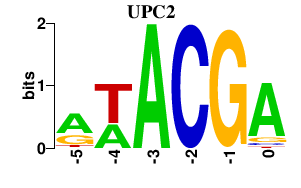
Results for UPC2
Z-value: 0.99
Transcription factors associated with UPC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
UPC2
|
S000002621 | Sterol regulatory element binding protein |
Activity-expression correlation:
Activity profile of UPC2 motif
Sorted Z-values of UPC2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YIL162W | 5.22 |
SUC2
|
Invertase, sucrose hydrolyzing enzyme; a secreted, glycosylated form is regulated by glucose repression, and an intracellular, nonglycosylated enzyme is produced constitutively |
|
| YDR541C | 2.54 |
Putative dihydrokaempferol 4-reductase |
||
| YOR382W | 2.47 |
FIT2
|
Mannoprotein that is incorporated into the cell wall via a glycosylphosphatidylinositol (GPI) anchor, involved in the retention of siderophore-iron in the cell wall |
|
| YOL154W | 2.42 |
ZPS1
|
Putative GPI-anchored protein; transcription is induced under low-zinc conditions, as mediated by the Zap1p transcription factor, and at alkaline pH |
|
| YLR295C | 2.40 |
ATP14
|
Subunit h of the F0 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YLR056W | 2.30 |
ERG3
|
C-5 sterol desaturase, catalyzes the introduction of a C-5(6) double bond into episterol, a precursor in ergosterol biosynthesis; mutants are viable, but cannot grow on non-fermentable carbon sources |
|
| YIL012W | 2.29 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR279C | 2.27 |
SCW4
|
Cell wall protein with similarity to glucanases; scw4 scw10 double mutants exhibit defects in mating |
|
| YFL051C | 2.04 |
Putative protein of unknown function; YFL051C is not an essential gene |
||
| YMR316C-B | 1.87 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YCR005C | 1.82 |
CIT2
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate, peroxisomal isozyme involved in glyoxylate cycle; expression is controlled by Rtg1p and Rtg2p transcription factors |
|
| YMR271C | 1.81 |
URA10
|
Minor orotate phosphoribosyltransferase (OPRTase) isozyme that catalyzes the fifth enzymatic step in the de novo biosynthesis of pyrimidines, converting orotate into orotidine-5'-phosphate; major OPRTase encoded by URA5 |
|
| YGR234W | 1.75 |
YHB1
|
Nitric oxide oxidoreductase, flavohemoglobin involved in nitric oxide detoxification; plays a role in the oxidative and nitrosative stress responses |
|
| YEL065W | 1.72 |
SIT1
|
Ferrioxamine B transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; transcription is induced during iron deprivation and diauxic shift; potentially phosphorylated by Cdc28p |
|
| YHL046W-A | 1.67 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOL136C | 1.65 |
PFK27
|
6-phosphofructo-2-kinase, catalyzes synthesis of fructose-2,6-bisphosphate; inhibited by phosphoenolpyruvate and sn-glycerol 3-phosphate, expression induced by glucose and sucrose, transcriptional regulation involves protein kinase A |
|
| YMR317W | 1.64 |
Putative protein of unknown function with some similarity to sialidase from Trypanosoma; YMR317W is not an essential gene |
||
| YOR010C | 1.59 |
TIR2
|
Putative cell wall mannoprotein of the Srp1p/Tip1p family of serine-alanine-rich proteins; transcription is induced by cold shock and anaerobiosis |
|
| YLR297W | 1.56 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YLR297W is not an essential gene; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YDR516C | 1.52 |
EMI2
|
Non-essential protein of unknown function required for transcriptional induction of the early meiotic-specific transcription factor IME1; required for sporulation; expression is regulated by glucose-repression transcription factors Mig1/2p |
|
| YER158C | 1.49 |
Protein of unknown function, has similarity to Afr1p; potentially phosphorylated by Cdc28p |
||
| YHR143W | 1.49 |
DSE2
|
Daughter cell-specific secreted protein with similarity to glucanases, degrades cell wall from the daughter side causing daughter to separate from mother; expression is repressed by cAMP |
|
| YHL047C | 1.48 |
ARN2
|
Transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; responsible for uptake of iron bound to the siderophore triacetylfusarinine C |
|
| YPL017C | 1.47 |
IRC15
|
Putative S-adenosylmethionine-dependent methyltransferase of the seven beta-strand family, required for accurate meiotic chromosome segregation; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YPR146C | 1.47 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNR016C | 1.44 |
ACC1
|
Acetyl-CoA carboxylase, biotin containing enzyme that catalyzes the carboxylation of acetyl-CoA to form malonyl-CoA; required for de novo biosynthesis of long-chain fatty acids |
|
| YBL082C | 1.43 |
ALG3
|
Dolichol-P-Man dependent alpha(1-3) mannosyltransferase, involved in the synthesis of dolichol-linked oligosaccharide donor for N-linked glycosylation of proteins |
|
| YPL271W | 1.43 |
ATP15
|
Epsilon subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; phosphorylated |
|
| YML083C | 1.38 |
Putative protein of unknown function; strong increase in transcript abundance during anaerobic growth compared to aerobic growth; cells deleted for YML083C do not exhibit growth defects in anerobic or anaerobic conditions |
||
| YOL128C | 1.37 |
YGK3
|
Protein kinase related to mammalian glycogen synthase kinases of the GSK-3 family; GSK-3 homologs (Mck1p, Rim11p, Mrk1p, Ygk3p) are involved in control of Msn2p-dependent transcription of stress responsive genes and in protein degradation |
|
| YMR082C | 1.37 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNL134C | 1.36 |
Putative protein of unknown function with similarity to dehydrogenases from other model organisms; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and nucleus and is induced by the DNA-damaging agent MMS |
||
| YNL327W | 1.33 |
EGT2
|
Glycosylphosphatidylinositol (GPI)-anchored cell wall endoglucanase required for proper cell separation after cytokinesis, expression is activated by Swi5p and tightly regulated in a cell cycle-dependent manner |
|
| YMR297W | 1.33 |
PRC1
|
Vacuolar carboxypeptidase Y (proteinase C), broad-specificity C-terminal exopeptidase involved in non-specific protein degradation in the vacuole; member of the serine carboxypeptidase family |
|
| YLR350W | 1.33 |
ORM2
|
Evolutionarily conserved protein with similarity to Orm1p, required for resistance to agents that induce the unfolded protein response; human ortholog is located in the endoplasmic reticulum |
|
| YML081C-A | 1.32 |
ATP18
|
Subunit of the mitochondrial F1F0 ATP synthase, which is a large enzyme complex required for ATP synthesis; termed subunit I or subunit j; does not correspond to known ATP synthase subunits in other organisms |
|
| YIR020C | 1.30 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YKL044W | 1.28 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR032C | 1.28 |
PST2
|
Protein with similarity to members of a family of flavodoxin-like proteins; induced by oxidative stress in a Yap1p dependent manner; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YGL012W | 1.26 |
ERG4
|
C-24(28) sterol reductase, catalyzes the final step in ergosterol biosynthesis; mutants are viable, but lack ergosterol |
|
| YPR145C-A | 1.24 |
Putative protein of unknown function |
||
| YGR191W | 1.20 |
HIP1
|
High-affinity histidine permease, also involved in the transport of manganese ions |
|
| YMR081C | 1.20 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YOL075C | 1.17 |
Putative ABC transporter |
||
| YMR319C | 1.13 |
FET4
|
Low-affinity Fe(II) transporter of the plasma membrane |
|
| YML131W | 1.11 |
Putative protein of unknown function with similarity to oxidoreductases; HOG1 and SKO1-dependent mRNA expression is induced after osmotic shock; GFP-fusion protein localizes to the cytoplasm and is induced by the DNA-damaging agent MMS |
||
| YPR157W | 1.08 |
Putative protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YLR178C | 1.08 |
TFS1
|
Carboxypeptidase Y inhibitor, function requires acetylation by the NatB N-terminal acetyltransferase; phosphatidylethanolamine-binding protein involved in protein kinase A signaling pathway |
|
| YKL103C | 1.07 |
LAP4
|
Vacuolar aminopeptidase, often used as a marker protein in studies of autophagy and cytosol to vacuole targeting (CVT) pathway |
|
| YOL082W | 1.06 |
ATG19
|
Receptor protein for the cytoplasm-to-vacuole targeting (Cvt) pathway; recognizes cargo proteins aminopeptidase I (Lap4p) and alpha-mannosidase (Ams1p) and delivers them to the preautophagosomal structure for packaging into Cvt vesicles |
|
| YMR092C | 1.06 |
AIP1
|
Actin cortical patch component, interacts with the actin depolymerizing factor cofilin; required to restrict cofilin localization to cortical patches; contains WD repeats |
|
| YLR327C | 1.05 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YPR196W | 1.04 |
Putative maltose activator |
||
| YBR182C | 1.04 |
SMP1
|
Putative transcription factor involved in regulating the response to osmotic stress; member of the MADS-box family of transcription factors |
|
| YEL008W | 1.03 |
Hypothetical protein predicted to be involved in metabolism |
||
| YNL160W | 1.02 |
YGP1
|
Cell wall-related secretory glycoprotein; induced by nutrient deprivation-associated growth arrest and upon entry into stationary phase; may be involved in adaptation prior to stationary phase entry; has similarity to Sps100p |
|
| YOL119C | 1.01 |
MCH4
|
Protein with similarity to mammalian monocarboxylate permeases, which are involved in transport of monocarboxylic acids across the plasma membrane; mutant is not deficient in monocarboxylate transport |
|
| YGR190C | 1.01 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene HIP1/YGR191W |
||
| YLR296W | 1.00 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR256W | 1.00 |
HAP1
|
Zinc finger transcription factor involved in the complex regulation of gene expression in response to levels of heme and oxygen; the S288C sequence differs from other strain backgrounds due to a Ty1 insertion in the carboxy terminus |
|
| YPR160W | 1.00 |
GPH1
|
Non-essential glycogen phosphorylase required for the mobilization of glycogen, activity is regulated by cyclic AMP-mediated phosphorylation, expression is regulated by stress-response elements and by the HOG MAP kinase pathway |
|
| YNL077W | 0.99 |
APJ1
|
Putative chaperone of the HSP40 (DNAJ) family; overexpression interferes with propagation of the [Psi+] prion; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YDR542W | 0.97 |
PAU10
|
Hypothetical protein |
|
| YHL026C | 0.96 |
Putative protein of unknown function; YHL026C is not an essential gene; in 2005 the start site was moved 141 nt upstream (see Locus History) |
||
| YGL255W | 0.96 |
ZRT1
|
High-affinity zinc transporter of the plasma membrane, responsible for the majority of zinc uptake; transcription is induced under low-zinc conditions by the Zap1p transcription factor |
|
| YPL240C | 0.96 |
HSP82
|
Hsp90 chaperone required for pheromone signaling and negative regulation of Hsf1p; docks with Tom70p for mitochondrial preprotein delivery; promotes telomerase DNA binding and nucleotide addition; interacts with Cns1p, Cpr6p, Cpr7p, Sti1p |
|
| YCL048W-A | 0.95 |
Putative protein of unknown function |
||
| YHR092C | 0.95 |
HXT4
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YDR476C | 0.94 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; YDR476C is not an essential gene |
||
| YBL049W | 0.94 |
MOH1
|
Protein of unknown function, has homology to kinase Snf7p; not required for growth on nonfermentable carbon sources; essential for viability in stationary phase |
|
| YDR453C | 0.94 |
TSA2
|
Stress inducible cytoplasmic thioredoxin peroxidase; cooperates with Tsa1p in the removal of reactive oxygen, nitrogen and sulfur species using thioredoxin as hydrogen donor; deletion enhances the mutator phenotype of tsa1 mutants |
|
| YPR184W | 0.93 |
GDB1
|
Glycogen debranching enzyme containing glucanotranferase and alpha-1,6-amyloglucosidase activities, required for glycogen degradation; phosphorylated in mitochondria |
|
| YIL108W | 0.93 |
Putative metalloprotease |
||
| YOR343C | 0.92 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YBR145W | 0.92 |
ADH5
|
Alcohol dehydrogenase isoenzyme V; involved in ethanol production |
|
| YFL031W | 0.92 |
HAC1
|
bZIP transcription factor (ATF/CREB1 homolog) that regulates the unfolded protein response, via UPRE binding, and membrane biogenesis; ER stress-induced splicing pathway utilizing Ire1p, Trl1p and Ada5p facilitates efficient Hac1p synthesis |
|
| YKL043W | 0.92 |
PHD1
|
Transcriptional activator that enhances pseudohyphal growth; regulates expression of FLO11, an adhesin required for pseudohyphal filament formation; similar to StuA, an A. nidulans developmental regulator; potential Cdc28p substrate |
|
| YPL171C | 0.91 |
OYE3
|
Widely conserved NADPH oxidoreductase containing flavin mononucleotide (FMN), homologous to Oye2p with slight differences in ligand binding and catalytic properties; may be involved in sterol metabolism |
|
| YOR317W | 0.90 |
FAA1
|
Long chain fatty acyl-CoA synthetase with a preference for C12:0-C16:0 fatty acids; involved in the activation of imported fatty acids; localized to both lipid particles and mitochondrial outer membrane; essential for stationary phase |
|
| YDR033W | 0.90 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YEL028W | 0.89 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YBL045C | 0.86 |
COR1
|
Core subunit of the ubiquinol-cytochrome c reductase complex (bc1 complex), which is a component of the mitochondrial inner membrane electron transport chain |
|
| YDR264C | 0.86 |
AKR1
|
Palmitoyl transferase involved in protein palmitoylation; acts as a negative regulator of pheromone response pathway; required for endocytosis of pheromone receptors; involved in cell shape control; contains ankyrin repeats |
|
| YEL027W | 0.85 |
CUP5
|
Proteolipid subunit of the vacuolar H(+)-ATPase V0 sector (subunit c; dicyclohexylcarbodiimide binding subunit); required for vacuolar acidification and important for copper and iron metal ion homeostasis |
|
| YNR050C | 0.84 |
LYS9
|
Saccharopine dehydrogenase (NADP+, L-glutamate-forming); catalyzes the formation of saccharopine from alpha-aminoadipate 6-semialdehyde, which is the seventh step in lysine biosynthesis pathway |
|
| YER010C | 0.84 |
Protein of unknown function, forms a ring-shaped homotrimer; has similarity to members of the prokaryotic rraA family; possibly involved in a phosphotransfer reaction |
||
| YER141W | 0.83 |
COX15
|
Protein required for the hydroxylation of heme O to form heme A, which is an essential prosthetic group for cytochrome c oxidase |
|
| YNL190W | 0.83 |
Cell wall protein of unknown function; proposed role as a hydrophilin induced by osmotic stress; contains a putative GPI-attachment site |
||
| YHR177W | 0.82 |
Putative protein of unknown function |
||
| YIR020W-A | 0.81 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YDR010C | 0.80 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLL053C | 0.79 |
Putative protein; in the Sigma 1278B strain background YLL053C is contiguous with AQY2 which encodes an aquaporin |
||
| YHL044W | 0.79 |
Putative integral membrane protein, member of DUP240 gene family; green fluorescent protein (GFP)-fusion protein localizes to the plasma membrane in a punctate pattern |
||
| YHL040C | 0.79 |
ARN1
|
Transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; responsible for uptake of iron bound to ferrirubin, ferrirhodin, and related siderophores |
|
| YER066W | 0.79 |
Putative protein of unknown function; YER066W is not an essential gene |
||
| YOR152C | 0.78 |
Putative protein of unknown function; has no similarity to any known protein; YOR152C is not an essential gene |
||
| YLR438W | 0.78 |
CAR2
|
L-ornithine transaminase (OTAse), catalyzes the second step of arginine degradation, expression is dually-regulated by allophanate induction and a specific arginine induction process; not nitrogen catabolite repression sensitive |
|
| YIL033C | 0.78 |
BCY1
|
Regulatory subunit of the cyclic AMP-dependent protein kinase (PKA), a component of a signaling pathway that controls a variety of cellular processes, including metabolism, cell cycle, stress response, stationary phase, and sporulation |
|
| YPL092W | 0.77 |
SSU1
|
Plasma membrane sulfite pump involved in sulfite metabolism and required for efficient sulfite efflux; major facilitator superfamily protein |
|
| YBR177C | 0.77 |
EHT1
|
Acyl-coenzymeA:ethanol O-acyltransferase that plays a minor role in medium-chain fatty acid ethyl ester biosynthesis; possesses short-chain esterase activity; localizes to lipid particles and the mitochondrial outer membrane |
|
| YFR053C | 0.76 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YIR039C | 0.76 |
YPS6
|
Putative GPI-anchored aspartic protease |
|
| YJL172W | 0.75 |
CPS1
|
Vacuolar carboxypeptidase yscS; expression is induced under low-nitrogen conditions |
|
| YJR116W | 0.73 |
Putative protein of unknown function |
||
| YMR210W | 0.73 |
Putative acyltransferase with similarity to Eeb1p and Eht1p, has a minor role in medium-chain fatty acid ethyl ester biosynthesis; may be involved in lipid metabolism and detoxification |
||
| YDL204W | 0.73 |
RTN2
|
Protein of unknown function; has similarity to mammalian reticulon proteins; member of the RTNLA (reticulon-like A) subfamily |
|
| YBR183W | 0.73 |
YPC1
|
Alkaline ceramidase that also has reverse (CoA-independent) ceramide synthase activity, catalyzes both breakdown and synthesis of phytoceramide; overexpression confers fumonisin B1 resistance |
|
| YGR204W | 0.73 |
ADE3
|
Cytoplasmic trifunctional enzyme C1-tetrahydrofolate synthase, involved in single carbon metabolism and required for biosynthesis of purines, thymidylate, methionine, and histidine |
|
| YDR345C | 0.72 |
HXT3
|
Low affinity glucose transporter of the major facilitator superfamily, expression is induced in low or high glucose conditions |
|
| YLR328W | 0.72 |
NMA1
|
Nicotinic acid mononucleotide adenylyltransferase, involved in pathways of NAD biosynthesis, including the de novo, NAD(+) salvage, and nicotinamide riboside salvage pathways |
|
| YMR316C-A | 0.72 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps the verified gene DIA1/YMR316W |
||
| YJR146W | 0.72 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene HMS2 |
||
| YBR162C | 0.72 |
TOS1
|
Covalently-bound cell wall protein of unknown function; identified as a cell cycle regulated SBF target gene; deletion mutants are highly resistant to treatment with beta-1,3-glucanase; has sequence similarity to YJL171C |
|
| YOR100C | 0.71 |
CRC1
|
Mitochondrial inner membrane carnitine transporter, required for carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria during fatty acid beta-oxidation |
|
| YFR047C | 0.71 |
BNA6
|
Quinolinate phosphoribosyl transferase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p |
|
| YGR144W | 0.71 |
THI4
|
Thiazole synthase, catalyzes formation of the thiazole moiety of thiamin pyrophosphate; required for thiamine biosynthesis and for mitochondrial genome stability |
|
| YGL053W | 0.70 |
PRM8
|
Pheromone-regulated protein with 2 predicted transmembrane segments and an FF sequence, a motif involved in COPII binding; forms a complex with Prp9p in the ER; member of DUP240 gene family |
|
| YGL258W-A | 0.70 |
Putative protein of unknown function |
||
| YEL009C | 0.68 |
GCN4
|
Transcriptional activator of amino acid biosynthetic genes in response to amino acid starvation; expression is tightly regulated at both the transcriptional and translational levels |
|
| YBR214W | 0.68 |
SDS24
|
One of two S. cerevisiae homologs (Sds23p and Sds24p) of the Schizosaccharomyces pombe Sds23 protein, which genetic studies have implicated in APC/cyclosome regulation; may play an indirect role in fluid-phase endocytosis |
|
| YLL052C | 0.68 |
AQY2
|
Water channel that mediates the transport of water across cell membranes, only expressed in proliferating cells, controlled by osmotic signals, may be involved in freeze tolerance; disrupted by a stop codon in many S. cerevisiae strains |
|
| YLR259C | 0.68 |
HSP60
|
Tetradecameric mitochondrial chaperonin required for ATP-dependent folding of precursor polypeptides and complex assembly; prevents aggregation and mediates protein refolding after heat shock; role in mtDNA transmission; phosphorylated |
|
| YER091C-A | 0.67 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YNL336W | 0.67 |
COS1
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YOL081W | 0.67 |
IRA2
|
GTPase-activating protein that negatively regulates RAS by converting it from the GTP- to the GDP-bound inactive form, required for reducing cAMP levels under nutrient limiting conditions, has similarity to Ira1p and human neurofibromin |
|
| YKL051W | 0.67 |
SFK1
|
Plasma membrane protein that may act together with or upstream of Stt4p to generate normal levels of the essential phospholipid PI4P, at least partially mediates proper localization of Stt4p to the plasma membrane |
|
| YLR437C-A | 0.67 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF CAR2/YLR438W |
||
| YAL061W | 0.66 |
BDH2
|
Putative medium-chain alcohol dehydrogenase with similarity to BDH1; transcription induced by constitutively active PDR1 and PDR3; BDH2 is an essential gene |
|
| YML126C | 0.65 |
ERG13
|
3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase, catalyzes the formation of HMG-CoA from acetyl-CoA and acetoacetyl-CoA; involved in the second step in mevalonate biosynthesis |
|
| YER044C | 0.65 |
ERG28
|
Endoplasmic reticulum membrane protein, may facilitate protein-protein interactions between the Erg26p dehydrogenase and the Erg27p 3-ketoreductase and/or tether these enzymes to the ER, also interacts with Erg6p |
|
| YHL025W | 0.64 |
SNF6
|
Subunit of the SWI/SNF chromatin remodeling complex involved in transcriptional regulation; functions interdependently in transcriptional activation with Snf2p and Snf5p |
|
| YPR010C-A | 0.64 |
Putative protein of unknown function; conserved among Saccharomyces sensu stricto species |
||
| YHR199C | 0.64 |
AIM46
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YCL035C | 0.63 |
GRX1
|
Hydroperoxide and superoxide-radical responsive heat-stable glutathione-dependent disulfide oxidoreductase with active site cysteine pair; protects cells from oxidative damage |
|
| YLR122C | 0.63 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR123C |
||
| YML132W | 0.63 |
COS3
|
Protein involved in salt resistance; interacts with sodium:hydrogen antiporter Nha1p; member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YGR284C | 0.63 |
ERV29
|
Protein localized to COPII-coated vesicles, involved in vesicle formation and incorporation of specific secretory cargo |
|
| YLR023C | 0.63 |
IZH3
|
Membrane protein involved in zinc metabolism, member of the four-protein IZH family, expression induced by zinc deficiency; deletion reduces sensitivity to elevated zinc and shortens lag phase, overexpression reduces Zap1p activity |
|
| YGR023W | 0.62 |
MTL1
|
Protein with both structural and functional similarity to Mid2p, which is a plasma membrane sensor required for cell integrity signaling during pheromone-induced morphogenesis; suppresses rgd1 null mutations |
|
| YMR206W | 0.62 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YNR014W | 0.62 |
Putative protein of unknown function; expression is cell-cycle regulated, Azf1p-dependent, and heat-inducible |
||
| YMR007W | 0.62 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YNL307C | 0.62 |
MCK1
|
Protein serine/threonine/tyrosine (dual-specificity) kinase involved in control of chromosome segregation and in regulating entry into meiosis; related to mammalian glycogen synthase kinases of the GSK-3 family |
|
| YMR170C | 0.62 |
ALD2
|
Cytoplasmic aldehyde dehydrogenase, involved in ethanol oxidation and beta-alanine biosynthesis; uses NAD+ as the preferred coenzyme; expression is stress induced and glucose repressed; very similar to Ald3p |
|
| YDL129W | 0.62 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and the nucleus; YDL129W is not an essential gene |
||
| YML058W | 0.62 |
SML1
|
Ribonucleotide reductase inhibitor involved in regulating dNTP production; regulated by Mec1p and Rad53p during DNA damage and S phase |
|
| YJL213W | 0.61 |
Protein of unknown function that may interact with ribosomes; periodically expressed during the yeast metabolic cycle; phosphorylated in vitro by the mitotic exit network (MEN) kinase complex, Dbf2p/Mob1p |
||
| YAL042W | 0.61 |
ERV46
|
Protein localized to COPII-coated vesicles, forms a complex with Erv41p; involved in the membrane fusion stage of transport |
|
| YOR204W | 0.61 |
DED1
|
ATP-dependent DEAD (Asp-Glu-Ala-Asp)-box RNA helicase, required for translation initiation of all yeast mRNAs; mutations in human DEAD-box DBY are a frequent cause of male infertility |
|
| YNR034W | 0.61 |
SOL1
|
Protein with a possible role in tRNA export; shows similarity to 6-phosphogluconolactonase non-catalytic domains but does not exhibit this enzymatic activity; homologous to Sol2p, Sol3p, and Sol4p |
|
| YJR153W | 0.61 |
PGU1
|
Endo-polygalacturonase, pectolytic enzyme that hydrolyzes the alpha-1,4-glycosidic bonds in the rhamnogalacturonan chains in pectins |
|
| YOR378W | 0.61 |
Putative protein of unknown function; belongs to the DHA2 family of drug:H+ antiporters; YOR378W is not an essential gene |
||
| YLR356W | 0.60 |
Putative protein of unknown function with similarity to SCM4; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR356W is not an essential gene |
||
| YAL034C | 0.60 |
FUN19
|
Non-essential protein of unknown function |
|
| YNL195C | 0.60 |
Putative protein of unknown function; shares a promoter with YNL194C; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YLR130C | 0.60 |
ZRT2
|
Low-affinity zinc transporter of the plasma membrane; transcription is induced under low-zinc conditions by the Zap1p transcription factor |
|
| YNL117W | 0.59 |
MLS1
|
Malate synthase, enzyme of the glyoxylate cycle, involved in utilization of non-fermentable carbon sources; expression is subject to carbon catabolite repression; localizes in peroxisomes during growth in oleic acid medium |
|
| YPL257W | 0.59 |
Putative protein of unknown function;; homozygous diploid deletion strain exhibits low budding index; physically interacts with Hsp82p; YPL257W is not an essential gene |
||
| YNR058W | 0.59 |
BIO3
|
7,8-diamino-pelargonic acid aminotransferase (DAPA), catalyzes the second step in the biotin biosynthesis pathway; BIO3 is in a cluster of 3 genes (BIO3, BIO4, and BIO5) that mediate biotin synthesis |
|
| YBR101C | 0.59 |
FES1
|
Hsp70 (Ssa1p) nucleotide exchange factor, cytosolic homolog of Sil1p, which is the nucleotide exchange factor for BiP (Kar2p) in the endoplasmic reticulum |
|
| YNR057C | 0.59 |
BIO4
|
Dethiobiotin synthetase, catalyzes the third step in the biotin biosynthesis pathway; BIO4 is in a cluster of 3 genes (BIO3, BIO4, and BIO5) that mediate biotin synthesis; expression appears to be repressed at low iron levels |
|
| YOR247W | 0.59 |
SRL1
|
Mannoprotein that exhibits a tight association with the cell wall, required for cell wall stability in the absence of GPI-anchored mannoproteins; has a high serine-threonine content; expression is induced in cell wall mutants |
|
| YNL305C | 0.58 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YNL305C is not an essential gene |
||
| YPL244C | 0.58 |
HUT1
|
Protein with a role in UDP-galactose transport to the Golgi lumen, has similarity to human UDP-galactose transporter UGTrel1, exhibits a genetic interaction with S. cerevisiae ERO1 |
|
| YIL121W | 0.58 |
QDR2
|
Multidrug transporter of the major facilitator superfamily, required for resistance to quinidine, barban, cisplatin, and bleomycin; may have a role in potassium uptake |
|
| YER124C | 0.57 |
DSE1
|
Daughter cell-specific protein, may participate in pathways regulating cell wall metabolism; deletion affects cell separation after division and sensitivity to drugs targeted against the cell wall |
|
| YPL179W | 0.57 |
PPQ1
|
Putative protein serine/threonine phosphatase; null mutation enhances efficiency of translational suppressors |
|
| YJR085C | 0.57 |
Putative protein of unknown function; GFP-fusion protein is induced in response to the DNA-damaging agent MMS; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YER065C | 0.57 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YJR048W | 0.57 |
CYC1
|
Cytochrome c, isoform 1; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YGR038W | 0.57 |
ORM1
|
Evolutionarily conserved protein with similarity to Orm2p, required for resistance to agents that induce the unfolded protein response; human ortholog is located in the endoplasmic reticulum |
|
| YGL055W | 0.56 |
OLE1
|
Delta(9) fatty acid desaturase, required for monounsaturated fatty acid synthesis and for normal distribution of mitochondria |
|
| YGL040C | 0.56 |
HEM2
|
Delta-aminolevulinate dehydratase, a homo-octameric enzyme, catalyzes the conversion of delta-aminolevulinic acid to porphobilinogen, the second step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus |
|
| YER060W | 0.56 |
FCY21
|
Putative purine-cytosine permease, very similar to Fcy2p but cannot substitute for its function |
|
| YLR121C | 0.56 |
YPS3
|
Aspartic protease, attached to the plasma membrane via a glycosylphosphatidylinositol (GPI) anchor |
|
| YOL047C | 0.56 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
||
| YPR156C | 0.56 |
TPO3
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; member of the major facilitator superfamily |
|
| YHR212C | 0.55 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR065C | 0.55 |
VHT1
|
High-affinity plasma membrane H+-biotin (vitamin H) symporter; mutation results in fatty acid auxotrophy; 12 transmembrane domain containing major facilitator subfamily member; mRNA levels negatively regulated by iron deprivation and biotin |
|
| YPL250C | 0.55 |
ICY2
|
Protein of unknown function; mobilized into polysomes upon a shift from a fermentable to nonfermentable carbon source; potential Cdc28p substrate |
|
| YDL160C | 0.55 |
DHH1
|
Cytoplasmic DExD/H-box helicase, stimulates mRNA decapping, coordinates distinct steps in mRNA function and decay, interacts with both the decapping and deadenylase complexes, may have a role in mRNA export and translation |
|
| YGL187C | 0.55 |
COX4
|
Subunit IV of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; N-terminal 25 residues of precursor are cleaved during mitochondrial import; phosphorylated |
|
| YDR533C | 0.54 |
HSP31
|
Possible chaperone and cysteine protease with similarity to E. coli Hsp31; member of the DJ-1/ThiJ/PfpI superfamily, which includes human DJ-1 involved in Parkinson's disease; exists as a dimer and contains a putative metal-binding site |
|
| YDR492W | 0.54 |
IZH1
|
Membrane protein involved in zinc metabolism, member of the four-protein IZH family; transcription is regulated directly by Zap1p, expression induced by zinc deficiency and fatty acids; deletion increases sensitivity to elevated zinc |
|
| YDL124W | 0.54 |
NADPH-dependent alpha-keto amide reductase; reduces aromatic alpha-keto amides, aliphatic alpha-keto esters, and aromatic alpha-keto esters; member of the aldo-keto reductase (AKR) family |
||
| YIL109C | 0.53 |
SEC24
|
Component of the Sec23p-Sec24p heterodimeric complex of the COPII vesicle coat; involved in ER to Golgi transport, cargo selection and autophagy; required for the binding of the Sec13 complex to ER membranes; homologous to Lst1p and Lss1p |
|
| YGR022C | 0.53 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF MTL1/YGR023W |
||
| YDL244W | 0.53 |
THI13
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YGR032W | 0.53 |
GSC2
|
Catalytic subunit of 1,3-beta-glucan synthase, involved in formation of the inner layer of the spore wall; activity positively regulated by Rho1p and negatively by Smk1p; has similarity to an alternate catalytic subunit, Fks1p (Gsc1p) |
|
| YBL099W | 0.53 |
ATP1
|
Alpha subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; phosphorylated |
|
| YJL152W | 0.52 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPL272C | 0.52 |
Putative protein of unknown function; gene expression induced in response to ketoconazole; YPL272C is not an essential gene |
||
| YOR099W | 0.52 |
KTR1
|
Alpha-1,2-mannosyltransferase involved in O- and N-linked protein glycosylation; type II membrane protein; member of the KRE2/MNT1 mannosyltransferase family |
|
| YJL158C | 0.52 |
CIS3
|
Mannose-containing glycoprotein constituent of the cell wall; member of the PIR (proteins with internal repeats) family |
|
| YGL256W | 0.52 |
ADH4
|
Alcohol dehydrogenase isoenzyme type IV, dimeric enzyme demonstrated to be zinc-dependent despite sequence similarity to iron-activated alcohol dehydrogenases; transcription is induced in response to zinc deficiency |
|
| YBL081W | 0.51 |
Non-essential protein of unknown function |
||
| YFL054C | 0.50 |
Putative channel-like protein; similar to Fps1p; mediates passive diffusion of glycerol in the presence of ethanol |
||
| YBR139W | 0.50 |
Putative serine type carboxypeptidase with a role in phytochelatin synthesis; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; expression induced by nitrogen limitation in a GLN3, GAT1-independent manner |
||
| YPL014W | 0.50 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and to the nucleus |
||
| YDL125C | 0.50 |
HNT1
|
Adenosine 5'-monophosphoramidase; interacts physically and genetically with Kin28p, a CDK and TFIIK subunit, and genetically with CAK1; member of the histidine triad (HIT) superfamily of nucleotide-binding proteins and similar to Hint |
Network of associatons between targets according to the STRING database.
First level regulatory network of UPC2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.3 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.7 | 5.5 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.6 | 1.9 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) regulation of membrane lipid metabolic process(GO:1905038) |
| 0.5 | 1.6 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.5 | 2.0 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.5 | 1.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.5 | 0.5 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.4 | 0.9 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.4 | 6.0 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.4 | 2.1 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.4 | 1.6 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.4 | 1.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 0.4 | 3.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.4 | 1.1 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.4 | 2.2 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.3 | 1.0 | GO:0046160 | heme a biosynthetic process(GO:0006784) heme a metabolic process(GO:0046160) |
| 0.3 | 1.0 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.3 | 0.3 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.3 | 1.7 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.3 | 0.6 | GO:0033559 | unsaturated fatty acid biosynthetic process(GO:0006636) unsaturated fatty acid metabolic process(GO:0033559) |
| 0.3 | 0.8 | GO:1900371 | regulation of cyclic nucleotide metabolic process(GO:0030799) regulation of cyclic nucleotide biosynthetic process(GO:0030802) regulation of nucleotide biosynthetic process(GO:0030808) regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of purine nucleotide biosynthetic process(GO:1900371) |
| 0.3 | 0.3 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.2 | 2.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 2.0 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.2 | 1.5 | GO:0010969 | regulation of pheromone-dependent signal transduction involved in conjugation with cellular fusion(GO:0010969) regulation of signal transduction involved in conjugation with cellular fusion(GO:0060238) |
| 0.2 | 0.7 | GO:0090110 | cargo loading into vesicle(GO:0035459) cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 0.4 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.2 | 0.9 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.2 | 0.6 | GO:0015856 | cytosine transport(GO:0015856) purine-containing compound transmembrane transport(GO:0072530) |
| 0.2 | 0.6 | GO:0015786 | pyrimidine nucleotide-sugar transport(GO:0015781) UDP-glucose transport(GO:0015786) |
| 0.2 | 0.8 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.2 | 0.8 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.2 | 0.8 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.2 | 1.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 3.6 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.2 | 1.1 | GO:0006768 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.2 | 0.9 | GO:0048209 | regulation of COPII vesicle coating(GO:0003400) regulation of vesicle targeting, to, from or within Golgi(GO:0048209) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.2 | 0.5 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.2 | 0.5 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.2 | 0.8 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.2 | 0.5 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.2 | 0.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.2 | 0.2 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) |
| 0.2 | 0.9 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.2 | 0.8 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.2 | 0.6 | GO:0010674 | negative regulation of transcription from RNA polymerase II promoter involved in meiotic cell cycle(GO:0010674) |
| 0.2 | 0.3 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.2 | 0.9 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.2 | 0.3 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.1 | 0.7 | GO:0043954 | cellular component maintenance(GO:0043954) |
| 0.1 | 0.9 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.8 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.7 | GO:0034487 | amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 0.1 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 4.0 | GO:0044108 | ergosterol biosynthetic process(GO:0006696) phytosteroid biosynthetic process(GO:0016129) cellular alcohol biosynthetic process(GO:0044108) cellular lipid biosynthetic process(GO:0097384) |
| 0.1 | 0.3 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.1 | 0.3 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.1 | 0.5 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.1 | 0.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 1.3 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.1 | 2.1 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 0.1 | GO:1902653 | secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 0.4 | GO:0050685 | positive regulation of mRNA processing(GO:0050685) |
| 0.1 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.7 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.4 | GO:0006672 | ceramide metabolic process(GO:0006672) ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.2 | GO:0034764 | positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) positive regulation of sterol transport(GO:0032373) positive regulation of transmembrane transport(GO:0034764) regulation of sterol import by regulation of transcription from RNA polymerase II promoter(GO:0035968) positive regulation of sterol import by positive regulation of transcription from RNA polymerase II promoter(GO:0035969) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.1 | 0.6 | GO:0008299 | isoprenoid metabolic process(GO:0006720) isoprenoid biosynthetic process(GO:0008299) |
| 0.1 | 0.6 | GO:0032105 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.1 | 0.3 | GO:0032365 | intracellular lipid transport(GO:0032365) |
| 0.1 | 0.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 1.4 | GO:0007135 | meiosis II(GO:0007135) meiotic sister chromatid segregation(GO:0045144) |
| 0.1 | 0.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.8 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.6 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.3 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) |
| 0.1 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.1 | GO:0019365 | nicotinate nucleotide biosynthetic process(GO:0019357) nicotinate nucleotide salvage(GO:0019358) pyridine nucleotide salvage(GO:0019365) nicotinate nucleotide metabolic process(GO:0046497) |
| 0.1 | 0.4 | GO:0009164 | nucleoside catabolic process(GO:0009164) glycosyl compound catabolic process(GO:1901658) |
| 0.1 | 0.7 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 0.3 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) |
| 0.1 | 0.5 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.1 | 0.5 | GO:0019563 | alditol catabolic process(GO:0019405) glycerol catabolic process(GO:0019563) |
| 0.1 | 0.4 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.1 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.2 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.1 | GO:0010672 | regulation of transcription from RNA polymerase II promoter involved in meiotic cell cycle(GO:0010672) |
| 0.1 | 0.4 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 2.6 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 0.3 | GO:0000296 | spermine transport(GO:0000296) |
| 0.1 | 0.3 | GO:1903513 | retrograde protein transport, ER to cytosol(GO:0030970) endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.1 | 0.2 | GO:0015851 | nucleobase transport(GO:0015851) |
| 0.1 | 0.2 | GO:0060988 | lipid tube assembly(GO:0060988) protein-lipid complex assembly(GO:0065005) protein-lipid complex subunit organization(GO:0071825) |
| 0.1 | 0.4 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.1 | 0.1 | GO:0031684 | heterotrimeric G-protein complex cycle(GO:0031684) |
| 0.1 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.2 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 1.2 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.1 | 0.3 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) |
| 0.1 | 0.1 | GO:0061414 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 0.1 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.1 | GO:0046323 | glucose import(GO:0046323) |
| 0.1 | 0.8 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 0.5 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.1 | 0.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.1 | 0.2 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 0.1 | 0.9 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.1 | 0.3 | GO:1901070 | guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.1 | 1.3 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.1 | 0.3 | GO:0071616 | thioester biosynthetic process(GO:0035384) acyl-CoA biosynthetic process(GO:0071616) |
| 0.1 | 0.1 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.4 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 1.0 | GO:0031503 | protein complex localization(GO:0031503) |
| 0.0 | 0.4 | GO:0048017 | inositol lipid-mediated signaling(GO:0048017) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.8 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.0 | GO:0042743 | hydrogen peroxide metabolic process(GO:0042743) |
| 0.0 | 0.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0016241 | regulation of macroautophagy(GO:0016241) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.3 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.2 | GO:0035753 | maintenance of DNA trinucleotide repeats(GO:0035753) |
| 0.0 | 0.3 | GO:0000272 | polysaccharide catabolic process(GO:0000272) |
| 0.0 | 0.1 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 0.0 | 0.3 | GO:0070096 | mitochondrial outer membrane translocase complex assembly(GO:0070096) |
| 0.0 | 0.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.0 | GO:0052652 | cAMP biosynthetic process(GO:0006171) cyclic nucleotide biosynthetic process(GO:0009190) cyclic purine nucleotide metabolic process(GO:0052652) |
| 0.0 | 0.3 | GO:0009262 | deoxyribonucleotide metabolic process(GO:0009262) deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.0 | GO:0001315 | age-dependent response to reactive oxygen species(GO:0001315) cellular age-dependent response to reactive oxygen species(GO:0072353) |
| 0.0 | 0.8 | GO:0097034 | mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0043649 | glutamate catabolic process(GO:0006538) dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.7 | GO:0008645 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.0 | 0.1 | GO:0034762 | regulation of transmembrane transport(GO:0034762) |
| 0.0 | 0.2 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.2 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.1 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:1901661 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) ketone biosynthetic process(GO:0042181) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.3 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.2 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0010388 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 | 0.1 | GO:0032186 | cellular bud neck septin ring organization(GO:0032186) |
| 0.0 | 0.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:0007119 | budding cell isotropic bud growth(GO:0007119) |
| 0.0 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0098630 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) aggregation of unicellular organisms(GO:0098630) cell aggregation(GO:0098743) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.3 | GO:0007155 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 0.1 | GO:0044107 | ergosterol metabolic process(GO:0008204) phytosteroid metabolic process(GO:0016128) cellular alcohol metabolic process(GO:0044107) |
| 0.0 | 0.1 | GO:0001881 | receptor recycling(GO:0001881) protein import into peroxisome matrix, receptor recycling(GO:0016562) receptor metabolic process(GO:0043112) |
| 0.0 | 0.1 | GO:0071041 | antisense RNA transcript catabolic process(GO:0071041) |
| 0.0 | 0.4 | GO:0042326 | negative regulation of phosphorylation(GO:0042326) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.0 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.0 | 0.2 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 2.7 | GO:0045229 | external encapsulating structure organization(GO:0045229) cell wall organization(GO:0071555) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.4 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.2 | GO:0090529 | barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.0 | 0.0 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.4 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.0 | 0.0 | GO:0031333 | negative regulation of protein complex assembly(GO:0031333) |
| 0.0 | 0.1 | GO:0000730 | meiotic DNA recombinase assembly(GO:0000707) DNA recombinase assembly(GO:0000730) |
| 0.0 | 0.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0000435 | regulation of transcription from RNA polymerase II promoter by galactose(GO:0000431) positive regulation of transcription from RNA polymerase II promoter by galactose(GO:0000435) |
| 0.0 | 0.1 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0006639 | neutral lipid metabolic process(GO:0006638) acylglycerol metabolic process(GO:0006639) triglyceride metabolic process(GO:0006641) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) pantothenate biosynthetic process(GO:0015940) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.0 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) maintenance of sister chromatid cohesion(GO:0034086) regulation of maintenance of sister chromatid cohesion(GO:0034091) |
| 0.0 | 0.2 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.0 | 0.1 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.0 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.1 | GO:0006101 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.0 | 0.1 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) AMP metabolic process(GO:0046033) |
| 0.0 | 0.0 | GO:0045980 | negative regulation of nucleotide metabolic process(GO:0045980) |
| 0.0 | 0.1 | GO:0034620 | endoplasmic reticulum unfolded protein response(GO:0030968) cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.0 | GO:0045744 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.0 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.0 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.0 | GO:0051303 | meiotic telomere clustering(GO:0045141) establishment of chromosome localization(GO:0051303) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 | 0.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.0 | GO:0006465 | signal peptide processing(GO:0006465) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0005756 | mitochondrial proton-transporting ATP synthase, central stalk(GO:0005756) proton-transporting ATP synthase, central stalk(GO:0045269) |
| 0.4 | 1.3 | GO:0030428 | cell septum(GO:0030428) |
| 0.4 | 3.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.4 | 3.7 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.3 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 0.5 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 0.5 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.2 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 15.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.2 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 0.6 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.2 | 0.7 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.2 | 1.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 4.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.2 | 1.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 1.0 | GO:0035339 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.2 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 3.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.7 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.4 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.8 | GO:0009277 | fungal-type cell wall(GO:0009277) |
| 0.1 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.3 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.1 | 0.4 | GO:0000837 | Doa10p ubiquitin ligase complex(GO:0000837) |
| 0.1 | 0.2 | GO:0071561 | nucleus-vacuole junction(GO:0071561) |
| 0.1 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.1 | GO:0031310 | intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.2 | GO:0001400 | mating projection base(GO:0001400) |
| 0.1 | 0.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 2.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 2.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.7 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0032126 | eisosome(GO:0032126) |
| 0.0 | 0.1 | GO:0071256 | translocon complex(GO:0071256) |
| 0.0 | 0.5 | GO:0098797 | plasma membrane protein complex(GO:0098797) |
| 0.0 | 0.2 | GO:0034044 | exomer complex(GO:0034044) |
| 0.0 | 3.0 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 2.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 25.5 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.2 | GO:0000500 | RNA polymerase I upstream activating factor complex(GO:0000500) |
| 0.0 | 0.2 | GO:0044440 | endosomal part(GO:0044440) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0004564 | beta-fructofuranosidase activity(GO:0004564) sucrose alpha-glucosidase activity(GO:0004575) |
| 1.0 | 3.9 | GO:0015343 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.7 | 3.0 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.6 | 1.9 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.6 | 1.9 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.6 | 2.4 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.5 | 6.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.5 | 2.2 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.5 | 1.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.4 | 1.2 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.4 | 1.6 | GO:0016885 | ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.4 | 1.2 | GO:0030414 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 0.4 | 1.9 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.3 | 1.0 | GO:0016872 | intramolecular lyase activity(GO:0016872) |
| 0.3 | 0.9 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.3 | 0.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 1.7 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.3 | 4.6 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.3 | 1.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 1.8 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.3 | 0.8 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.2 | 2.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 0.7 | GO:0015489 | putrescine transmembrane transporter activity(GO:0015489) |
| 0.2 | 0.7 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 1.6 | GO:0010181 | FMN binding(GO:0010181) |
| 0.2 | 0.6 | GO:0033592 | RNA strand annealing activity(GO:0033592) annealing activity(GO:0097617) |
| 0.2 | 2.4 | GO:0022838 | substrate-specific channel activity(GO:0022838) |
| 0.2 | 1.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 0.6 | GO:0016880 | AMP binding(GO:0016208) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 1.7 | GO:0016723 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.2 | 0.9 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.2 | 1.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 1.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 0.8 | GO:0034318 | alcohol O-acetyltransferase activity(GO:0004026) alcohol O-acyltransferase activity(GO:0034318) |
| 0.2 | 1.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 0.5 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.2 | 0.8 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.2 | 1.1 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.2 | 0.6 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.2 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 0.5 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 0.6 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 1.7 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 2.5 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 0.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.4 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.1 | 0.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 2.2 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 1.1 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.4 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.1 | 2.4 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.1 | 0.4 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.1 | 0.7 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.7 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.9 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 0.6 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.7 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.4 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 1.7 | GO:0016769 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.3 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.4 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.1 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.4 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 0.1 | 1.1 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.9 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.1 | 0.2 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.1 | 2.1 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.1 | 0.1 | GO:0008106 | aldo-keto reductase (NADP) activity(GO:0004033) alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 0.7 | GO:0004601 | peroxidase activity(GO:0004601) oxidoreductase activity, acting on peroxide as acceptor(GO:0016684) |
| 0.1 | 0.1 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 0.2 | GO:0015216 | purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.1 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.7 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.5 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.1 | 0.8 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.1 | 0.1 | GO:0015085 | calcium ion transmembrane transporter activity(GO:0015085) |
| 0.1 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.7 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 0.1 | 0.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.3 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.1 | 0.4 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 0.2 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.2 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.0 | 0.1 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 1.0 | GO:0016830 | carbon-carbon lyase activity(GO:0016830) |
| 0.0 | 0.2 | GO:0001168 | transcription factor activity, RNA polymerase I upstream control element sequence-specific binding(GO:0001168) |
| 0.0 | 0.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.2 | GO:0019212 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.2 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.6 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.0 | 0.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.3 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.4 | GO:0015578 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.4 | GO:0038023 | transmembrane signaling receptor activity(GO:0004888) signaling receptor activity(GO:0038023) transmembrane receptor activity(GO:0099600) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.2 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.0 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0016898 | lactate dehydrogenase activity(GO:0004457) D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.0 | 0.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0016859 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.0 | 0.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0008144 | drug binding(GO:0008144) |
| 0.0 | 0.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.8 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.0 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.3 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 0.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 36.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.0 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.2 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.3 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.1 | 1.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 0.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 0.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.0 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.0 | 36.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | Genes involved in Transmembrane transport of small molecules |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |