
HIP1
|
High-affinity histidine permease, also involved in the transport of manganese ions |
|
| YJR147W | 0.55 |
HMS2
|
Protein with similarity to heat shock transcription factors; overexpression suppresses the pseudohyphal filamentation defect of a diploid mep1 mep2 homozygous null mutant |
|
| YCR022C | 0.55 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YCR022C is not an essential gene |
||
| YOL052C-A | 0.55 |
DDR2
|
Multistress response protein, expression is activated by a variety of xenobiotic agents and environmental or physiological stresses |
|
| YDR277C | 0.55 |
MTH1
|
Negative regulator of the glucose-sensing signal transduction pathway, required for repression of transcription by Rgt1p; interacts with Rgt1p and the Snf3p and Rgt2p glucose sensors; phosphorylated by Yck1p, triggering Mth1p degradation |
|
| YLR347C | 0.55 |
KAP95
|
Karyopherin beta, forms a dimeric complex with Srp1p (Kap60p) that mediates nuclear import of cargo proteins via a nuclear localization signal (NLS), interacts with nucleoporins to guide transport across the nuclear pore complex |
|
| YPL036W | 0.54 |
PMA2
|
Plasma membrane H+-ATPase, isoform of Pma1p, involved in pumping protons out of the cell; regulator of cytoplasmic pH and plasma membrane potential |
|
| YML131W | 0.53 |
Putative protein of unknown function with similarity to oxidoreductases; HOG1 and SKO1-dependent mRNA expression is induced after osmotic shock; GFP-fusion protein localizes to the cytoplasm and is induced by the DNA-damaging agent MMS |
||
| YPL247C | 0.53 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; similar to the petunia WD repeat protein an11; YPL247C is not an essential gene |
||
| YBR117C | 0.53 |
TKL2
|
Transketolase, similar to Tkl1p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YLL023C | 0.52 |
Protein of unknown function; highly conserved across species and orthologous to human TMEM33; may interact with ribosomes, based on co-purification experiments; GFP-fusion protein localizes to the endoplasmic reticulum |
||
| YNL077W | 0.52 |
APJ1
|
Putative chaperone of the HSP40 (DNAJ) family; overexpression interferes with propagation of the [Psi+] prion; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YBL044W | 0.50 |
Putative protein of unknown function; YBL044W is not an essential protein |
||
| YKL056C | 0.50 |
TMA19
|
Protein that associates with ribosomes; homolog of translationally controlled tumor protein; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and relocates to the mitochondrial outer surface upon oxidative stress |
|
| YJL220W | 0.49 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YJL221C/FSP2 |
||
| YGR282C | 0.48 |
BGL2
|
Endo-beta-1,3-glucanase, major protein of the cell wall, involved in cell wall maintenance |
|
| YNL194C | 0.48 |
Integral membrane protein localized to eisosomes; sporulation and plasma membrane sphingolipid content are altered in mutants; has homologs SUR7 and FMP45; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YPR074C | 0.47 |
TKL1
|
Transketolase, similar to Tkl2p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YDR453C | 0.47 |
TSA2
|
Stress inducible cytoplasmic thioredoxin peroxidase; cooperates with Tsa1p in the removal of reactive oxygen, nitrogen and sulfur species using thioredoxin as hydrogen donor; deletion enhances the mutator phenotype of tsa1 mutants |
|
| YGR288W | 0.47 |
MAL13
|
MAL-activator protein, part of complex locus MAL1; nonfunctional in genomic reference strain S288C |
|
| YDR171W | 0.47 |
HSP42
|
Small heat shock protein (sHSP) with chaperone activity; forms barrel-shaped oligomers that suppress unfolded protein aggregation; involved in cytoskeleton reorganization after heat shock |
|
| YOR049C | 0.46 |
RSB1
|
Suppressor of sphingoid long chain base (LCB) sensitivity of an LCB-lyase mutation; putative integral membrane transporter or flippase that may transport LCBs from the cytoplasmic side toward the extracytoplasmic side of the membrane |
|
| YOR247W | 0.46 |
SRL1
|
Mannoprotein that exhibits a tight association with the cell wall, required for cell wall stability in the absence of GPI-anchored mannoproteins; has a high serine-threonine content; expression is induced in cell wall mutants |
|
| YDL130W-A | 0.46 |
STF1
|
Protein involved in regulation of the mitochondrial F1F0-ATP synthase; Stf1p and Stf2p may act as stabilizing factors that enhance inhibitory action of the Inh1p protein |
|
| YJL152W | 0.45 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBL049W | 0.45 |
MOH1
|
Protein of unknown function, has homology to kinase Snf7p; not required for growth on nonfermentable carbon sources; essential for viability in stationary phase |
|
| YGR190C | 0.44 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene HIP1/YGR191W |
||
| YGR234W | 0.44 |
YHB1
|
Nitric oxide oxidoreductase, flavohemoglobin involved in nitric oxide detoxification; plays a role in the oxidative and nitrosative stress responses |
|
| YGL187C | 0.44 |
COX4
|
Subunit IV of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; N-terminal 25 residues of precursor are cleaved during mitochondrial import; phosphorylated |
|
| YJL221C | 0.44 |
FSP2
|
Protein of unknown function, expression is induced during nitrogen limitation |
|
| YIL059C | 0.43 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YIL060W |
||
| YMR250W | 0.43 |
GAD1
|
Glutamate decarboxylase, converts glutamate into gamma-aminobutyric acid (GABA) during glutamate catabolism; involved in response to oxidative stress |
|
| YHL036W | 0.42 |
MUP3
|
Low affinity methionine permease, similar to Mup1p |
|
| YGR088W | 0.42 |
CTT1
|
Cytosolic catalase T, has a role in protection from oxidative damage by hydrogen peroxide |
|
| YPL154C | 0.41 |
PEP4
|
Vacuolar aspartyl protease (proteinase A), required for the posttranslational precursor maturation of vacuolar proteinases; important for protein turnover after oxidative damage; synthesized as a zymogen, self-activates |
|
| YKL103C | 0.41 |
LAP4
|
Vacuolar aminopeptidase, often used as a marker protein in studies of autophagy and cytosol to vacuole targeting (CVT) pathway |
|
| YDR129C | 0.41 |
SAC6
|
Fimbrin, actin-bundling protein; cooperates with Scp1p (calponin/transgelin) in the organization and maintenance of the actin cytoskeleton |
|
| YNL028W | 0.41 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YKR067W | 0.40 |
GPT2
|
Glycerol-3-phosphate/dihydroxyacetone phosphate dual substrate-specific sn-1 acyltransferase located in lipid particles and the ER; involved in the stepwise acylation of glycerol-3-phosphate and dihydroxyacetone in lipid biosynthesis |
|
| YGL202W | 0.39 |
ARO8
|
Aromatic aminotransferase I, expression is regulated by general control of amino acid biosynthesis |
|
| YJR025C | 0.39 |
BNA1
|
3-hydroxyanthranilic acid dioxygenase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p |
|
| YKL044W | 0.39 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBR214W | 0.38 |
SDS24
|
One of two S. cerevisiae homologs (Sds23p and Sds24p) of the Schizosaccharomyces pombe Sds23 protein, which genetic studies have implicated in APC/cyclosome regulation; may play an indirect role in fluid-phase endocytosis |
|
| YJL213W | 0.38 |
Protein of unknown function that may interact with ribosomes; periodically expressed during the yeast metabolic cycle; phosphorylated in vitro by the mitotic exit network (MEN) kinase complex, Dbf2p/Mob1p |
||
| YNL130C | 0.38 |
CPT1
|
Cholinephosphotransferase, required for phosphatidylcholine biosynthesis and for inositol-dependent regulation of EPT1 transcription |
|
| YLR450W | 0.37 |
HMG2
|
One of two isozymes of HMG-CoA reductase that convert HMG-CoA to mevalonate, a rate-limiting step in sterol biosynthesis; overproduction induces assembly of peripheral ER membrane arrays and short nuclear-associated membrane stacks |
|
| YDR234W | 0.37 |
LYS4
|
Homoaconitase, catalyzes the conversion of homocitrate to homoisocitrate, which is a step in the lysine biosynthesis pathway |
|
| YFL051C | 0.36 |
Putative protein of unknown function; YFL051C is not an essential gene |
||
| YDR309C | 0.36 |
GIC2
|
Redundant rho-like GTPase Cdc42p effector; homolog of Gic1p; involved in initiation of budding and cellular polarization; interacts with Cdc42p via the Cdc42/Rac-interactive binding (CRIB) domain and with PI(4,5)P2 via a polybasic region |
|
| YPL014W | 0.36 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and to the nucleus |
||
| YKL150W | 0.34 |
MCR1
|
Mitochondrial NADH-cytochrome b5 reductase, involved in ergosterol biosynthesis |
|
| YGR250C | 0.34 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YDR185C | 0.34 |
Mitochondrial protein of unknown function; has similarity to Ups1p, which is involved in regulation of alternative topogenesis of the dynamin-related GTPase Mgm1p |
||
| YER053C | 0.33 |
PIC2
|
Mitochondrial phosphate carrier, imports inorganic phosphate into mitochondria; functionally redundant with Mir1p but less abundant than Mir1p under normal conditions; expression is induced at high temperature |
|
| YOR393W | 0.33 |
ERR1
|
Protein of unknown function, has similarity to enolases |
|
| YIL087C | 0.32 |
LRC2
|
Putative protein of unknown function; protein is detected in purified mitochondria in high-throughput studies; null mutant displays decreased mitochondrial genome loss (petite formation) and severe growth defect in minimal glycerol media |
|
| YLR177W | 0.32 |
Putative protein of unknown function; phosphorylated by Dbf2p-Mob1p in vitro; some strains contain microsatellite polymophisms at this locus; YLR177W is not an essential gene |
||
| YBR054W | 0.32 |
YRO2
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in a phosphorylated state in highly purified mitochondria in high-throughput studies; transcriptionally regulated by Haa1p |
|
| YMR251W-A | 0.31 |
HOR7
|
Protein of unknown function; overexpression suppresses Ca2+ sensitivity of mutants lacking inositol phosphorylceramide mannosyltransferases Csg1p and Csh1p; transcription is induced under hyperosmotic stress and repressed by alpha factor |
|
| YPL271W | 0.31 |
ATP15
|
Epsilon subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; phosphorylated |
|
| YOR378W | 0.30 |
Putative protein of unknown function; belongs to the DHA2 family of drug:H+ antiporters; YOR378W is not an essential gene |
||
| YJR149W | 0.30 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YLL026W | 0.30 |
HSP104
|
Heat shock protein that cooperates with Ydj1p (Hsp40) and Ssa1p (Hsp70) to refold and reactivate previously denatured, aggregated proteins; responsive to stresses including: heat, ethanol, and sodium arsenite; involved in [PSI+] propagation |
|
| YKL151C | 0.30 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YIR027C | 0.30 |
DAL1
|
Allantoinase, converts allantoin to allantoate in the first step of allantoin degradation; expression sensitive to nitrogen catabolite repression |
|
| YOL052C | 0.29 |
SPE2
|
S-adenosylmethionine decarboxylase, required for the biosynthesis of spermidine and spermine; cells lacking Spe2p require spermine or spermidine for growth in the presence of oxygen but not when grown anaerobically |
|
| YAL005C | 0.29 |
SSA1
|
ATPase involved in protein folding and nuclear localization signal (NLS)-directed nuclear transport; member of heat shock protein 70 (HSP70) family; forms a chaperone complex with Ydj1p; localized to the nucleus, cytoplasm, and cell wall |
|
| YKL043W | 0.28 |
PHD1
|
Transcriptional activator that enhances pseudohyphal growth; regulates expression of FLO11, an adhesin required for pseudohyphal filament formation; similar to StuA, an A. nidulans developmental regulator; potential Cdc28p substrate |
|
| YKL215C | 0.28 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YNL066W | 0.28 |
SUN4
|
Cell wall protein related to glucanases, possibly involved in cell wall septation; member of the SUN family |
|
| YDL146W | 0.28 |
LDB17
|
Protein of unknown function; GFP-fusion protein localizes to the cell periphery, cytoplasm, bud, and bud neck; null mutant shows a reduced affinity for the alcian blue dye suggesting a decreased net negative charge of the cell surface |
|
| YHR001W-A | 0.28 |
QCR10
|
Subunit of the ubiqunol-cytochrome c oxidoreductase complex which includes Cobp, Rip1p, Cyt1p, Cor1p, Qcr2p, Qcr6p, Qcr7p, Qcr8p, Qcr9p, and Qcr10p and comprises part of the mitochondrial respiratory chain |
|
| YJL153C | 0.27 |
INO1
|
Inositol 1-phosphate synthase, involved in synthesis of inositol phosphates and inositol-containing phospholipids; transcription is coregulated with other phospholipid biosynthetic genes by Ino2p and Ino4p, which bind the UASINO DNA element |
|
| YOL157C | 0.27 |
Putative protein of unknown function |
||
| YKL037W | 0.26 |
AIM26
|
Putative protein of unknown function; null mutant is viable and displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YGR111W | 0.26 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and the nucleus |
||
| YDR535C | 0.26 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YDR535C is not an essential gene. |
||
| YOR386W | 0.26 |
PHR1
|
DNA photolyase involved in photoreactivation, repairs pyrimidine dimers in the presence of visible light; induced by DNA damage; regulated by transcriptional repressor Rph1p |
|
| YMR261C | 0.25 |
TPS3
|
Regulatory subunit of trehalose-6-phosphate synthase/phosphatase complex, which synthesizes the storage carbohydrate trehalose; expression is induced by stress conditions and repressed by the Ras-cAMP pathway |
|
| YGR157W | 0.25 |
CHO2
|
Phosphatidylethanolamine methyltransferase (PEMT), catalyzes the first step in the conversion of phosphatidylethanolamine to phosphatidylcholine during the methylation pathway of phosphatidylcholine biosynthesis |
|
| YDR542W | 0.25 |
PAU10
|
Hypothetical protein |
|
| YLR286C | 0.25 |
CTS1
|
Endochitinase, required for cell separation after mitosis; transcriptional activation during late G and early M cell cycle phases is mediated by transcription factor Ace2p |
|
| YJR148W | 0.25 |
BAT2
|
Cytosolic branched-chain amino acid aminotransferase, homolog of murine ECA39; highly expressed during stationary phase and repressed during logarithmic phase |
|
| YBR139W | 0.25 |
Putative serine type carboxypeptidase with a role in phytochelatin synthesis; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; expression induced by nitrogen limitation in a GLN3, GAT1-independent manner |
||
| YPL240C | 0.24 |
HSP82
|
Hsp90 chaperone required for pheromone signaling and negative regulation of Hsf1p; docks with Tom70p for mitochondrial preprotein delivery; promotes telomerase DNA binding and nucleotide addition; interacts with Cns1p, Cpr6p, Cpr7p, Sti1p |
|
| YDR540C | 0.24 |
IRC4
|
Putative protein of unknown function; null mutant displays increased levels of spontaneous Rad52p foci; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
|
| YLR154W-B | 0.24 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YDL199C | 0.23 |
Putative transporter, member of the sugar porter family |
||
| YPR007C | 0.23 |
REC8
|
Meiosis-specific component of sister chromatid cohesion complex; maintains cohesion between sister chromatids during meiosis I; maintains cohesion between centromeres of sister chromatids until meiosis II; homolog of S. pombe Rec8p |
|
| YCR030C | 0.23 |
SYP1
|
Protein with a potential role in actin cytoskeletal organization; overexpression suppresses a pfy1 (profilin) null mutation |
|
| YIL123W | 0.23 |
SIM1
|
Protein of the SUN family (Sim1p, Uth1p, Nca3p, Sun4p) that may participate in DNA replication, promoter contains SCB regulation box at -300 bp indicating that expression may be cell cycle-regulated |
|
| YMR243C | 0.23 |
ZRC1
|
Vacuolar membrane zinc transporter, transports zinc from the cytosol into the vacuole for storage; also has a role in resistance to zinc shock resulting from a sudden influx of zinc into the cytoplasm |
|
| YJR045C | 0.23 |
SSC1
|
Mitochondrial matrix ATPase, subunit of the presequence translocase-associated protein import motor (PAM) and of SceI endonuclease; involved in protein folding and translocation into the matrix; phosphorylated; member of HSP70 family |
|
| YHR198C | 0.22 |
AIM18
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YDR148C | 0.22 |
KGD2
|
Dihydrolipoyl transsuccinylase, component of the mitochondrial alpha-ketoglutarate dehydrogenase complex, which catalyzes the oxidative decarboxylation of alpha-ketoglutarate to succinyl-CoA in the TCA cycle; phosphorylated |
|
| YLR174W | 0.22 |
IDP2
|
Cytosolic NADP-specific isocitrate dehydrogenase, catalyzes oxidation of isocitrate to alpha-ketoglutarate; levels are elevated during growth on non-fermentable carbon sources and reduced during growth on glucose |
|
| YMR181C | 0.22 |
Protein of unknown function; mRNA transcribed as part of a bicistronic transcript with a predicted transcriptional repressor RGM1/YMR182C; mRNA is destroyed by nonsense-mediated decay (NMD); YMR181C is not an essential gene |
||
| YOR134W | 0.21 |
BAG7
|
Rho GTPase activating protein (RhoGAP), stimulates the intrinsic GTPase activity of Rho1p, which plays a role in actin cytoskeleton organization and control of cell wall synthesis; structurally and functionally related to Sac7p |
|
| YPR026W | 0.21 |
ATH1
|
Acid trehalase required for utilization of extracellular trehalose |
|
| YBR053C | 0.21 |
Putative protein of unknown function; induced by cell wall perturbation |
||
| YLR054C | 0.21 |
OSW2
|
Protein of unknown function proposed to be involved in the assembly of the spore wall |
|
| YER101C | 0.21 |
AST2
|
Protein that may have a role in targeting of plasma membrane [H+]ATPase (Pma1p) to the plasma membrane, as suggested by analysis of genetic interactions |
|
| YFL044C | 0.21 |
OTU1
|
Deubiquitylation enzyme that binds to the chaperone-ATPase Cdc48p; may contribute to regulation of protein degradation by deubiquitylating substrates that have been ubiquitylated by Ufd2p; member of the Ovarian Tumor (OTU) family |
|
| YPL087W | 0.20 |
YDC1
|
Alkaline dihydroceramidase, involved in sphingolipid metabolism; preferentially hydrolyzes dihydroceramide to a free fatty acid and dihydrosphingosine; has a minor reverse activity |
|
| YLR233C | 0.20 |
EST1
|
TLC1 RNA-associated factor involved in telomere length regulation as the recruitment subunit of the telomerase holoenzyme, has a possible role in activating Est2p-TLC1-RNA bound to the telomere |
|
| YGR279C | 0.20 |
SCW4
|
Cell wall protein with similarity to glucanases; scw4 scw10 double mutants exhibit defects in mating |
|
| YAL034C | 0.20 |
FUN19
|
Non-essential protein of unknown function |
|
| YJR127C | 0.20 |
RSF2
|
Zinc-finger protein involved in transcriptional control of both nuclear and mitochondrial genes, many of which specify products required for glycerol-based growth, respiration, and other functions |
|
| YLR332W | 0.20 |
MID2
|
O-glycosylated plasma membrane protein that acts as a sensor for cell wall integrity signaling and activates the pathway; interacts with Rom2p, a guanine nucleotide exchange factor for Rho1p, and with cell integrity pathway protein Zeo1p |
|
| YEL013W | 0.20 |
VAC8
|
Phosphorylated vacuolar membrane protein that interacts with Atg13p, required for the cytoplasm-to-vacuole targeting (Cvt) pathway; interacts with Nvj1p to form nucleus-vacuole junctions |
|
| YPL015C | 0.20 |
HST2
|
Cytoplasmic member of the silencing information regulator 2 (Sir2) family of NAD(+)-dependent protein deacetylases; modulates nucleolar (rDNA) and telomeric silencing; possesses NAD(+)-dependent histone deacetylase activity in vitro |
|
| YJR121W | 0.20 |
ATP2
|
Beta subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; phosphorylated |
|
| YJR115W | 0.19 |
Putative protein of unknown function |
||
| YJR046W | 0.19 |
TAH11
|
DNA replication licensing factor, required for pre-replication complex assembly |
|
| YKR102W | 0.19 |
FLO10
|
Lectin-like protein with similarity to Flo1p, thought to be involved in flocculation |
|
| YLR436C | 0.19 |
ECM30
|
Non-essential protein of unknown function |
|
| YNL305C | 0.19 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YNL305C is not an essential gene |
||
| YLR331C | 0.18 |
JIP3
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 98% of ORF overlaps the verified gene MID2 |
|
| YHR162W | 0.18 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the mitochondrion |
||
| YLR446W | 0.18 |
Putative protein of unknown function with similarity to hexokinases; transcript is upregulated during sporulation and the unfolded protein response; YLR446W is not an essential gene |
||
| YEL017W | 0.18 |
GTT3
|
Protein of unknown function with a possible role in glutathione metabolism, as suggested by computational analysis of large-scale protein-protein interaction data; GFP-fusion protein localizes to the nuclear periphery |
|
| YGL236C | 0.18 |
MTO1
|
Mitochondrial protein, forms a heterodimer complex with Mss1p that performs the 5-carboxymethylaminomethyl modification of the wobble uridine base in mitochondrial tRNAs; required for respiration in paromomycin-resistant 15S rRNA mutants |
|
| YJL133C-A | 0.18 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YJL077C | 0.18 |
ICS3
|
Protein of unknown function |
|
| YMR175W | 0.18 |
SIP18
|
Protein of unknown function whose expression is induced by osmotic stress |
|
| YPL171C | 0.18 |
OYE3
|
Widely conserved NADPH oxidoreductase containing flavin mononucleotide (FMN), homologous to Oye2p with slight differences in ligand binding and catalytic properties; may be involved in sterol metabolism |
|
| YDR215C | 0.17 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; null mutant displays elevated sensitivity to expression of a mutant huntingtin fragment or of alpha-synuclein |
||
| YLR259C | 0.17 |
HSP60
|
Tetradecameric mitochondrial chaperonin required for ATP-dependent folding of precursor polypeptides and complex assembly; prevents aggregation and mediates protein refolding after heat shock; role in mtDNA transmission; phosphorylated |
|
| YNL274C | 0.17 |
GOR1
|
Glyoxylate reductase; null mutation results in increased biomass after diauxic shift; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YLR110C | 0.17 |
CCW12
|
Cell wall mannoprotein, mutants are defective in mating and agglutination, expression is downregulated by alpha-factor |
|
| YIR036C | 0.17 |
IRC24
|
Putative benzil reductase;(GFP)-fusion protein localizes to the cytoplasm and is induced by the DNA-damaging agent MMS; sequence similarity with short-chain dehydrogenase/reductases; null mutant has increased spontaneous Rad52p foci |
|
| YJR153W | 0.17 |
PGU1
|
Endo-polygalacturonase, pectolytic enzyme that hydrolyzes the alpha-1,4-glycosidic bonds in the rhamnogalacturonan chains in pectins |
|
| YNL190W | 0.16 |
Cell wall protein of unknown function; proposed role as a hydrophilin induced by osmotic stress; contains a putative GPI-attachment site |
||
| YIR036W-A | 0.16 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR072C | 0.16 |
IPT1
|
Inositolphosphotransferase 1, involved in synthesis of mannose-(inositol-P)2-ceramide (M(IP)2C), which is the most abundant sphingolipid in cells, mutation confers resistance to the antifungals syringomycin E and DmAMP1 in some growth media |
|
| YJL158C | 0.16 |
CIS3
|
Mannose-containing glycoprotein constituent of the cell wall; member of the PIR (proteins with internal repeats) family |
|
| YAR027W | 0.16 |
UIP3
|
Putative integral membrane protein of unknown function; interacts with Ulp1p at the nuclear periphery; member of DUP240 gene family |
|
| YIL111W | 0.16 |
COX5B
|
Subunit Vb of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; predominantly expressed during anaerobic growth while its isoform Va (Cox5Ap) is expressed during aerobic growth |
|
| YOR377W | 0.16 |
ATF1
|
Alcohol acetyltransferase with potential roles in lipid and sterol metabolism; responsible for the major part of volatile acetate ester production during fermentation |
Network of associatons between targets according to the STRING database.
First level regulatory network of USV1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0015755 | fructose transport(GO:0015755) |
| 0.9 | 4.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.7 | 2.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.5 | 1.0 | GO:0045980 | negative regulation of nucleotide metabolic process(GO:0045980) |
| 0.5 | 1.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.4 | 1.2 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.3 | 2.7 | GO:0005987 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.3 | 0.9 | GO:0043065 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.3 | 5.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.3 | 2.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 1.3 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.2 | 1.0 | GO:0015848 | spermidine transport(GO:0015848) |
| 0.2 | 0.9 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.2 | 1.3 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.2 | 0.5 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.2 | 0.5 | GO:0046039 | GTP biosynthetic process(GO:0006183) GTP metabolic process(GO:0046039) |
| 0.2 | 0.9 | GO:0019563 | alditol catabolic process(GO:0019405) glycerol catabolic process(GO:0019563) |
| 0.2 | 0.6 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.2 | 2.7 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 0.3 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.1 | 0.4 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.6 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.4 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.1 | 1.0 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.1 | 0.4 | GO:1902223 | tyrosine biosynthetic process(GO:0006571) L-phenylalanine biosynthetic process(GO:0009094) erythrose 4-phosphate/phosphoenolpyruvate family amino acid biosynthetic process(GO:1902223) |
| 0.1 | 1.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.5 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.1 | 0.6 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.4 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.1 | 0.7 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.1 | 1.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.5 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.3 | GO:0044088 | regulation of vacuole fusion, non-autophagic(GO:0032889) regulation of vacuole organization(GO:0044088) |
| 0.1 | 0.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.6 | GO:0045117 | azole transport(GO:0045117) |
| 0.1 | 0.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.2 | GO:0071462 | response to water(GO:0009415) cellular response to water stimulus(GO:0071462) |
| 0.1 | 1.3 | GO:0006754 | ATP biosynthetic process(GO:0006754) purine nucleoside triphosphate biosynthetic process(GO:0009145) purine ribonucleoside triphosphate biosynthetic process(GO:0009206) |
| 0.1 | 0.4 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.1 | 0.2 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 0.1 | 0.3 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.1 | 0.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.8 | GO:0072593 | reactive oxygen species metabolic process(GO:0072593) |
| 0.1 | 0.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.4 | GO:0000272 | polysaccharide catabolic process(GO:0000272) |
| 0.1 | 0.5 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.2 | GO:0070054 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) IRE1-mediated unfolded protein response(GO:0036498) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) |
| 0.0 | 0.2 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.0 | 0.1 | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.2 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.2 | GO:0000435 | regulation of transcription from RNA polymerase II promoter by galactose(GO:0000431) positive regulation of transcription from RNA polymerase II promoter by galactose(GO:0000435) |
| 0.0 | 0.2 | GO:0098743 | agglutination involved in conjugation with cellular fusion(GO:0000752) agglutination involved in conjugation(GO:0000771) heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) cell-cell adhesion(GO:0098609) adhesion between unicellular organisms(GO:0098610) aggregation of unicellular organisms(GO:0098630) multi organism cell adhesion(GO:0098740) cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) cell aggregation(GO:0098743) |
| 0.0 | 0.1 | GO:0048024 | regulation of RNA splicing(GO:0043484) regulation of mRNA splicing, via spliceosome(GO:0048024) positive regulation of mRNA processing(GO:0050685) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.8 | GO:0008645 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.0 | 0.2 | GO:0071041 | antisense RNA transcript catabolic process(GO:0071041) |
| 0.0 | 0.3 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 2.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.2 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.4 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) allantoin catabolic process(GO:0000256) |
| 0.0 | 0.4 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.0 | 0.4 | GO:0008299 | isoprenoid metabolic process(GO:0006720) isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.2 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0046320 | ethanol catabolic process(GO:0006068) regulation of fatty acid beta-oxidation(GO:0031998) positive regulation of fatty acid beta-oxidation(GO:0032000) primary alcohol catabolic process(GO:0034310) regulation of fatty acid oxidation(GO:0046320) positive regulation of fatty acid oxidation(GO:0046321) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.3 | GO:0007009 | plasma membrane organization(GO:0007009) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.1 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0044396 | actin cortical patch assembly(GO:0000147) actin cortical patch organization(GO:0044396) |
| 0.0 | 0.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.0 | 0.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.1 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.0 | GO:0031684 | heterotrimeric G-protein complex cycle(GO:0031684) |
| 0.0 | 0.2 | GO:0000011 | vacuole inheritance(GO:0000011) |
| 0.0 | 0.0 | GO:0015691 | cadmium ion transport(GO:0015691) |
| 0.0 | 0.9 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) pantothenate biosynthetic process(GO:0015940) |
| 0.0 | 0.0 | GO:0010039 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.0 | 0.1 | GO:0001009 | transcription from RNA polymerase III type 2 promoter(GO:0001009) transcription from a RNA polymerase III hybrid type promoter(GO:0001041) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.1 | GO:0051318 | mitotic G1 phase(GO:0000080) G1 phase(GO:0051318) |
| 0.0 | 0.0 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.0 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 1.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 0.6 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.1 | 4.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.4 | GO:0005756 | mitochondrial proton-transporting ATP synthase, central stalk(GO:0005756) proton-transporting ATP synthase, central stalk(GO:0045269) |
| 0.1 | 0.3 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.0 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial oxoglutarate dehydrogenase complex(GO:0009353) dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.5 | GO:0072379 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 0.1 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.3 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.1 | 0.2 | GO:0071561 | nucleus-vacuole junction(GO:0071561) |
| 0.1 | 1.8 | GO:0009277 | fungal-type cell wall(GO:0009277) |
| 0.1 | 4.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 0.4 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.1 | 0.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 0.2 | GO:0001400 | mating projection base(GO:0001400) |
| 0.1 | 1.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.8 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.5 | GO:0033698 | Rpd3L complex(GO:0033698) |
| 0.0 | 0.1 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.0 | 0.4 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.0 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 12.9 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.9 | 2.7 | GO:0004575 | beta-fructofuranosidase activity(GO:0004564) sucrose alpha-glucosidase activity(GO:0004575) |
| 0.5 | 1.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 1.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.4 | 4.8 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 2.6 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.3 | 1.9 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.3 | 0.9 | GO:0008137 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 0.8 | GO:0015151 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.2 | 0.7 | GO:0016208 | AMP binding(GO:0016208) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 0.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.2 | 3.2 | GO:0046906 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.2 | 0.6 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.2 | 0.7 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.2 | 0.7 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 0.2 | 1.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.2 | 1.7 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.2 | 0.5 | GO:0016872 | intramolecular lyase activity(GO:0016872) |
| 0.2 | 0.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 0.9 | GO:0004030 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 0.8 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 0.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 1.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.5 | GO:0042973 | glucan endo-1,3-beta-D-glucosidase activity(GO:0042973) |
| 0.1 | 0.5 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.3 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 0.5 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 2.2 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 0.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 0.5 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.5 | GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism(GO:0015662) |
| 0.1 | 0.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.2 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.1 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 3.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.5 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 3.1 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 0.2 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.6 | GO:0019205 | nucleobase-containing compound kinase activity(GO:0019205) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0008242 | gamma-glutamyltransferase activity(GO:0003840) omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.4 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.1 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.0 | GO:0008106 | aldo-keto reductase (NADP) activity(GO:0004033) alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.2 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.1 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:1901618 | organic hydroxy compound transmembrane transporter activity(GO:1901618) |
| 0.0 | 0.1 | GO:0001031 | RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) transcription factor activity, RNA polymerase III promoter sequence-specific binding, TFIIIB recruiting(GO:0001004) transcription factor activity, RNA polymerase III type 1 promoter sequence-specific binding, TFIIIB recruiting(GO:0001005) transcription factor activity, RNA polymerase III type 2 promoter sequence-specific binding, TFIIIB recruiting(GO:0001008) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.0 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.2 | GO:0008483 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.1 | GO:0005471 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.0 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.0 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 1.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 0.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 0.2 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |