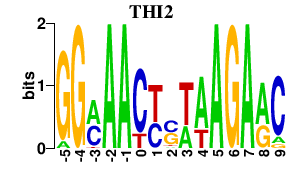
Results for THI2
Z-value: 1.17
Transcription factors associated with THI2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THI2
|
S000000444 | Transcriptional activator of thiamine biosynthetic genes |
Activity-expression correlation:
Activity profile of THI2 motif
Sorted Z-values of THI2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YFR055W | 23.12 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YFR056C | 20.15 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YER131W | 7.37 |
RPS26B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps26Ap and has similarity to rat S26 ribosomal protein |
|
| YOL127W | 7.15 |
RPL25
|
Primary rRNA-binding ribosomal protein component of the large (60S) ribosomal subunit, has similarity to E. coli L23 and rat L23a ribosomal proteins; binds to 26S rRNA via a conserved C-terminal motif |
|
| YJL115W | 5.95 |
ASF1
|
Nucleosome assembly factor, involved in chromatin assembly and disassembly, anti-silencing protein that causes derepression of silent loci when overexpressed; plays a role in regulating Ty1 transposition |
|
| YPL250W-A | 5.82 |
Identified by fungal homology and RT-PCR |
||
| YLR154C | 5.47 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YLR154W-B | 4.73 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR154W-A | 4.73 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YMR290W-A | 4.67 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; overlaps 5' end of essential HAS1 gene which encodes an ATP-dependent RNA helicase |
||
| YDR033W | 4.62 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YMR290C | 4.62 |
HAS1
|
ATP-dependent RNA helicase; localizes to both the nuclear periphery and nucleolus; highly enriched in nuclear pore complex fractions; constituent of 66S pre-ribosomal particles |
|
| YBR092C | 4.49 |
PHO3
|
Constitutively expressed acid phosphatase similar to Pho5p; brought to the cell surface by transport vesicles; hydrolyzes thiamin phosphates in the periplasmic space, increasing cellular thiamin uptake; expression is repressed by thiamin |
|
| YCR031C | 4.29 |
RPS14A
|
Ribosomal protein 59 of the small subunit, required for ribosome assembly and 20S pre-rRNA processing; mutations confer cryptopleurine resistance; nearly identical to Rps14Bp and similar to E. coli S11 and rat S14 ribosomal proteins |
|
| YKR092C | 4.29 |
SRP40
|
Nucleolar, serine-rich protein with a role in preribosome assembly or transport; may function as a chaperone of small nucleolar ribonucleoprotein particles (snoRNPs); immunologically and structurally to rat Nopp140 |
|
| YFR054C | 4.17 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YFR031C-A | 4.00 |
RPL2A
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl2Bp and has similarity to E. coli L2 and rat L8 ribosomal proteins |
|
| YGR108W | 3.96 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YLR150W | 3.92 |
STM1
|
Protein that binds G4 quadruplex and purine motif triplex nucleic acid; acts with Cdc13p to maintain telomere structure; interacts with ribosomes and subtelomeric Y' DNA; multicopy suppressor of tom1 and pop2 mutations |
|
| YNL248C | 3.74 |
RPA49
|
RNA polymerase I subunit A49 |
|
| YGL031C | 3.41 |
RPL24A
|
Ribosomal protein L30 of the large (60S) ribosomal subunit, nearly identical to Rpl24Bp and has similarity to rat L24 ribosomal protein; not essential for translation but may be required for normal translation rate |
|
| YNR018W | 3.34 |
AIM38
|
Putative protein of unknown function; non-tagged protein is detected in purified mitochondria; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) and severe growth defect in minimal glycerol media |
|
| YOR008C-A | 3.26 |
Putative protein of unknown function, includes a potential transmembrane domain; deletion results in slightly lengthened telomeres |
||
| YOR143C | 3.25 |
THI80
|
Thiamine pyrophosphokinase, phosphorylates thiamine to produce the coenzyme thiamine pyrophosphate (thiamine diphosphate) |
|
| YKL219W | 3.21 |
COS9
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YGL077C | 3.15 |
HNM1
|
Choline/ethanolamine transporter; involved in the uptake of nitrogen mustard and the uptake of glycine betaine during hypersaline stress; co-regulated with phospholipid biosynthetic genes and negatively regulated by choline and myo-inositol |
|
| YMR123W | 3.01 |
PKR1
|
V-ATPase assembly factor, functions with other V-ATPase assembly factors in the ER to efficiently assemble the V-ATPase membrane sector (V0); overproduction confers resistance to Pichia farinosa killer toxin |
|
| YGL030W | 2.88 |
RPL30
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L30 ribosomal protein; involved in pre-rRNA processing in the nucleolus; autoregulates splicing of its transcript |
|
| YGR020C | 2.87 |
VMA7
|
Subunit F of the eight-subunit V1 peripheral membrane domain of vacuolar H+-ATPase (V-ATPase), an electrogenic proton pump found throughout the endomembrane system; required for the V1 domain to assemble onto the vacuolar membrane |
|
| YMR246W | 2.87 |
FAA4
|
Long chain fatty acyl-CoA synthetase, regulates protein modification during growth in the presence of ethanol, functions to incorporate palmitic acid into phospholipids and neutral lipids |
|
| YPL177C | 2.84 |
CUP9
|
Homeodomain-containing transcriptional repressor of PTR2, which encodes a major peptide transporter; imported peptides activate ubiquitin-dependent proteolysis, resulting in degradation of Cup9p and de-repression of PTR2 transcription |
|
| YBR084W | 2.82 |
MIS1
|
Mitochondrial C1-tetrahydrofolate synthase, involved in interconversion between different oxidation states of tetrahydrofolate (THF); provides activities of formyl-THF synthetase, methenyl-THF cyclohydrolase, and methylene-THF dehydrogenase |
|
| YGL105W | 2.78 |
ARC1
|
Protein that binds tRNA and methionyl- and glutamyl-tRNA synthetases (Mes1p and Gus1p), delivering tRNA to them, stimulating catalysis, and ensuring their localization to the cytoplasm; also binds quadruplex nucleic acids |
|
| YKL110C | 2.75 |
KTI12
|
Protein that plays a role, with Elongator complex, in modification of wobble nucleosides in tRNA; involved in sensitivity to G1 arrest induced by zymocin; interacts with chromatin throughout the genome; also interacts with Cdc19p |
|
| YOR272W | 2.74 |
YTM1
|
Constituent of 66S pre-ribosomal particles, required for maturation of the large ribosomal subunit |
|
| YNL231C | 2.70 |
PDR16
|
Phosphatidylinositol transfer protein (PITP) controlled by the multiple drug resistance regulator Pdr1p, localizes to lipid particles and microsomes, controls levels of various lipids, may regulate lipid synthesis, homologous to Pdr17p |
|
| YGR040W | 2.65 |
KSS1
|
Mitogen-activated protein kinase (MAPK) involved in signal transduction pathways that control filamentous growth and pheromone response; the KSS1 gene is nonfunctional in S288C strains and functional in W303 strains |
|
| YMR177W | 2.63 |
MMT1
|
Putative metal transporter involved in mitochondrial iron accumulation; closely related to Mmt2p |
|
| YAL038W | 2.60 |
CDC19
|
Pyruvate kinase, functions as a homotetramer in glycolysis to convert phosphoenolpyruvate to pyruvate, the input for aerobic (TCA cycle) or anaerobic (glucose fermentation) respiration |
|
| YBL077W | 2.59 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene ILS1/YBL076C |
||
| YLR154W-C | 2.59 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YER146W | 2.56 |
LSM5
|
Lsm (Like Sm) protein; part of heteroheptameric complexes (Lsm2p-7p and either Lsm1p or 8p): cytoplasmic Lsm1p complex involved in mRNA decay; nuclear Lsm8p complex part of U6 snRNP and possibly involved in processing tRNA, snoRNA, and rRNA |
|
| YHR193C | 2.55 |
EGD2
|
Alpha subunit of the heteromeric nascent polypeptide-associated complex (NAC) involved in protein sorting and translocation, associated with cytoplasmic ribosomes |
|
| YER130C | 2.42 |
Hypothetical protein |
||
| YHR070C-A | 2.38 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene TRM5/YHR070W |
||
| YHR181W | 2.32 |
SVP26
|
Integral membrane protein of the early Golgi apparatus and endoplasmic reticulum, involved in COP II vesicle transport; may also function to promote retention of proteins in the early Golgi compartment |
|
| YOR271C | 2.32 |
FSF1
|
Putative protein, predicted to be an alpha-isopropylmalate carrier; belongs to the sideroblastic-associated protein family; non-tagged protein is detected in purified mitochondria; likely to play a role in iron homeostasis |
|
| YPR074C | 2.28 |
TKL1
|
Transketolase, similar to Tkl2p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YBL003C | 2.25 |
HTA2
|
One of two nearly identical (see also HTA1) histone H2A subtypes; core histone required for chromatin assembly and chromosome function; DNA damage-dependent phosphorylation by Mec1p facilitates DNA repair; acetylated by Nat4p |
|
| YJL177W | 2.22 |
RPL17B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl17Ap and has similarity to E. coli L22 and rat L17 ribosomal proteins |
|
| YPL043W | 2.18 |
NOP4
|
Nucleolar protein, essential for processing and maturation of 27S pre-rRNA and large ribosomal subunit biogenesis; constituent of 66S pre-ribosomal particles; contains four RNA recognition motifs (RRMs) |
|
| YMR116C | 2.18 |
ASC1
|
G-protein beta subunit and guanine nucleotide dissociation inhibitor for Gpa2p; ortholog of RACK1 that inhibits translation; core component of the small (40S) ribosomal subunit; represses Gcn4p in the absence of amino acid starvation |
|
| YHR007C | 2.17 |
ERG11
|
Lanosterol 14-alpha-demethylase, catalyzes the C-14 demethylation of lanosterol to form 4,4''-dimethyl cholesta-8,14,24-triene-3-beta-ol in the ergosterol biosynthesis pathway; member of the cytochrome P450 family |
|
| YKL063C | 2.16 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the Golgi |
||
| YDR133C | 2.16 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR134C |
||
| YDR509W | 2.13 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR508C | 2.12 |
GNP1
|
High-affinity glutamine permease, also transports Leu, Ser, Thr, Cys, Met and Asn; expression is fully dependent on Grr1p and modulated by the Ssy1p-Ptr3p-Ssy5p (SPS) sensor of extracellular amino acids |
|
| YIL118W | 2.12 |
RHO3
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins involved in the establishment of cell polarity; GTPase activity positively regulated by the GTPase activating protein (GAP) Rgd1p |
|
| YNL030W | 2.08 |
HHF2
|
One of two identical histone H4 proteins (see also HHF1); core histone required for chromatin assembly and chromosome function; contributes to telomeric silencing; N-terminal domain involved in maintaining genomic integrity |
|
| YGR078C | 2.04 |
PAC10
|
Part of the heteromeric co-chaperone GimC/prefoldin complex, which promotes efficient protein folding |
|
| YLR262C-A | 2.02 |
TMA7
|
Protein of unknown that associates with ribosomes; null mutant exhibits translation defects, altered polyribosome profiles, and resistance to the translation inhibitor anisomcyin |
|
| YNL247W | 2.01 |
Essential protein of unknown function; may interact with ribosomes, based on co-purification experiments |
||
| YMR009W | 2.00 |
ADI1
|
Acireductone dioxygenease involved in the methionine salvage pathway; ortholog of human MTCBP-1; transcribed with YMR010W and regulated post-transcriptionally by RNase III (Rnt1p) cleavage; ADI1 mRNA is induced in heat shock conditions |
|
| YML111W | 1.99 |
BUL2
|
Component of the Rsp5p E3-ubiquitin ligase complex, involved in intracellular amino acid permease sorting, functions in heat shock element mediated gene expression, essential for growth in stress conditions, functional homolog of BUL1 |
|
| YDL055C | 1.99 |
PSA1
|
GDP-mannose pyrophosphorylase (mannose-1-phosphate guanyltransferase), synthesizes GDP-mannose from GTP and mannose-1-phosphate in cell wall biosynthesis; required for normal cell wall structure |
|
| YOL124C | 1.96 |
TRM11
|
Catalytic subunit of an adoMet-dependent tRNA methyltransferase complex (Trm11p-Trm112p), required for the methylation of the guanosine nucleotide at position 10 (m2G10) in tRNAs; contains a THUMP domain and a methyltransferase domain |
|
| YMR037C | 1.91 |
MSN2
|
Transcriptional activator related to Msn4p; activated in stress conditions, which results in translocation from the cytoplasm to the nucleus; binds DNA at stress response elements of responsive genes, inducing gene expression |
|
| YML075C | 1.88 |
HMG1
|
One of two isozymes of HMG-CoA reductase that catalyzes the conversion of HMG-CoA to mevalonate, which is a rate-limiting step in sterol biosynthesis; localizes to the nuclear envelope; overproduction induces the formation of karmellae |
|
| YKL096W-A | 1.82 |
CWP2
|
Covalently linked cell wall mannoprotein, major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored |
|
| YPR063C | 1.81 |
ER-localized protein of unknown function |
||
| YOL130W | 1.79 |
ALR1
|
Plasma membrane Mg(2+) transporter, expression and turnover are regulated by Mg(2+) concentration; overexpression confers increased tolerance to Al(3+) and Ga(3+) ions |
|
| YCR016W | 1.79 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus and nucleus; YCR016W is not an essential gene |
||
| YNL290W | 1.78 |
RFC3
|
Subunit of heteropentameric Replication factor C (RF-C), which is a DNA binding protein and ATPase that acts as a clamp loader of the proliferating cell nuclear antigen (PCNA) processivity factor for DNA polymerases delta and epsilon |
|
| YLR109W | 1.76 |
AHP1
|
Thiol-specific peroxiredoxin, reduces hydroperoxides to protect against oxidative damage; function in vivo requires covalent conjugation to Urm1p |
|
| YOR248W | 1.72 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YLR257W | 1.72 |
Putative protein of unknown function |
||
| YBL002W | 1.71 |
HTB2
|
One of two nearly identical (see HTB1) histone H2B subtypes required for chromatin assembly and chromosome function; Rad6p-Bre1p-Lge1p mediated ubiquitination regulates transcriptional activation, meiotic DSB formation and H3 methylation |
|
| YGR195W | 1.71 |
SKI6
|
3'-to-5' phosphorolytic exoribonuclease that is a subunit of the exosome; required for 3' processing of the 5.8S rRNA; involved in 3' to 5' mRNA degradation and translation inhibition of non-poly(A) mRNAs |
|
| YDR050C | 1.70 |
TPI1
|
Triose phosphate isomerase, abundant glycolytic enzyme; mRNA half-life is regulated by iron availability; transcription is controlled by activators Reb1p, Gcr1p, and Rap1p through binding sites in the 5' non-coding region |
|
| YEL054C | 1.70 |
RPL12A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl12Bp; rpl12a rpl12b double mutant exhibits slow growth and slow translation; has similarity to E. coli L11 and rat L12 ribosomal proteins |
|
| YJR118C | 1.69 |
ILM1
|
Protein of unknown function; may be involved in mitochondrial DNA maintenance; required for slowed DNA synthesis-induced filamentous growth |
|
| YHR163W | 1.69 |
SOL3
|
6-phosphogluconolactonase, catalyzes the second step of the pentose phosphate pathway; weak multicopy suppressor of los1-1 mutation; homologous to Sol2p and Sol1p |
|
| YNL301C | 1.68 |
RPL18B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl18Ap and has similarity to rat L18 ribosomal protein |
|
| YNR001W-A | 1.66 |
Dubious open reading frame unlikely to encode a functional protein; identified by homology |
||
| YLR437C | 1.66 |
Putative protein of unknown function; epitope tagged protein localizes to the cytoplasm |
||
| YDR041W | 1.65 |
RSM10
|
Mitochondrial ribosomal protein of the small subunit, has similarity to E. coli S10 ribosomal protein; essential for viability, unlike most other mitoribosomal proteins |
|
| YKR013W | 1.64 |
PRY2
|
Protein of unknown function, has similarity to Pry1p and Pry3p and to the plant PR-1 class of pathogen related proteins |
|
| YOR107W | 1.58 |
RGS2
|
Negative regulator of glucose-induced cAMP signaling; directly activates the GTPase activity of the heterotrimeric G protein alpha subunit Gpa2p |
|
| YHR094C | 1.56 |
HXT1
|
Low-affinity glucose transporter of the major facilitator superfamily, expression is induced by Hxk2p in the presence of glucose and repressed by Rgt1p when glucose is limiting |
|
| YDR094W | 1.56 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified ORF DNF2 |
||
| YBR084C-A | 1.55 |
RPL19A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl19Bp and has similarity to rat L19 ribosomal protein; rpl19a and rpl19b single null mutations result in slow growth, while the double null mutation is lethal |
|
| YML088W | 1.54 |
UFO1
|
F-box receptor protein, subunit of the Skp1-Cdc53-F-box receptor (SCF) E3 ubiquitin ligase complex; binds to phosphorylated Ho endonuclease, allowing its ubiquitylation by SCF and subsequent degradation |
|
| YKR093W | 1.53 |
PTR2
|
Integral membrane peptide transporter, mediates transport of di- and tri-peptides; conserved protein that contains 12 transmembrane domains; PTR2 expression is regulated by the N-end rule pathway via repression by Cup9p |
|
| YOL123W | 1.53 |
HRP1
|
Subunit of cleavage factor I, a five-subunit complex required for the cleavage and polyadenylation of pre-mRNA 3' ends; RRM-containing heteronuclear RNA binding protein and hnRNPA/B family member that binds to poly (A) signal sequences |
|
| YGL201C | 1.52 |
MCM6
|
Protein involved in DNA replication; component of the Mcm2-7 hexameric complex that binds chromatin as a part of the pre-replicative complex |
|
| YIL069C | 1.50 |
RPS24B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps24Ap and has similarity to rat S24 ribosomal protein |
|
| YBR017C | 1.49 |
KAP104
|
Transportin, cytosolic karyopherin beta 2 involved in delivery of heterogeneous nuclear ribonucleoproteins to the nucleoplasm, binds rg-nuclear localization signals on Nab2p and Hrp1p, plays a role in cell-cycle progression |
|
| YDR344C | 1.46 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR060W | 1.45 |
ERG25
|
C-4 methyl sterol oxidase, catalyzes the first of three steps required to remove two C-4 methyl groups from an intermediate in ergosterol biosynthesis; mutants accumulate the sterol intermediate 4,4-dimethylzymosterol |
|
| YPR119W | 1.43 |
CLB2
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YJL222W | 1.41 |
VTH2
|
Putative membrane glycoprotein with strong similarity to Vth1p and Pep1p/Vps10p, may be involved in vacuolar protein sorting |
|
| YJL167W | 1.41 |
ERG20
|
Farnesyl pyrophosphate synthetase, has both dimethylallyltranstransferase and geranyltranstransferase activities; catalyzes the formation of C15 farnesyl pyrophosphate units for isoprenoid and sterol biosynthesis |
|
| YDR040C | 1.40 |
ENA1
|
P-type ATPase sodium pump, involved in Na+ and Li+ efflux to allow salt tolerance |
|
| YKR075C | 1.40 |
Protein of unknown function; similar to YOR062Cp and Reg1p; expression regulated by glucose and Rgt1p; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YCR087C-A | 1.40 |
LUG1
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus; YCR087C-A is not an essential gene |
|
| YLR108C | 1.39 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus; YLR108C is not an esssential gene |
||
| YHR128W | 1.39 |
FUR1
|
Uracil phosphoribosyltransferase, synthesizes UMP from uracil; involved in the pyrimidine salvage pathway |
|
| YLL045C | 1.37 |
RPL8B
|
Ribosomal protein L4 of the large (60S) ribosomal subunit, nearly identical to Rpl8Ap and has similarity to rat L7a ribosomal protein; mutation results in decreased amounts of free 60S subunits |
|
| YOR009W | 1.35 |
TIR4
|
Cell wall mannoprotein of the Srp1p/Tip1p family of serine-alanine-rich proteins; expressed under anaerobic conditions and required for anaerobic growth; transcription is also induced by cold shock |
|
| YDL211C | 1.35 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YBR048W | 1.35 |
RPS11B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps11Ap and has similarity to E. coli S17 and rat S11 ribosomal proteins |
|
| YPR165W | 1.33 |
RHO1
|
GTP-binding protein of the rho subfamily of Ras-like proteins, involved in establishment of cell polarity; regulates protein kinase C (Pkc1p) and the cell wall synthesizing enzyme 1,3-beta-glucan synthase (Fks1p and Gsc2p) |
|
| YLR406C | 1.32 |
RPL31B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl31Ap and has similarity to rat L31 ribosomal protein; associates with the karyopherin Sxm1p |
|
| YLR287C-A | 1.30 |
RPS30A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps30Bp and has similarity to rat S30 ribosomal protein |
|
| YHR180W-A | 1.30 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps dubious ORF YHR180C-B and long terminal repeat YHRCsigma3 |
||
| YDR095C | 1.30 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJR094W-A | 1.28 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YGR280C | 1.28 |
PXR1
|
Essential protein involved in rRNA and snoRNA maturation; competes with TLC1 RNA for binding to Est2p, suggesting a role in negative regulation of telomerase; human homolog inhibits telomerase; contains a G-patch RNA interacting domain |
|
| YGL179C | 1.25 |
TOS3
|
Protein kinase, related to and functionally redundant with Elm1p and Sak1p for the phosphorylation and activation of Snf1p; functionally orthologous to LKB1, a mammalian kinase associated with Peutz-Jeghers cancer-susceptibility syndrome |
|
| YKR012C | 1.23 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene PRY2 |
||
| YDR418W | 1.22 |
RPL12B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl12Ap; rpl12a rpl12b double mutant exhibits slow growth and slow translation; has similarity to E. coli L11 and rat L12 ribosomal proteins |
|
| YOR320C | 1.22 |
GNT1
|
N-acetylglucosaminyltransferase capable of modification of N-linked glycans in the Golgi apparatus |
|
| YCR028C-A | 1.21 |
RIM1
|
Single-stranded DNA-binding protein essential for mitochondrial genome maintenance; involved in mitochondrial DNA replication |
|
| YHL028W | 1.21 |
WSC4
|
ER membrane protein involved in the translocation of soluble secretory proteins and insertion of membrane proteins into the ER membrane; may also have a role in the stress response but has only partial functional overlap with WSC1-3 |
|
| YJL080C | 1.21 |
SCP160
|
Essential RNA-binding G protein effector of mating response pathway, mainly associated with nuclear envelope and ER, interacts in mRNA-dependent manner with translating ribosomes via multiple KH domains, similar to vertebrate vigilins |
|
| YPL112C | 1.18 |
PEX25
|
Peripheral peroxisomal membrane peroxin required for the regulation of peroxisome size and maintenance, recruits GTPase Rho1p to peroxisomes, induced by oleate, interacts with homologous protein Pex27p |
|
| YOR263C | 1.18 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF DES3/YOR264W |
||
| YNR028W | 1.17 |
CPR8
|
Peptidyl-prolyl cis-trans isomerase (cyclophilin), catalyzes the cis-trans isomerization of peptide bonds N-terminal to proline residues; similarity to Cpr4p suggests a potential role in the secretory pathway |
|
| YER109C | 1.15 |
FLO8
|
Transcription factor required for flocculation, diploid filamentous growth, and haploid invasive growth; genome reference strain S288C and most laboratory strains have a mutation in this gene |
|
| YPL263C | 1.14 |
KEL3
|
Cytoplasmic protein of unknown function |
|
| YJL168C | 1.13 |
SET2
|
Histone methyltransferase with a role in transcriptional elongation, methylates a lysine residue of histone H3; associates with the C-terminal domain of Rpo21p; histone methylation activity is regulated by phosphorylation status of Rpo21p |
|
| YJL193W | 1.12 |
Putative protein of unknown function, predicted to encode a triose phosphate transporter subfamily member based on phylogenetic analysis; similar to YOR307C/SLY41; deletion mutant has a respiratory growth defect |
||
| YPL014W | 1.12 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and to the nucleus |
||
| YLR060W | 1.10 |
FRS1
|
Beta subunit of cytoplasmic phenylalanyl-tRNA synthetase, forms a tetramer with Frs2p to generate active enzyme; sequence is evolutionarily distant from mitochondrial phenylalanyl-tRNA synthetase (Msf1p), but substrate binding is similar |
|
| YNL031C | 1.08 |
HHT2
|
One of two identical histone H3 proteins (see also HHT1); core histone required for chromatin assembly, involved in heterochromatin-mediated telomeric and HM silencing; regulated by acetylation, methylation, and mitotic phosphorylation |
|
| YPL137C | 1.07 |
GIP3
|
Glc7-interacting protein whose overexpression relocalizes Glc7p from the nucleus and prevents chromosome segregation; may interact with ribosomes, based on co-purification experiments |
|
| YOL064C | 1.07 |
MET22
|
Bisphosphate-3'-nucleotidase, involved in salt tolerance and methionine biogenesis; dephosphorylates 3'-phosphoadenosine-5'-phosphate and 3'-phosphoadenosine-5'-phosphosulfate, intermediates of the sulfate assimilation pathway |
|
| YDR044W | 1.06 |
HEM13
|
Coproporphyrinogen III oxidase, an oxygen requiring enzyme that catalyzes the sixth step in the heme biosynthetic pathway; localizes to the mitochondrial inner membrane; transcription is repressed by oxygen and heme (via Rox1p and Hap1p) |
|
| YMR230W | 1.06 |
RPS10B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps10Ap and has similarity to rat ribosomal protein S10 |
|
| YBR283C | 1.06 |
SSH1
|
Subunit of the Ssh1 translocon complex; Sec61p homolog involved in co-translational pathway of protein translocation; not essential |
|
| YGR123C | 1.05 |
PPT1
|
Protein serine/threonine phosphatase with similarity to human phosphatase PP5; present in both the nucleus and cytoplasm; expressed during logarithmic growth; computational analyses suggest roles in phosphate metabolism and rRNA processing |
|
| YOR362C | 1.04 |
PRE10
|
Alpha 7 subunit of the 20S proteasome |
|
| YJL223C | 1.04 |
PAU1
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YPL254W | 1.03 |
HFI1
|
Adaptor protein required for structural integrity of the SAGA complex, a histone acetyltransferase-coactivator complex that is involved in global regulation of gene expression through acetylation and transcription functions |
|
| YLR413W | 1.02 |
Putative protein of unknown function; YLR413W is not an essential gene |
||
| YBR233W-A | 1.02 |
DAD3
|
Essential subunit of the Dam1 complex (aka DASH complex), couples kinetochores to the force produced by MT depolymerization thereby aiding in chromosome segregation; is transferred to the kinetochore prior to mitosis |
|
| YML052W | 1.01 |
SUR7
|
Putative integral membrane protein; component of eisosomes; associated with endocytosis, along with Pil1p and Lsp1p; sporulation and plasma membrane sphingolipid content are altered in mutants |
|
| YJL222W-A | 1.01 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR138C | 1.00 |
TPO2
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; transcription of TPO2 is regulated by Haa1p; member of the major facilitator superfamily |
|
| YOR264W | 1.00 |
DSE3
|
Daughter cell-specific protein, may help establish daughter fate |
|
| YHR162W | 1.00 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the mitochondrion |
||
| YGR249W | 1.00 |
MGA1
|
Protein similar to heat shock transcription factor; multicopy suppressor of pseudohyphal growth defects of ammonium permease mutants |
|
| YGL097W | 1.00 |
SRM1
|
Nucleotide exchange factor for Gsp1p, localizes to the nucleus, required for nucleocytoplasmic trafficking of macromolecules; suppressor of the pheromone response pathway; potentially phosphorylated by Cdc28p |
|
| YGL035C | 1.00 |
MIG1
|
Transcription factor involved in glucose repression; sequence specific DNA binding protein containing two Cys2His2 zinc finger motifs; regulated by the SNF1 kinase and the GLC7 phosphatase |
|
| YDL121C | 0.99 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic retiuculum; YDL121C is not an essential protein |
||
| YIL164C | 0.99 |
NIT1
|
Nitrilase, member of the nitrilase branch of the nitrilase superfamily; in closely related species and other S. cerevisiae strain backgrounds YIL164C and adjacent ORF, YIL165C, likely constitute a single ORF encoding a nitrilase gene |
|
| YKL152C | 0.98 |
GPM1
|
Tetrameric phosphoglycerate mutase, mediates the conversion of 3-phosphoglycerate to 2-phosphoglycerate during glycolysis and the reverse reaction during gluconeogenesis |
|
| YBR286W | 0.97 |
APE3
|
Vacuolar aminopeptidase Y, processed to mature form by Prb1p |
|
| YEL051W | 0.95 |
VMA8
|
Subunit D of the eight-subunit V1 peripheral membrane domain of the vacuolar H+-ATPase (V-ATPase), an electrogenic proton pump found throughout the endomembrane system; plays a role in the coupling of proton transport and ATP hydrolysis |
|
| YPR074W-A | 0.93 |
Hypothetical protein identified by homology |
||
| YMR194C-A | 0.92 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDL240W | 0.92 |
LRG1
|
Putative GTPase-activating protein (GAP) involved in the Pkc1p-mediated signaling pathway that controls cell wall integrity; appears to specifically regulate 1,3-beta-glucan synthesis |
|
| YNL300W | 0.92 |
Glycosylphosphatidylinositol-dependent cell wall protein, expression is periodic and decreases in respone to ergosterol perturbation or upon entry into stationary phase; depletion increases resistance to lactic acid |
||
| YKL205W | 0.92 |
LOS1
|
Nuclear pore protein involved in nuclear export of pre-tRNA |
|
| YJL173C | 0.91 |
RFA3
|
Subunit of heterotrimeric Replication Protein A (RPA), which is a highly conserved single-stranded DNA binding protein involved in DNA replication, repair, and recombination |
|
| YIL011W | 0.90 |
TIR3
|
Cell wall mannoprotein of the Srp1p/Tip1p family of serine-alanine-rich proteins; expressed under anaerobic conditions and required for anaerobic growth |
|
| YMR300C | 0.90 |
ADE4
|
Phosphoribosylpyrophosphate amidotransferase (PRPPAT; amidophosphoribosyltransferase), catalyzes first step of the 'de novo' purine nucleotide biosynthetic pathway |
|
| YIL114C | 0.89 |
POR2
|
Putative mitochondrial porin (voltage-dependent anion channel), related to Por1p but not required for mitochondrial membrane permeability or mitochondrial osmotic stability |
|
| YLR198C | 0.89 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene SIK1/YLR197W |
||
| YML103C | 0.88 |
NUP188
|
Subunit of the nuclear pore complex (NPC), involved in the structural organization of the complex and of the nuclear envelope, also involved in nuclear envelope permeability, interacts with Pom152p and Nic96p |
|
| YPR170W-B | 0.88 |
Putative protein of unknown function, conserved in fungi; partially overlaps the dubious genes YPR169W-A, YPR170W-A and YRP170C |
||
| YOR181W | 0.86 |
LAS17
|
Actin assembly factor, activates the Arp2/3 protein complex that nucleates branched actin filaments; localizes with the Arp2/3 complex to actin patches; homolog of the human Wiskott-Aldrich syndrome protein (WASP) |
|
| YGR140W | 0.86 |
CBF2
|
Essential kinetochore protein, component of the CBF3 multisubunit complex that binds to the CDEIII region of the centromere; Cbf2p also binds to the CDEII region possibly forming a different multimeric complex, ubiquitinated in vivo |
|
| YBR158W | 0.86 |
AMN1
|
Protein required for daughter cell separation, multiple mitotic checkpoints, and chromosome stability; contains 12 degenerate leucine-rich repeat motifs; expression is induced by the Mitotic Exit Network (MEN) |
|
| YGR130C | 0.85 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; specifically phosphorylated in vitro by mammalian diphosphoinositol pentakisphosphate (IP7) |
||
| YGR124W | 0.85 |
ASN2
|
Asparagine synthetase, isozyme of Asn1p; catalyzes the synthesis of L-asparagine from L-aspartate in the asparagine biosynthetic pathway |
|
| YLR048W | 0.84 |
RPS0B
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps0Ap; required for maturation of 18S rRNA along with Rps0Ap; deletion of either RPS0 gene reduces growth rate, deletion of both genes is lethal |
|
| YGL009C | 0.83 |
LEU1
|
Isopropylmalate isomerase, catalyzes the second step in the leucine biosynthesis pathway |
|
| YKR074W | 0.83 |
AIM29
|
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm; null mutant displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YGL202W | 0.83 |
ARO8
|
Aromatic aminotransferase I, expression is regulated by general control of amino acid biosynthesis |
|
| YLR224W | 0.83 |
F-box protein and component of SCF ubiquitin ligase complexes involved in ubiquitin-dependent protein catabolism; readily monoubiquitinated in vitro by SCF-Ubc4 complexes; YLR224W is not an essential gene |
||
| YLR009W | 0.82 |
RLP24
|
Essential protein with similarity to Rpl24Ap and Rpl24Bp, associated with pre-60S ribosomal subunits and required for ribosomal large subunit biogenesis |
|
| YJR097W | 0.81 |
JJJ3
|
Protein of unknown function, contains a J-domain, which is a region with homology to the E. coli DnaJ protein |
|
| YKL065C | 0.81 |
YET1
|
Endoplasmic reticulum transmembrane protein; may interact with ribosomes, based on co-purification experiments; homolog of human BAP31 protein |
|
| YDR510W | 0.80 |
SMT3
|
Ubiquitin-like protein of the SUMO family, conjugated to lysine residues of target proteins; regulates chromatid cohesion, chromosome segregation, APC-mediated proteolysis, DNA replication and septin ring dynamics |
|
| YOL161C | 0.80 |
PAU20
|
Hypothetical protein |
|
| YDR502C | 0.79 |
SAM2
|
S-adenosylmethionine synthetase, catalyzes transfer of the adenosyl group of ATP to the sulfur atom of methionine; one of two differentially regulated isozymes (Sam1p and Sam2p) |
|
| YGL157W | 0.79 |
Oxidoreductase, catalyzes NADPH-dependent reduction of the bicyclic diketone bicyclo[2.2.2]octane-2,6-dione (BCO2,6D) to the chiral ketoalcohol (1R,4S,6S)-6-hydroxybicyclo[2.2.2]octane-2-one (BCO2one6ol) |
||
| YJR105W | 0.79 |
ADO1
|
Adenosine kinase, required for the utilization of S-adenosylmethionine (AdoMet); may be involved in recycling adenosine produced through the methyl cycle |
|
| YDR023W | 0.78 |
SES1
|
Cytosolic seryl-tRNA synthetase, class II aminoacyl-tRNA synthetase that aminoacylates tRNA(Ser), displays tRNA-dependent amino acid recognition which enhances discrimination of the serine substrate, interacts with peroxin Pex21p |
|
| YGR106C | 0.78 |
VOA1
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuolar memebrane |
|
| YFL015W-A | 0.78 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDL166C | 0.77 |
FAP7
|
Essential NTPase required for small ribosome subunit synthesis, mediates processing of the 20S pre-rRNA at site D in the cytoplasm but associates only transiently with 43S preribosomes via Rps14p, may be the endonuclease for site D |
|
| YGR180C | 0.77 |
RNR4
|
Ribonucleotide-diphosphate reductase (RNR), small subunit; the RNR complex catalyzes the rate-limiting step in dNTP synthesis and is regulated by DNA replication and DNA damage checkpoint pathways via localization of the small subunits |
|
| YFL017C | 0.77 |
GNA1
|
Evolutionarily conserved glucosamine-6-phosphate acetyltransferase required for multiple cell cycle events including passage through START, DNA synthesis, and mitosis; involved in UDP-N-acetylglucosamine synthesis, forms GlcNAc6P from AcCoA |
|
| YOR180C | 0.76 |
DCI1
|
Peroxisomal delta(3,5)-delta(2,4)-dienoyl-CoA isomerase, involved in fatty acid metabolism, contains peroxisome targeting signals at amino and carboxy termini |
|
| YFL015C | 0.76 |
Dubious open reading frame unlikely to encode a protein; partially overlaps dubious ORF YFL015W-A; YFL015C is not an essential gene |
||
| YGR155W | 0.75 |
CYS4
|
Cystathionine beta-synthase, catalyzes the synthesis of cystathionine from serine and homocysteine, the first committed step in cysteine biosynthesis |
|
| YDL047W | 0.75 |
SIT4
|
Type 2A-related serine-threonine phosphatase that functions in the G1/S transition of the mitotic cycle; cytoplasmic and nuclear protein that modulates functions mediated by Pkc1p including cell wall and actin cytoskeleton organization |
|
| YOR043W | 0.74 |
WHI2
|
Protein required, with binding partner Psr1p, for full activation of the general stress response, possibly through Msn2p dephosphorylation; regulates growth during the diauxic shift; negative regulator of G1 cyclin expression |
Network of associatons between targets according to the STRING database.
First level regulatory network of THI2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 23.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 1.4 | 7.1 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 1.3 | 3.9 | GO:0043069 | negative regulation of apoptotic process(GO:0043066) negative regulation of programmed cell death(GO:0043069) negative regulation of cell death(GO:0060548) |
| 0.9 | 2.8 | GO:0035955 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 0.9 | 2.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.9 | 2.6 | GO:0034225 | cellular response to zinc ion starvation(GO:0034224) regulation of transcription from RNA polymerase II promoter in response to zinc ion starvation(GO:0034225) positive regulation of transcription from RNA polymerase II promoter in response to ethanol(GO:0061410) positive regulation of transcription from RNA polymerase II promoter in response to alkaline pH(GO:0061422) cellular response to ethanol(GO:0071361) regulation of cell aging(GO:0090342) cellular response to alcohol(GO:0097306) regulation of replicative cell aging(GO:1900062) |
| 0.8 | 5.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.8 | 3.2 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.8 | 3.0 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.7 | 4.9 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 0.7 | 2.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.6 | 10.9 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.6 | 1.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.6 | 3.0 | GO:0032233 | regulation of actin filament bundle assembly(GO:0032231) positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.6 | 1.8 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.6 | 2.8 | GO:0046655 | folic acid metabolic process(GO:0046655) folic acid biosynthetic process(GO:0046656) |
| 0.6 | 0.6 | GO:0031564 | transcriptional attenuation(GO:0031555) transcription antitermination(GO:0031564) |
| 0.6 | 2.8 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.5 | 2.5 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.5 | 7.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.5 | 1.5 | GO:0038032 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.5 | 1.9 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.5 | 1.4 | GO:0016114 | terpenoid metabolic process(GO:0006721) terpenoid biosynthetic process(GO:0016114) farnesyl diphosphate biosynthetic process(GO:0045337) farnesyl diphosphate metabolic process(GO:0045338) |
| 0.5 | 1.4 | GO:0006571 | tyrosine biosynthetic process(GO:0006571) |
| 0.4 | 2.2 | GO:0010182 | cellular glucose homeostasis(GO:0001678) carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.4 | 0.4 | GO:0010695 | regulation of spindle pole body separation(GO:0010695) |
| 0.4 | 1.7 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) |
| 0.4 | 2.5 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.4 | 1.1 | GO:2000906 | starch metabolic process(GO:0005982) starch catabolic process(GO:0005983) regulation of starch catabolic process by regulation of transcription from RNA polymerase II promoter(GO:0035956) positive regulation of starch catabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0035957) positive regulation of multi-organism process(GO:0043902) regulation of starch catabolic process(GO:2000881) positive regulation of starch catabolic process(GO:2000883) regulation of starch metabolic process(GO:2000904) positive regulation of starch metabolic process(GO:2000906) |
| 0.4 | 1.5 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.4 | 8.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.4 | 1.4 | GO:0010138 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.4 | 3.6 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.3 | 3.0 | GO:0001402 | signal transduction involved in filamentous growth(GO:0001402) |
| 0.3 | 10.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 1.3 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.3 | 3.4 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.3 | 1.2 | GO:0051303 | meiotic telomere clustering(GO:0045141) establishment of chromosome localization(GO:0051303) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.3 | 0.9 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.3 | 1.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.3 | 0.9 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.3 | 2.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.3 | 1.2 | GO:0060277 | obsolete negative regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0060277) |
| 0.3 | 0.9 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.3 | 4.2 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.3 | 1.9 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.3 | 1.1 | GO:0007535 | donor selection(GO:0007535) |
| 0.3 | 2.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 0.8 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.3 | 0.8 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.3 | 1.0 | GO:0000296 | spermine transport(GO:0000296) |
| 0.2 | 2.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 1.0 | GO:0036170 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.2 | 1.2 | GO:0051597 | response to methylmercury(GO:0051597) cellular response to methylmercury(GO:0071406) |
| 0.2 | 1.9 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.2 | 0.7 | GO:0046136 | regulation of vitamin metabolic process(GO:0030656) positive regulation of vitamin metabolic process(GO:0046136) regulation of thiamine biosynthetic process(GO:0070623) positive regulation of thiamine biosynthetic process(GO:0090180) |
| 0.2 | 0.9 | GO:0036213 | actomyosin contractile ring contraction(GO:0000916) contractile ring contraction(GO:0036213) |
| 0.2 | 2.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.2 | 23.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 2.0 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.2 | 0.6 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of metal ion(GO:0051238) sequestering of iron ion(GO:0097577) |
| 0.2 | 0.8 | GO:0009226 | UDP-N-acetylglucosamine metabolic process(GO:0006047) UDP-N-acetylglucosamine biosynthetic process(GO:0006048) nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.2 | 6.0 | GO:0006694 | steroid biosynthetic process(GO:0006694) sterol biosynthetic process(GO:0016126) |
| 0.2 | 1.5 | GO:0042991 | transcription factor import into nucleus(GO:0042991) |
| 0.2 | 0.7 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.2 | 0.9 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.2 | 0.5 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.2 | 0.7 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.9 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.2 | 0.7 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.2 | 0.8 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.2 | 0.8 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 1.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.2 | 0.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.6 | GO:0019346 | transsulfuration(GO:0019346) homocysteine metabolic process(GO:0050667) |
| 0.2 | 0.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 1.8 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.4 | GO:0031047 | gene silencing involved in chronological cell aging(GO:0010978) gene silencing by RNA(GO:0031047) |
| 0.1 | 0.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.4 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.1 | 2.4 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.1 | 1.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.4 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 0.1 | 0.9 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 1.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.6 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 2.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) peptidyl-proline modification(GO:0018208) |
| 0.1 | 0.5 | GO:0043007 | maintenance of rDNA(GO:0043007) |
| 0.1 | 0.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.4 | GO:0032079 | positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.1 | 0.4 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.1 | 4.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.9 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 7.0 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.1 | 0.4 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 1.0 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 0.1 | GO:0043407 | inactivation of MAPK activity(GO:0000188) negative regulation of MAP kinase activity(GO:0043407) |
| 0.1 | 0.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 2.2 | GO:0010038 | response to metal ion(GO:0010038) |
| 0.1 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.2 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.1 | 0.9 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.1 | 0.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.3 | GO:0031684 | heterotrimeric G-protein complex cycle(GO:0031684) |
| 0.1 | 0.6 | GO:0071039 | nuclear polyadenylation-dependent CUT catabolic process(GO:0071039) |
| 0.1 | 0.8 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.9 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.1 | 0.2 | GO:0000304 | response to singlet oxygen(GO:0000304) positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.1 | 0.4 | GO:0046219 | tryptophan biosynthetic process(GO:0000162) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.1 | 0.3 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.1 | 0.1 | GO:1901351 | regulation of phosphatidylglycerol biosynthetic process(GO:1901351) negative regulation of phosphatidylglycerol biosynthetic process(GO:1901352) |
| 0.1 | 0.2 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.1 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.6 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 0.7 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 0.9 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.1 | 1.6 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.5 | GO:0030031 | cell projection organization(GO:0030030) cell projection assembly(GO:0030031) |
| 0.1 | 0.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.5 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.1 | 0.2 | GO:0009090 | homoserine biosynthetic process(GO:0009090) |
| 0.1 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.3 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.1 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 1.2 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 1.1 | GO:0015918 | sterol transport(GO:0015918) |
| 0.1 | 0.2 | GO:0009162 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) pyrimidine deoxyribonucleotide metabolic process(GO:0009219) pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) 2'-deoxyribonucleotide biosynthetic process(GO:0009265) 2'-deoxyribonucleotide metabolic process(GO:0009394) deoxyribose phosphate biosynthetic process(GO:0046385) |
| 0.1 | 0.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) fatty acid oxidation(GO:0019395) lipid oxidation(GO:0034440) |
| 0.1 | 8.8 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.1 | 0.4 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.1 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 1.4 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.3 | GO:0007119 | budding cell isotropic bud growth(GO:0007119) |
| 0.1 | 5.1 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) |
| 0.1 | 0.4 | GO:0000032 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) |
| 0.1 | 0.4 | GO:0044091 | ascospore-type prospore membrane assembly(GO:0032120) membrane biogenesis(GO:0044091) membrane assembly(GO:0071709) |
| 0.1 | 1.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.5 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.6 | GO:0051666 | actin cortical patch localization(GO:0051666) |
| 0.1 | 0.2 | GO:0010674 | negative regulation of transcription from RNA polymerase II promoter involved in meiotic cell cycle(GO:0010674) |
| 0.1 | 0.3 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.1 | 0.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.1 | 0.2 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.1 | 0.1 | GO:0072353 | cellular age-dependent response to reactive oxygen species(GO:0072353) |
| 0.1 | 0.6 | GO:0001100 | negative regulation of exit from mitosis(GO:0001100) |
| 0.0 | 0.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.2 | GO:0000084 | mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.0 | 0.3 | GO:0070096 | mitochondrial outer membrane translocase complex assembly(GO:0070096) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0015883 | FAD transport(GO:0015883) |
| 0.0 | 0.2 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.0 | 0.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.3 | GO:0070589 | cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0006952 | immune effector process(GO:0002252) immune system process(GO:0002376) defense response(GO:0006952) response to biotic stimulus(GO:0009607) response to virus(GO:0009615) response to external biotic stimulus(GO:0043207) defense response to virus(GO:0051607) response to other organism(GO:0051707) defense response to other organism(GO:0098542) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 1.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:1900182 | endoplasmic reticulum membrane fusion(GO:0016320) ribophagy(GO:0034517) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) ER-associated misfolded protein catabolic process(GO:0071712) regulation of protein localization to nucleus(GO:1900180) positive regulation of protein localization to nucleus(GO:1900182) positive regulation of cellular protein localization(GO:1903829) |
| 0.0 | 0.8 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.4 | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress(GO:0036003) |
| 0.0 | 0.2 | GO:0009749 | response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.4 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.2 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.2 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.2 | GO:0007323 | peptide pheromone maturation(GO:0007323) |
| 0.0 | 0.8 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.0 | GO:1900461 | positive regulation of cell growth(GO:0030307) positive regulation of pseudohyphal growth by positive regulation of transcription from RNA polymerase II promoter(GO:1900461) positive regulation of pseudohyphal growth(GO:2000222) |
| 0.0 | 0.2 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.3 | GO:0043094 | cellular metabolic compound salvage(GO:0043094) |
| 0.0 | 0.2 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.2 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.2 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.0 | 0.4 | GO:0019319 | gluconeogenesis(GO:0006094) hexose biosynthetic process(GO:0019319) |
| 0.0 | 0.1 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.0 | 0.5 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.0 | 0.2 | GO:0072503 | cellular divalent inorganic cation homeostasis(GO:0072503) divalent inorganic cation homeostasis(GO:0072507) |
| 0.0 | 0.2 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.5 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.2 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.3 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.1 | 4.5 | GO:0030287 | cell wall-bounded periplasmic space(GO:0030287) |
| 0.9 | 2.8 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.9 | 2.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.8 | 2.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.8 | 0.8 | GO:0030684 | preribosome(GO:0030684) |
| 0.7 | 8.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.6 | 2.6 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.5 | 3.8 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.4 | 33.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 2.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.4 | 2.9 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.4 | 1.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) Rad17 RFC-like complex(GO:0031389) Elg1 RFC-like complex(GO:0031391) |
| 0.4 | 1.1 | GO:0071261 | Ssh1 translocon complex(GO:0071261) |
| 0.4 | 21.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 2.0 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.3 | 0.6 | GO:0030689 | Noc complex(GO:0030689) |
| 0.3 | 0.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.3 | 1.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 0.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 0.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.3 | 1.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 4.7 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.3 | 3.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 0.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 0.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 0.9 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 0.9 | GO:0031518 | CBF3 complex(GO:0031518) |
| 0.2 | 0.8 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.2 | 0.6 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.2 | 1.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 1.1 | GO:0034456 | UTP-C complex(GO:0034456) |
| 0.2 | 2.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 0.4 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.2 | 1.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 0.7 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.2 | 1.9 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.2 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 0.7 | GO:0033179 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 1.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 6.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.3 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.4 | GO:0070823 | HDA1 complex(GO:0070823) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:0043529 | GET complex(GO:0043529) |
| 0.1 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.5 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.5 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.6 | GO:0035339 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.1 | 5.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 2.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.4 | GO:0044462 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 0.1 | 1.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 3.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 0.4 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.1 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.3 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 1.3 | GO:0005940 | septin ring(GO:0005940) |
| 0.1 | 0.3 | GO:0000399 | cellular bud neck septin structure(GO:0000399) cleavage apparatus septin structure(GO:0032161) |
| 0.1 | 0.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.0 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.3 | GO:0000500 | RNA polymerase I upstream activating factor complex(GO:0000500) |
| 0.1 | 0.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.3 | GO:0034044 | exomer complex(GO:0034044) |
| 0.1 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 1.5 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.5 | GO:0000142 | cellular bud neck contractile ring(GO:0000142) |
| 0.1 | 0.4 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.6 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 7.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.2 | GO:0033309 | SBF transcription complex(GO:0033309) |
| 0.0 | 1.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 1.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) nuclear telomeric heterochromatin(GO:0005724) telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) multi-eIF complex(GO:0043614) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 4.2 | GO:0005933 | cellular bud(GO:0005933) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.8 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.2 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0090544 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.0 | 0.2 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 20.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 1.4 | 5.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.0 | 5.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.9 | 2.8 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.7 | 2.9 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.7 | 2.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.7 | 2.0 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.6 | 3.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.6 | 1.8 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.6 | 2.8 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.6 | 2.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.5 | 3.8 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.5 | 3.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.5 | 2.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.5 | 5.2 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.5 | 2.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.5 | 3.3 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.4 | 2.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.4 | 1.7 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.4 | 1.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.4 | 2.3 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.4 | 1.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.4 | 1.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.4 | 2.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.3 | 1.0 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 0.3 | 3.1 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.3 | 1.2 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.3 | 1.8 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 13.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.3 | 3.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.3 | 1.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 2.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 1.6 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.3 | 0.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 1.5 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.3 | 7.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.3 | 1.0 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.2 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 0.7 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.2 | 0.9 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 0.8 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.2 | 4.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 0.8 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 34.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 0.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 1.3 | GO:1901681 | sulfur compound binding(GO:1901681) |
| 0.2 | 0.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 1.5 | GO:0032934 | steroid binding(GO:0005496) sterol binding(GO:0032934) |
| 0.2 | 0.7 | GO:0005471 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 0.5 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.2 | 1.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 5.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.2 | 6.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.2 | 0.8 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.2 | 3.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 1.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.5 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.2 | 1.4 | GO:0015079 | potassium ion transmembrane transporter activity(GO:0015079) |
| 0.2 | 1.2 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.2 | 0.5 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.1 | 0.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 1.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.6 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.1 | 0.5 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 4.1 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.4 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.1 | 0.6 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.9 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 2.0 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 0.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.4 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.1 | 1.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.5 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 1.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.2 | GO:0016859 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.1 | 2.0 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.1 | 1.0 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 1.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.4 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.3 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) transcription factor activity, RNA polymerase I upstream control element sequence-specific binding(GO:0001168) |
| 0.1 | 3.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 3.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.1 | 0.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 2.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.7 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.7 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.2 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 0.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) flap endonuclease activity(GO:0048256) |
| 0.1 | 0.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.1 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 2.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 3.2 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.3 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 3.2 | GO:0005525 | GTP binding(GO:0005525) guanyl nucleotide binding(GO:0019001) guanyl ribonucleotide binding(GO:0032561) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 1.2 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.6 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.5 | GO:0043566 | structure-specific DNA binding(GO:0043566) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.2 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 5.5 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.7 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0004559 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.4 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.8 | GO:0001159 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.1 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.1 | GO:0005319 | lipid transporter activity(GO:0005319) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.3 | 1.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 2.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 0.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 0.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 0.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.5 | 2.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 1.8 | REACTOME NUCLEOTIDE EXCISION REPAIR | Genes involved in Nucleotide Excision Repair |
| 0.4 | 2.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 0.9 | REACTOME SIGNALLING TO ERKS | Genes involved in Signalling to ERKs |
| 0.3 | 0.9 | REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT | Genes involved in SLC-mediated transmembrane transport |
| 0.3 | 2.0 | REACTOME SIGNALING BY GPCR | Genes involved in Signaling by GPCR |
| 0.3 | 1.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 3.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | Genes involved in Cytokine Signaling in Immune system |
| 0.0 | 0.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.1 | REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | Genes involved in Platelet activation, signaling and aggregation |
| 0.0 | 0.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |