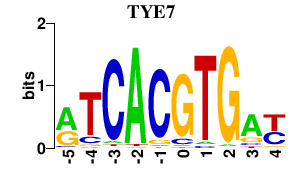
Results for TYE7
Z-value: 0.39
Transcription factors associated with TYE7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TYE7
|
S000005871 | Serine-rich protein that contains a bHLH DNA binding motif |
Activity-expression correlation:
Activity profile of TYE7 motif
Sorted Z-values of TYE7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YCR021C | 2.28 |
HSP30
|
Hydrophobic plasma membrane localized, stress-responsive protein that negatively regulates the H(+)-ATPase Pma1p; induced by heat shock, ethanol treatment, weak organic acid, glucose limitation, and entry into stationary phase |
|
| YJL133C-A | 2.25 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YKL016C | 2.19 |
ATP7
|
Subunit d of the stator stalk of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YML089C | 2.07 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression induced by calcium shortage |
||
| YJR095W | 1.99 |
SFC1
|
Mitochondrial succinate-fumarate transporter, transports succinate into and fumarate out of the mitochondrion; required for ethanol and acetate utilization |
|
| YJR115W | 1.98 |
Putative protein of unknown function |
||
| YPR001W | 1.71 |
CIT3
|
Dual specificity mitochondrial citrate and methylcitrate synthase; catalyzes the condensation of acetyl-CoA and oxaloacetate to form citrate and that of propionyl-CoA and oxaloacetate to form 2-methylcitrate |
|
| YJR138W | 1.70 |
IML1
|
Protein of unknown function, green fluorescent protein (GFP)-fusion protein localizes to the vacuolar membrane |
|
| YNL036W | 1.60 |
NCE103
|
Carbonic anhydrase; poorly transcribed under aerobic conditions and at an undetectable level under anaerobic conditions; involved in non-classical protein export pathway |
|
| YKL163W | 1.56 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YFL024C | 1.54 |
EPL1
|
Component of NuA4, which is an essential histone H4/H2A acetyltransferase complex; homologous to Drosophila Enhancer of Polycomb |
|
| YPL250C | 1.53 |
ICY2
|
Protein of unknown function; mobilized into polysomes upon a shift from a fermentable to nonfermentable carbon source; potential Cdc28p substrate |
|
| YMR206W | 1.46 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YNL063W | 1.39 |
MTQ1
|
S-adenosylmethionine-dependent methyltransferase; methylates translational release factor Mrf1p; similar to E.coli PrmC; is not an essential gene |
|
| YHL036W | 1.35 |
MUP3
|
Low affinity methionine permease, similar to Mup1p |
|
| YAL054C | 1.34 |
ACS1
|
Acetyl-coA synthetase isoform which, along with Acs2p, is the nuclear source of acetyl-coA for histone acetlyation; expressed during growth on nonfermentable carbon sources and under aerobic conditions |
|
| YCL042W | 1.32 |
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm |
||
| YCL040W | 1.30 |
GLK1
|
Glucokinase, catalyzes the phosphorylation of glucose at C6 in the first irreversible step of glucose metabolism; one of three glucose phosphorylating enzymes; expression regulated by non-fermentable carbon sources |
|
| YBR296C | 1.26 |
PHO89
|
Na+/Pi cotransporter, active in early growth phase; similar to phosphate transporters of Neurospora crassa; transcription regulated by inorganic phosphate concentrations and Pho4p |
|
| YHR001W-A | 1.23 |
QCR10
|
Subunit of the ubiqunol-cytochrome c oxidoreductase complex which includes Cobp, Rip1p, Cyt1p, Cor1p, Qcr2p, Qcr6p, Qcr7p, Qcr8p, Qcr9p, and Qcr10p and comprises part of the mitochondrial respiratory chain |
|
| YML090W | 1.22 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YML089C; exhibits growth defect on a non-fermentable (respiratory) carbon source |
||
| YJR137C | 1.22 |
ECM17
|
Sulfite reductase beta subunit, involved in amino acid biosynthesis, transcription repressed by methionine |
|
| YIL107C | 1.21 |
PFK26
|
6-phosphofructo-2-kinase, inhibited by phosphoenolpyruvate and sn-glycerol 3-phosphate, has negligible fructose-2,6-bisphosphatase activity, transcriptional regulation involves protein kinase A |
|
| YML120C | 1.21 |
NDI1
|
NADH:ubiquinone oxidoreductase, transfers electrons from NADH to ubiquinone in the respiratory chain but does not pump protons, in contrast to the higher eukaryotic multisubunit respiratory complex I; phosphorylated; homolog of human AMID |
|
| YLR307C-A | 1.15 |
Putative protein of unknown function |
||
| YJR146W | 1.12 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene HMS2 |
||
| YCL001W-B | 1.11 |
Putative protein of unknown function; identified by homology |
||
| YMR081C | 1.08 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YJL116C | 1.06 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YOR152C | 1.04 |
Putative protein of unknown function; has no similarity to any known protein; YOR152C is not an essential gene |
||
| YNL037C | 1.03 |
IDH1
|
Subunit of mitochondrial NAD(+)-dependent isocitrate dehydrogenase, which catalyzes the oxidation of isocitrate to alpha-ketoglutarate in the TCA cycle |
|
| YAL062W | 1.00 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YLR149C | 0.99 |
Putative protein of unknown function; YLR149C is not an essential gene |
||
| YLR356W | 0.91 |
Putative protein of unknown function with similarity to SCM4; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR356W is not an essential gene |
||
| YAL018C | 0.91 |
Putative protein of unknown function |
||
| YBR145W | 0.90 |
ADH5
|
Alcohol dehydrogenase isoenzyme V; involved in ethanol production |
|
| YKL085W | 0.88 |
MDH1
|
Mitochondrial malate dehydrogenase, catalyzes interconversion of malate and oxaloacetate; involved in the tricarboxylic acid (TCA) cycle; phosphorylated |
|
| YOR346W | 0.86 |
REV1
|
Deoxycytidyl transferase, forms a complex with the subunits of DNA polymerase zeta, Rev3p and Rev7p; involved in repair of abasic sites in damaged DNA |
|
| YPL024W | 0.86 |
RMI1
|
Involved in response to DNA damage; null mutants have increased rates of recombination and delayed S phase; interacts physically and genetically with Sgs1p (RecQ family member) and Top3p (topoisomerase III) |
|
| YGR065C | 0.85 |
VHT1
|
High-affinity plasma membrane H+-biotin (vitamin H) symporter; mutation results in fatty acid auxotrophy; 12 transmembrane domain containing major facilitator subfamily member; mRNA levels negatively regulated by iron deprivation and biotin |
|
| YML091C | 0.84 |
RPM2
|
Protein subunit of mitochondrial RNase P, has roles in nuclear transcription, cytoplasmic and mitochondrial RNA processing, and mitochondrial translation; distributed to mitochondria, cytoplasmic processing bodies, and the nucleus |
|
| YEL024W | 0.84 |
RIP1
|
Ubiquinol-cytochrome-c reductase, a Rieske iron-sulfur protein of the mitochondrial cytochrome bc1 complex; transfers electrons from ubiquinol to cytochrome c1 during respiration |
|
| YGR289C | 0.84 |
MAL11
|
Inducible high-affinity maltose transporter (alpha-glucoside transporter); encoded in the MAL1 complex locus; member of the 12 transmembrane domain superfamily of sugar transporters; broad substrate specificity that includes maltotriose |
|
| YLR092W | 0.83 |
SUL2
|
High affinity sulfate permease; sulfate uptake is mediated by specific sulfate transporters Sul1p and Sul2p, which control the concentration of endogenous activated sulfate intermediates |
|
| YOR328W | 0.83 |
PDR10
|
ATP-binding cassette (ABC) transporter, multidrug transporter involved in the pleiotropic drug resistance network; regulated by Pdr1p and Pdr3p |
|
| YGL187C | 0.81 |
COX4
|
Subunit IV of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; N-terminal 25 residues of precursor are cleaved during mitochondrial import; phosphorylated |
|
| YLR267W | 0.80 |
BOP2
|
Protein of unknown function, overproduction suppresses a pam1 slv3 double null mutation |
|
| YMR271C | 0.80 |
URA10
|
Minor orotate phosphoribosyltransferase (OPRTase) isozyme that catalyzes the fifth enzymatic step in the de novo biosynthesis of pyrimidines, converting orotate into orotidine-5'-phosphate; major OPRTase encoded by URA5 |
|
| YPL026C | 0.80 |
SKS1
|
Putative serine/threonine protein kinase; involved in the adaptation to low concentrations of glucose independent of the SNF3 regulated pathway |
|
| YOR345C | 0.78 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene REV1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YCR007C | 0.77 |
Putative integral membrane protein, member of DUP240 gene family; YCR007C is not an essential gene |
||
| YJL166W | 0.76 |
QCR8
|
Subunit 8 of ubiquinol cytochrome-c reductase complex, which is a component of the mitochondrial inner membrane electron transport chain; oriented facing the intermembrane space; expression is regulated by Abf1p and Cpf1p |
|
| YPL018W | 0.76 |
CTF19
|
Outer kinetochore protein, required for accurate mitotic chromosome segregation; component of the kinetochore sub-complex COMA (Ctf19p, Okp1p, Mcm21p, Ame1p) that functions as a platform for kinetochore assembly |
|
| YER004W | 0.75 |
FMP52
|
Protein of unknown function, localized to the mitochondrial outer membrane; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YDR022C | 0.75 |
CIS1
|
Autophagy-specific protein required for autophagosome formation; may form a complex with Atg17p and Atg29p that localizes other proteins to the pre-autophagosomal structure; high-copy suppressor of CIK1 deletion |
|
| YOR378W | 0.75 |
Putative protein of unknown function; belongs to the DHA2 family of drug:H+ antiporters; YOR378W is not an essential gene |
||
| YMR165C | 0.74 |
PAH1
|
Mg2+-dependent phosphatidate (PA) phosphatase, catalyzes the dephosphorylation of PA to yield diacylglycerol and Pi, responsible for de novo lipid synthesis; homologous to mammalian lipin 1 |
|
| YDR281C | 0.73 |
PHM6
|
Protein of unknown function, expression is regulated by phosphate levels |
|
| YGR290W | 0.73 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; putative HLH protein; partially overlaps the verified ORF MAL11/YGR289C (a high-affinity maltose transporter) |
||
| YJR150C | 0.73 |
DAN1
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YOL081W | 0.71 |
IRA2
|
GTPase-activating protein that negatively regulates RAS by converting it from the GTP- to the GDP-bound inactive form, required for reducing cAMP levels under nutrient limiting conditions, has similarity to Ira1p and human neurofibromin |
|
| YGL073W | 0.71 |
HSF1
|
Trimeric heat shock transcription factor, activates multiple genes in response to stresses that include hyperthermia; recognizes variable heat shock elements (HSEs) consisting of inverted NGAAN repeats; posttranslationally regulated |
|
| YDR504C | 0.70 |
SPG3
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YHR211W | 0.69 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YFR049W | 0.69 |
YMR31
|
Mitochondrial ribosomal protein of the small subunit, has similarity to human mitochondrial ribosomal protein MRP-S36 |
|
| YDR034C | 0.67 |
LYS14
|
Transcriptional activator involved in regulation of genes of the lysine biosynthesis pathway; requires 2-aminoadipate semialdehyde as co-inducer |
|
| YDR381C-A | 0.67 |
Protein of unknown function, localized to the mitochondrial outer membrane |
||
| YCR011C | 0.67 |
ADP1
|
Putative ATP-dependent permease of the ABC transporter family of proteins |
|
| YFR030W | 0.67 |
MET10
|
Subunit alpha of assimilatory sulfite reductase, which is responsible for the conversion of sulfite into sulfide |
|
| YOR054C | 0.67 |
VHS3
|
Functionally redundant (see also SIS2) inhibitory subunit of Ppz1p, a PP1c-related ser/thr protein phosphatase Z isoform; synthetically lethal with sis2; putative phosphopantothenoylcysteine decarboxylase involved in coenzyme A biosynthesis |
|
| YAL039C | 0.66 |
CYC3
|
Cytochrome c heme lyase (holocytochrome c synthase), attaches heme to apo-cytochrome c (Cyc1p or Cyc7p) in the mitochondrial intermembrane space; human ortholog may have a role in microphthalmia with linear skin defects (MLS) |
|
| YOR065W | 0.65 |
CYT1
|
Cytochrome c1, component of the mitochondrial respiratory chain; expression is regulated by the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex |
|
| YAR069C | 0.64 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YAR070C | 0.61 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YBL078C | 0.61 |
ATG8
|
Conserved protein that is a component of autophagosomes and Cvt vesicles; undergoes C-terminal conjugation to phosphatidylethanolamine (PE), Atg8p-PE is anchored to membranes and may mediate membrane fusion during autophagosome formation |
|
| YDR530C | 0.60 |
APA2
|
Diadenosine 5',5''-P1,P4-tetraphosphate phosphorylase II (AP4A phosphorylase), involved in catabolism of bis(5'-nucleosidyl) tetraphosphates; has similarity to Apa1p |
|
| YHR176W | 0.59 |
FMO1
|
Flavin-containing monooxygenase, localized to the cytoplasmic face of the ER membrane; catalyzes oxidation of biological thiols to maintain the ER redox buffer ratio for correct folding of disulfide-bonded proteins |
|
| YEL044W | 0.59 |
IES6
|
Protein that associates with the INO80 chromatin remodeling complex under low-salt conditions |
|
| YPL206C | 0.59 |
PGC1
|
Phosphatidyl Glycerol phospholipase C; regulates the phosphatidylglycerol (PG) content via a phospholipase C-type degradation mechanism; contains glycerophosphodiester phosphodiesterase motifs |
|
| YIL033C | 0.59 |
BCY1
|
Regulatory subunit of the cyclic AMP-dependent protein kinase (PKA), a component of a signaling pathway that controls a variety of cellular processes, including metabolism, cell cycle, stress response, stationary phase, and sporulation |
|
| YBR255C-A | 0.58 |
Putative protein of unknown function; identified by sequence comparison with hemiascomycetous yeast species |
||
| YBR250W | 0.58 |
SPO23
|
Protein of unknown function; associates with meiosis-specific protein Spo1p |
|
| YKL192C | 0.58 |
ACP1
|
Mitochondrial matrix acyl carrier protein, involved in biosynthesis of octanoate, which is a precursor to lipoic acid; activated by phosphopantetheinylation catalyzed by Ppt2p |
|
| YDL113C | 0.58 |
ATG20
|
Sorting nexin family member required for the cytoplasm-to-vacuole targeting (Cvt) pathway and for endosomal sorting; has a Phox homology domain that binds phosphatidylinositol-3-phosphate; interacts with Snx4p; potential Cdc28p substrate |
|
| YIL047C | 0.58 |
SYG1
|
Plasma membrane protein of unknown function; truncation and overexpression suppresses lethality of G-alpha protein deficiency |
|
| YPR023C | 0.57 |
EAF3
|
Esa1p-associated factor, nonessential component of the NuA4 acetyltransferase complex, homologous to Drosophila dosage compensation protein MSL3; plays a role in regulating Ty1 transposition |
|
| YOR298C-A | 0.57 |
MBF1
|
Transcriptional coactivator that bridges the DNA-binding region of Gcn4p and TATA-binding protein Spt15p; suppressor of frameshift mutations |
|
| YLR023C | 0.57 |
IZH3
|
Membrane protein involved in zinc metabolism, member of the four-protein IZH family, expression induced by zinc deficiency; deletion reduces sensitivity to elevated zinc and shortens lag phase, overexpression reduces Zap1p activity |
|
| YOR055W | 0.57 |
Dubious open reading frame unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YGR045C | 0.56 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YKR011C | 0.55 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus |
||
| YFL055W | 0.54 |
AGP3
|
Low-affinity amino acid permease, may act to supply the cell with amino acids as nitrogen source in nitrogen-poor conditions; transcription is induced under conditions of sulfur limitation; plays a role in regulating Ty1 transposition |
|
| YPR196W | 0.54 |
Putative maltose activator |
||
| YOR147W | 0.54 |
MDM32
|
Mitochondrial inner membrane protein with similarity to Mdm31p, required for normal mitochondrial morphology and inheritance; interacts genetically with MMM1, MDM10, MDM12, and MDM34 |
|
| YIR039C | 0.53 |
YPS6
|
Putative GPI-anchored aspartic protease |
|
| YKL109W | 0.53 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YOR208W | 0.53 |
PTP2
|
Phosphotyrosine-specific protein phosphatase involved in the inactivation of mitogen-activated protein kinase (MAPK) during osmolarity sensing; dephosporylates Hog1p MAPK and regulates its localization; localized to the nucleus |
|
| YGR110W | 0.53 |
Putative protein of unknown function; transcription is increased in response to genotoxic stress; plays a role in restricting Ty1 transposition |
||
| YNL220W | 0.50 |
ADE12
|
Adenylosuccinate synthase, catalyzes the first step in synthesis of adenosine monophosphate from inosine 5'monophosphate during purine nucleotide biosynthesis; exhibits binding to single-stranded autonomously replicating (ARS) core sequence |
|
| YJR147W | 0.49 |
HMS2
|
Protein with similarity to heat shock transcription factors; overexpression suppresses the pseudohyphal filamentation defect of a diploid mep1 mep2 homozygous null mutant |
|
| YPL189C-A | 0.49 |
COA2
|
Putative cytochrome oxidase assembly factor; identified by homology to Ashbya gossypii; null mutation results in respiratory deficiency with specific loss of cytochrome oxidase activity |
|
| YGR292W | 0.48 |
MAL12
|
Maltase (alpha-D-glucosidase), inducible protein involved in maltose catabolism; encoded in the MAL1 complex locus |
|
| YBR039W | 0.48 |
ATP3
|
Gamma subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YGL072C | 0.48 |
Dubious open reading frame unlikely to encode a protein; partially overlaps the verified gene HSF1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YAR053W | 0.47 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YNL277W | 0.47 |
MET2
|
L-homoserine-O-acetyltransferase, catalyzes the conversion of homoserine to O-acetyl homoserine which is the first step of the methionine biosynthetic pathway |
|
| YGL074C | 0.47 |
Dubious open reading frame unlikely to encode a functional protein; overlaps 5' end of essential HSF1 gene encoding heat shock transcription factor |
||
| YDR277C | 0.46 |
MTH1
|
Negative regulator of the glucose-sensing signal transduction pathway, required for repression of transcription by Rgt1p; interacts with Rgt1p and the Snf3p and Rgt2p glucose sensors; phosphorylated by Yck1p, triggering Mth1p degradation |
|
| YFR026C | 0.46 |
Putative protein of unknown function |
||
| YHR082C | 0.46 |
KSP1
|
Nonessential putative serine/threonine protein kinase of unknown cellular role; overproduction causes allele-specific suppression of the prp20-10 mutation |
|
| YDR114C | 0.46 |
Putative protein of unknown function; deletion mutant exhibits poor growth at elevated pH and calcium |
||
| YPR010C-A | 0.46 |
Putative protein of unknown function; conserved among Saccharomyces sensu stricto species |
||
| YNL283C | 0.45 |
WSC2
|
Partially redundant sensor-transducer of the stress-activated PKC1-MPK1 signaling pathway involved in maintenance of cell wall integrity and recovery from heat shock; secretory pathway Wsc2p is required for the arrest of secretion response |
|
| YCL057C-A | 0.45 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YAR068W | 0.45 |
Fungal-specific protein of unknown function; induced in respiratory-deficient cells |
||
| YCR088W | 0.45 |
ABP1
|
Actin-binding protein of the cortical actin cytoskeleton, important for activation of the Arp2/3 complex that plays a key role actin in cytoskeleton organization |
|
| YKL198C | 0.45 |
PTK1
|
Putative serine/threonine protein kinase that regulates spermine uptake; involved in polyamine transport; possible mitochondrial protein |
|
| YLR374C | 0.45 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF STP3/YLR375W |
||
| YLR375W | 0.44 |
STP3
|
Zinc-finger protein of unknown function, possibly involved in pre-tRNA splicing and in uptake of branched-chain amino acids |
|
| YER091C-A | 0.44 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPL236C | 0.44 |
Putative protein kinase that exhibits Akr1p-dependent palmitoylation |
||
| YNL104C | 0.44 |
LEU4
|
Alpha-isopropylmalate synthase (2-isopropylmalate synthase); the main isozyme responsible for the first step in the leucine biosynthesis pathway |
|
| YLL056C | 0.43 |
Putative protein of unknown function, transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in pleiotropic drug resistance (PDR) phenomenon; YLL056C is not an essential gene |
||
| YIL125W | 0.43 |
KGD1
|
Component of the mitochondrial alpha-ketoglutarate dehydrogenase complex, which catalyzes a key step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation of alpha-ketoglutarate to form succinyl-CoA |
|
| YDR542W | 0.43 |
PAU10
|
Hypothetical protein |
|
| YMR216C | 0.43 |
SKY1
|
SR protein kinase (SRPK) involved in regulating proteins involved in mRNA metabolism and cation homeostasis; similar to human SRPK1 |
|
| YDR055W | 0.42 |
PST1
|
Cell wall protein that contains a putative GPI-attachment site; secreted by regenerating protoplasts; up-regulated by activation of the cell integrity pathway, as mediated by Rlm1p; upregulated by cell wall damage via disruption of FKS1 |
|
| YHR096C | 0.42 |
HXT5
|
Hexose transporter with moderate affinity for glucose, induced in the presence of non-fermentable carbon sources, induced by a decrease in growth rate, contains an extended N-terminal domain relative to other HXTs |
|
| YCL048W-A | 0.42 |
Putative protein of unknown function |
||
| YEL059W | 0.42 |
Dubious open reading frame unlikely to encode a functional protein |
||
| YER060W-A | 0.42 |
FCY22
|
Putative purine-cytosine permease, very similar to Fcy2p but cannot substitute for its function |
|
| YMR196W | 0.42 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YMR196W is not an essential gene |
||
| YBR284W | 0.42 |
Putative protein of unknown function; YBR284W is not an essential gene; null mutant exhibits decreased resistance to rapamycin and wortmannin |
||
| YPR007C | 0.42 |
REC8
|
Meiosis-specific component of sister chromatid cohesion complex; maintains cohesion between sister chromatids during meiosis I; maintains cohesion between centromeres of sister chromatids until meiosis II; homolog of S. pombe Rec8p |
|
| YJR120W | 0.42 |
Protein of unknown function; essential for growth under anaerobic conditions; mutation causes decreased expression of ATP2, impaired respiration, defective sterol uptake, and altered levels/localization of ABC transporters Aus1p and Pdr11p |
||
| YPL223C | 0.42 |
GRE1
|
Hydrophilin of unknown function; stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YKL031W | 0.41 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species |
||
| YOR381W | 0.41 |
FRE3
|
Ferric reductase, reduces siderophore-bound iron prior to uptake by transporters; expression induced by low iron levels |
|
| YJL045W | 0.41 |
Minor succinate dehydrogenase isozyme; homologous to Sdh1p, the major isozyme reponsible for the oxidation of succinate and transfer of electrons to ubiquinone; induced during the diauxic shift in a Cat8p-dependent manner |
||
| YLR326W | 0.41 |
Hypothetical protein |
||
| YCL041C | 0.41 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps both the verified gene PDI1/YCL043C and the uncharacterized gene YCL042W |
||
| YMR017W | 0.41 |
SPO20
|
Meiosis-specific subunit of the t-SNARE complex, required for prospore membrane formation during sporulation; similar to but not functionally redundant with Sec9p; SNAP-25 homolog |
|
| YLL055W | 0.40 |
YCT1
|
High-affinity cysteine-specific transporter with similarity to the Dal5p family of transporters; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; YCT1 is not an essential gene |
|
| YGR087C | 0.39 |
PDC6
|
Minor isoform of pyruvate decarboxylase, decarboxylates pyruvate to acetaldehyde, involved in amino acid catabolism; transcription is glucose- and ethanol-dependent, and is strongly induced during sulfur limitation |
|
| YDR124W | 0.39 |
Putative protein of unknown function; non-essential gene; expression is strongly induced by alpha factor |
||
| YLL060C | 0.39 |
GTT2
|
Glutathione S-transferase capable of homodimerization; functional overlap with Gtt2p, Grx1p, and Grx2p |
|
| YPR013C | 0.39 |
Putative zinc finger protein; YPR013C is not an essential gene |
||
| YDR115W | 0.39 |
Putative mitochondrial ribosomal protein of the large subunit, has similarity to E. coli L34 ribosomal protein; required for respiratory growth, as are most mitochondrial ribosomal proteins |
||
| YDR014W-A | 0.39 |
HED1
|
Meiosis-specific protein that down-regulates Rad51p-mediated mitotic recombination when the meiotic recombination machinery is impaired; early meiotic gene, transcribed specifically during meiotic prophase |
|
| YPL187W | 0.38 |
MF(ALPHA)1
|
Mating pheromone alpha-factor, made by alpha cells; interacts with mating type a cells to induce cell cycle arrest and other responses leading to mating; also encoded by MF(ALPHA)2, although MF(ALPHA)1 produces most alpha-factor |
|
| YOR072W | 0.38 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOR072W-A; diploid deletion strains are methotrexate, paraquat and wortmannin sensitive |
||
| YDL026W | 0.37 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YER111C | 0.37 |
SWI4
|
DNA binding component of the SBF complex (Swi4p-Swi6p), a transcriptional activator that in concert with MBF (Mbp1-Swi6p) regulates late G1-specific transcription of targets including cyclins and genes required for DNA synthesis and repair |
|
| YDR441C | 0.37 |
APT2
|
Apparent pseudogene, not transcribed or translated under normal conditions; encodes a protein with similarity to adenine phosphoribosyltransferase, but artificially expressed protein exhibits no enzymatic activity |
|
| YOL163W | 0.37 |
Putative protein of unknown function; member of the Dal5p subfamily of the major facilitator family |
||
| YOR289W | 0.36 |
Putative protein of unknown function; transcription induced by the unfolded protein response; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and the nucleus |
||
| YGL256W | 0.36 |
ADH4
|
Alcohol dehydrogenase isoenzyme type IV, dimeric enzyme demonstrated to be zinc-dependent despite sequence similarity to iron-activated alcohol dehydrogenases; transcription is induced in response to zinc deficiency |
|
| YNL240C | 0.36 |
NAR1
|
Nuclear architecture related protein; component of the cytosolic iron-sulfur (FeS) protein assembly machinery, required for maturation of cytosolic and nuclear FeS proteins; homologous to human Narf |
|
| YEL028W | 0.36 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDL079C | 0.36 |
MRK1
|
Glycogen synthase kinase 3 (GSK-3) homolog; one of four GSK-3 homologs in S. cerevisiae that function to activate Msn2p-dependent transcription of stress responsive genes and that function in protein degradation |
|
| YNL094W | 0.36 |
APP1
|
Protein of unknown function, interacts with Rvs161p and Rvs167p; computational analysis of protein-protein interactions in large-scale studies suggests a possible role in actin filament organization |
|
| YCL035C | 0.35 |
GRX1
|
Hydroperoxide and superoxide-radical responsive heat-stable glutathione-dependent disulfide oxidoreductase with active site cysteine pair; protects cells from oxidative damage |
|
| YAL017W | 0.35 |
PSK1
|
One of two (see also PSK2) PAS domain containing S/T protein kinases; coordinately regulates protein synthesis and carbohydrate metabolism and storage in response to a unknown metabolite that reflects nutritional status |
|
| YDR541C | 0.35 |
Putative dihydrokaempferol 4-reductase |
||
| YHR204W | 0.35 |
MNL1
|
Alpha mannosidase-like protein of the endoplasmic reticulum required for degradation of glycoproteins but not for processing of N-linked oligosaccharides |
|
| YIL088C | 0.34 |
AVT7
|
Putative transporter, member of a family of seven S. cerevisiae genes (AVT1-7) related to vesicular GABA-glycine transporters |
|
| YML017W | 0.34 |
PSP2
|
Asn rich cytoplasmic protein that contains RGG motifs; high-copy suppressor of group II intron-splicing defects of a mutation in MRS2 and of a conditional mutation in POL1 (DNA polymerase alpha); possible role in mitochondrial mRNA splicing |
|
| YNL282W | 0.34 |
POP3
|
Subunit of both RNase MRP, which cleaves pre-rRNA, and nuclear RNase P, which cleaves tRNA precursors to generate mature 5' ends |
|
| YHR159W | 0.34 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; potential Cdc28p substrate |
||
| YML012C-A | 0.33 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene SEL1 |
||
| YAR060C | 0.33 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YNL055C | 0.33 |
POR1
|
Mitochondrial porin (voltage-dependent anion channel), outer membrane protein required for the maintenance of mitochondrial osmotic stability and mitochondrial membrane permeability; phosphorylated |
|
| YOR071C | 0.33 |
NRT1
|
High-affinity nicotinamide riboside transporter; also transports thiamine with low affinity; shares sequence similarity with Thi7p and Thi72p; proposed to be involved in 5-fluorocytosine sensitivity |
|
| YNL180C | 0.33 |
RHO5
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely involved in protein kinase C (Pkc1p)-dependent signal transduction pathway that controls cell integrity |
|
| YBR117C | 0.33 |
TKL2
|
Transketolase, similar to Tkl1p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YDR406W | 0.32 |
PDR15
|
Plasma membrane ATP binding cassette (ABC) transporter, multidrug transporter and general stress response factor implicated in cellular detoxification; regulated by Pdr1p, Pdr3p and Pdr8p; promoter contains a PDR responsive element |
|
| YDL020C | 0.32 |
RPN4
|
Transcription factor that stimulates expression of proteasome genes; Rpn4p levels are in turn regulated by the 26S proteasome in a negative feedback control mechanism; RPN4 is transcriptionally regulated by various stress responses |
|
| YPR103W | 0.32 |
PRE2
|
Beta 5 subunit of the 20S proteasome, responsible for the chymotryptic activity of the proteasome |
|
| YLR438W | 0.32 |
CAR2
|
L-ornithine transaminase (OTAse), catalyzes the second step of arginine degradation, expression is dually-regulated by allophanate induction and a specific arginine induction process; not nitrogen catabolite repression sensitive |
|
| YHR210C | 0.32 |
Putative protein of unknown function; non-essential gene; highly expressed under anaeorbic conditions; sequence similarity to aldose 1-epimerases such as GAL10 |
||
| YNL179C | 0.31 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YLR410W | 0.31 |
VIP1
|
Inositol hexakisphosphate (IP6) and inositol heptakisphosphate (IP7) kinase; generation of IP7 by Vip1p is important for phosphate signaling; likely involved in cortical actin cytoskeleton function, by analogy with S. pombe ortholog asp1 |
|
| YLR402W | 0.31 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDL027C | 0.30 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YDL027C is not an essential gene |
||
| YER187W | 0.30 |
Putative protein of unknown function; induced in respiratory-deficient cells |
||
| YDR338C | 0.30 |
Hypothetical protein |
||
| YHR138C | 0.30 |
Putative protein of unknown function; has similarity to Pbi2p; double null mutant lacking Pbi2p and Yhr138p exhibits highly fragmented vacuoles |
||
| YLR403W | 0.30 |
SFP1
|
Transcription factor that controls expression of many ribosome biogenesis genes in response to nutrients and stress, regulates G2/M transitions during mitotic cell cycle and DNA-damage response, involved in cell size modulation |
|
| YMR291W | 0.29 |
Putative kinase of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YMR291W is not an essential gene |
||
| YNL095C | 0.29 |
Putative protein of unknown function predicted to contain a transmembrane domain; YNL095C is not an essential gene |
||
| YEL043W | 0.29 |
Predicted cytoskeleton protein involved in intracellular signalling based on quantitative analysis of protein-protein interaction maps; may interact with ribosomes, based on co-purification studies; contains fibronectin type III domain fold |
||
| YER125W | 0.29 |
RSP5
|
Ubiquitin-protein ligase involved in ubiquitin-mediated protein degradation; functions in multivesicular body sorting, heat shock response and ubiquitylation of arrested RNAPII; contains a hect (homologous to E6-AP carboxyl terminus) domain |
|
| YJR152W | 0.29 |
DAL5
|
Allantoin permease; ureidosuccinate permease; expression is constitutive but sensitive to nitrogen catabolite repression |
|
| YFL018C | 0.29 |
LPD1
|
Dihydrolipoamide dehydrogenase, the lipoamide dehydrogenase component (E3) of the pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase multi-enzyme complexes |
|
| YOR072W-A | 0.28 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YOR072W; originally identified by fungal homology and RT-PCR |
||
| YLR004C | 0.28 |
THI73
|
Putative plasma membrane permease proposed to be involved in carboxylic acid uptake and repressed by thiamine; substrate of Dbf2p/Mob1p kinase; transcription is altered if mitochondrial dysfunction occurs |
|
| YPL190C | 0.28 |
NAB3
|
Single stranded RNA binding protein; acidic ribonucleoprotein; required for termination of non-poly(A) transcripts and efficient splicing; interacts with Nrd1p |
|
| YKR009C | 0.28 |
FOX2
|
Multifunctional enzyme of the peroxisomal fatty acid beta-oxidation pathway; has 3-hydroxyacyl-CoA dehydrogenase and enoyl-CoA hydratase activities |
|
| YJR089W | 0.28 |
BIR1
|
Essential chromosomal passenger protein involved in coordinating cell cycle events for proper chromosome segregation; C-terminal region binds Sli15p, and the middle region, upon phosphorylation, localizes Cbf2p to the spindle at anaphase |
|
| YGR256W | 0.27 |
GND2
|
6-phosphogluconate dehydrogenase (decarboxylating), catalyzes an NADPH regenerating reaction in the pentose phosphate pathway; required for growth on D-glucono-delta-lactone |
Network of associatons between targets according to the STRING database.
First level regulatory network of TYE7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.7 | 2.0 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.6 | 1.7 | GO:0019629 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 0.4 | 1.3 | GO:0010942 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.4 | 0.8 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.4 | 1.6 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.4 | 0.8 | GO:0045980 | negative regulation of nucleotide metabolic process(GO:0045980) |
| 0.4 | 1.5 | GO:0046323 | glucose import(GO:0046323) |
| 0.4 | 3.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.3 | 0.9 | GO:0006000 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 1.1 | GO:0051101 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.2 | 0.7 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.2 | 0.7 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.2 | 1.6 | GO:0015758 | glucose transport(GO:0015758) |
| 0.2 | 1.3 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.2 | 3.1 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 0.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 1.3 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.2 | 1.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 0.4 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.2 | 0.6 | GO:0009106 | lipoate metabolic process(GO:0009106) lipoate biosynthetic process(GO:0009107) |
| 0.2 | 0.6 | GO:0060195 | antisense RNA transcription(GO:0009300) regulation of antisense RNA transcription(GO:0060194) negative regulation of antisense RNA transcription(GO:0060195) |
| 0.2 | 1.3 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.2 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.3 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.2 | 0.7 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.2 | 0.9 | GO:0006167 | AMP biosynthetic process(GO:0006167) AMP metabolic process(GO:0046033) |
| 0.2 | 0.5 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.2 | 1.2 | GO:0019660 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.2 | 1.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.6 | GO:2000284 | positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 1.2 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.4 | GO:0051222 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 0.1 | 0.4 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.4 | GO:0072530 | cytosine transport(GO:0015856) purine-containing compound transmembrane transport(GO:0072530) |
| 0.1 | 0.3 | GO:2000219 | positive regulation of invasive growth in response to glucose limitation by positive regulation of transcription from RNA polymerase II promoter(GO:0036095) positive regulation of invasive growth in response to glucose limitation(GO:2000219) |
| 0.1 | 0.8 | GO:0044205 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.4 | GO:0015804 | sulfur amino acid transport(GO:0000101) neutral amino acid transport(GO:0015804) |
| 0.1 | 0.5 | GO:0000200 | inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.1 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 0.4 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 3.0 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 0.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.5 | GO:0032105 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.1 | 1.6 | GO:0034087 | establishment of sister chromatid cohesion(GO:0034085) establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.1 | 0.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 0.8 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 0.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.9 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.4 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.1 | 0.4 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.6 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.2 | GO:0042843 | D-xylose catabolic process(GO:0042843) |
| 0.1 | 2.2 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.1 | 0.1 | GO:0031564 | transcriptional attenuation(GO:0031555) transcription antitermination(GO:0031564) |
| 0.1 | 0.6 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.1 | 0.2 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.1 | 0.2 | GO:0042743 | hydrogen peroxide metabolic process(GO:0042743) |
| 0.1 | 0.2 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 0.1 | 0.4 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.1 | GO:0051099 | positive regulation of binding(GO:0051099) |
| 0.1 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.3 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.1 | 0.3 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.0 | 0.3 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.3 | GO:0031134 | sister chromatid biorientation(GO:0031134) |
| 0.0 | 0.2 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.0 | 0.1 | GO:1900544 | positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.0 | 0.2 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) |
| 0.0 | 0.4 | GO:0000272 | polysaccharide catabolic process(GO:0000272) |
| 0.0 | 0.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:0034395 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.0 | 0.2 | GO:0032079 | positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 0.0 | 0.1 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.7 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.5 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.2 | GO:0006771 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.0 | 0.0 | GO:0033523 | histone H2B ubiquitination(GO:0033523) histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.3 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.5 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 0.0 | 0.1 | GO:0000709 | meiotic joint molecule formation(GO:0000709) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0046503 | glycerolipid catabolic process(GO:0046503) |
| 0.0 | 0.1 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.8 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.4 | GO:0000147 | actin cortical patch assembly(GO:0000147) actin cortical patch organization(GO:0044396) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.2 | GO:1901658 | nucleoside catabolic process(GO:0009164) glycosyl compound catabolic process(GO:1901658) |
| 0.0 | 0.2 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.2 | GO:0006078 | (1->6)-beta-D-glucan biosynthetic process(GO:0006078) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) threonine catabolic process(GO:0006567) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.1 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.0 | 0.2 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0010994 | free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.8 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.0 | 0.2 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 0.1 | GO:1902532 | negative regulation of intracellular signal transduction(GO:1902532) |
| 0.0 | 0.8 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.5 | GO:0035966 | response to topologically incorrect protein(GO:0035966) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.1 | GO:0071241 | cellular response to inorganic substance(GO:0071241) |
| 0.0 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.3 | GO:0042168 | porphyrin-containing compound metabolic process(GO:0006778) tetrapyrrole metabolic process(GO:0033013) heme metabolic process(GO:0042168) |
| 0.0 | 0.2 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.0 | 0.4 | GO:0000001 | mitochondrion inheritance(GO:0000001) mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0005987 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0000114 | obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.0 | 0.0 | GO:0019627 | urea metabolic process(GO:0019627) |
| 0.0 | 0.1 | GO:0007534 | gene conversion at mating-type locus(GO:0007534) |
| 0.0 | 0.1 | GO:0045912 | negative regulation of cellular carbohydrate metabolic process(GO:0010677) negative regulation of gluconeogenesis(GO:0045721) negative regulation of carbohydrate metabolic process(GO:0045912) |
| 0.0 | 0.1 | GO:0034965 | intronic snoRNA processing(GO:0031070) intronic box C/D snoRNA processing(GO:0034965) |
| 0.0 | 0.2 | GO:0044088 | regulation of vacuole fusion, non-autophagic(GO:0032889) regulation of vacuole organization(GO:0044088) |
| 0.0 | 0.1 | GO:0001080 | nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) nitrogen catabolite activation of transcription(GO:0090294) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.0 | GO:1903727 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) positive regulation of phospholipid biosynthetic process(GO:0071073) positive regulation of phospholipid metabolic process(GO:1903727) |
| 0.0 | 0.0 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.0 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:0000321 | re-entry into mitotic cell cycle after pheromone arrest(GO:0000321) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) |
| 0.0 | 0.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.0 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0000274 | mitochondrial proton-transporting ATP synthase, stator stalk(GO:0000274) proton-transporting ATP synthase, stator stalk(GO:0045265) |
| 0.4 | 1.7 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.4 | 3.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.4 | 1.8 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.3 | 0.9 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.3 | 1.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 0.8 | GO:0000817 | COMA complex(GO:0000817) |
| 0.2 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.5 | GO:0005756 | mitochondrial proton-transporting ATP synthase, central stalk(GO:0005756) proton-transporting ATP synthase, central stalk(GO:0045269) |
| 0.1 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.1 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 0.6 | GO:0032221 | Rpd3S complex(GO:0032221) |
| 0.1 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.0 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.1 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.3 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.1 | 0.4 | GO:0033309 | SBF transcription complex(GO:0033309) |
| 0.1 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.2 | GO:0044426 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 0.1 | 2.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0032126 | eisosome(GO:0032126) |
| 0.0 | 4.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0070775 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.0 | 0.1 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.0 | 0.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.0 | 0.1 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.9 | GO:0030479 | actin cortical patch(GO:0030479) endocytic patch(GO:0061645) |
| 0.0 | 0.4 | GO:0042763 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.0 | 1.3 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.5 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.0 | GO:0005740 | mitochondrial envelope(GO:0005740) |
| 0.0 | 0.6 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.0 | 0.0 | GO:0030874 | nucleolar chromatin(GO:0030874) |
| 0.0 | 0.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 3.5 | GO:0005886 | plasma membrane(GO:0005886) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.5 | 2.0 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.5 | 1.5 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 1.3 | GO:0016208 | AMP binding(GO:0016208) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.4 | 0.8 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.4 | 1.6 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.4 | 2.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.4 | 1.8 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.3 | 1.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.3 | 0.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 0.9 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.3 | 3.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 1.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 0.8 | GO:0015151 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.2 | 2.3 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.2 | 0.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 1.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.2 | 0.2 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.2 | 0.6 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.2 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 0.6 | GO:0004564 | beta-fructofuranosidase activity(GO:0004564) sucrose alpha-glucosidase activity(GO:0004575) |
| 0.2 | 0.7 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 0.8 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.6 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.1 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 0.4 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 0.1 | 0.4 | GO:0005186 | mating pheromone activity(GO:0000772) pheromone activity(GO:0005186) |
| 0.1 | 1.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.7 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.6 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.3 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 2.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.1 | 0.6 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.1 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 0.3 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.1 | 0.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.7 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.1 | 0.2 | GO:0005471 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.4 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.2 | GO:0004691 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.5 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 1.0 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0042887 | amide transmembrane transporter activity(GO:0042887) |
| 0.0 | 0.1 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.0 | 0.9 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0008902 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.0 | 0.2 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.2 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.1 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.5 | GO:0004888 | transmembrane signaling receptor activity(GO:0004888) signaling receptor activity(GO:0038023) transmembrane receptor activity(GO:0099600) |
| 0.0 | 0.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.0 | 1.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.0 | 0.1 | GO:0001139 | transcription factor activity, core RNA polymerase II recruiting(GO:0001139) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.4 | GO:0020037 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.0 | 0.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0097372 | NAD-dependent histone deacetylase activity (H3-K18 specific)(GO:0097372) |
| 0.0 | 0.1 | GO:0050253 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.4 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.0 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 0.0 | 0.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0001004 | RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) transcription factor activity, RNA polymerase III promoter sequence-specific binding, TFIIIB recruiting(GO:0001004) transcription factor activity, RNA polymerase III type 1 promoter sequence-specific binding, TFIIIB recruiting(GO:0001005) transcription factor activity, RNA polymerase III type 2 promoter sequence-specific binding, TFIIIB recruiting(GO:0001008) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.0 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.0 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.2 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 0.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 0.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 0.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 0.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 19.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.2 | 1.0 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.1 | 0.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.1 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 19.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | Genes involved in Transmembrane transport of small molecules |