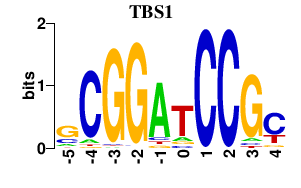
Results for TBS1
Z-value: 0.99
Transcription factors associated with TBS1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBS1
|
S000000354 | Putative protein of unknown function |
Activity-expression correlation:
Activity profile of TBS1 motif
Sorted Z-values of TBS1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YLR349W | 2.61 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified ORF DIC1/YLR348C |
||
| YNL134C | 2.59 |
Putative protein of unknown function with similarity to dehydrogenases from other model organisms; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and nucleus and is induced by the DNA-damaging agent MMS |
||
| YLR348C | 2.42 |
DIC1
|
Mitochondrial dicarboxylate carrier, integral membrane protein, catalyzes a dicarboxylate-phosphate exchange across the inner mitochondrial membrane, transports cytoplasmic dicarboxylates into the mitochondrial matrix |
|
| YER088C-A | 2.34 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YEL001C | 2.20 |
IRC22
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion localizes to the ER; YEL001C is non-essential; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YDR345C | 1.94 |
HXT3
|
Low affinity glucose transporter of the major facilitator superfamily, expression is induced in low or high glucose conditions |
|
| YCL018W | 1.93 |
LEU2
|
Beta-isopropylmalate dehydrogenase (IMDH), catalyzes the third step in the leucine biosynthesis pathway |
|
| YLR350W | 1.74 |
ORM2
|
Evolutionarily conserved protein with similarity to Orm1p, required for resistance to agents that induce the unfolded protein response; human ortholog is located in the endoplasmic reticulum |
|
| YOL086C | 1.70 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YBR177C | 1.68 |
EHT1
|
Acyl-coenzymeA:ethanol O-acyltransferase that plays a minor role in medium-chain fatty acid ethyl ester biosynthesis; possesses short-chain esterase activity; localizes to lipid particles and the mitochondrial outer membrane |
|
| YGL157W | 1.53 |
Oxidoreductase, catalyzes NADPH-dependent reduction of the bicyclic diketone bicyclo[2.2.2]octane-2,6-dione (BCO2,6D) to the chiral ketoalcohol (1R,4S,6S)-6-hydroxybicyclo[2.2.2]octane-2-one (BCO2one6ol) |
||
| YPR157W | 1.52 |
Putative protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YOR028C | 1.46 |
CIN5
|
Basic leucine zipper transcriptional factor of the yAP-1 family that mediates pleiotropic drug resistance and salt tolerance; localizes constitutively to the nucleus |
|
| YFR055W | 1.45 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YJR016C | 1.40 |
ILV3
|
Dihydroxyacid dehydratase, catalyzes third step in the common pathway leading to biosynthesis of branched-chain amino acids |
|
| YOL085C | 1.40 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOL085W-A |
||
| YLR110C | 1.37 |
CCW12
|
Cell wall mannoprotein, mutants are defective in mating and agglutination, expression is downregulated by alpha-factor |
|
| YLL028W | 1.35 |
TPO1
|
Polyamine transporter that recognizes spermine, putrescine, and spermidine; catalyzes uptake of polyamines at alkaline pH and excretion at acidic pH; phosphorylation enhances activity and sorting to the plasma membrane |
|
| YDR133C | 1.34 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR134C |
||
| YOR273C | 1.31 |
TPO4
|
Polyamine transport protein, recognizes spermine, putrescine, and spermidine; localizes to the plasma membrane; member of the major facilitator superfamily |
|
| YKL096W | 1.26 |
CWP1
|
Cell wall mannoprotein, linked to a beta-1,3- and beta-1,6-glucan heteropolymer through a phosphodiester bond; involved in cell wall organization |
|
| YNR014W | 1.25 |
Putative protein of unknown function; expression is cell-cycle regulated, Azf1p-dependent, and heat-inducible |
||
| YDR046C | 1.19 |
BAP3
|
Amino acid permease involved in the uptake of cysteine, leucine, isoleucine and valine |
|
| YDR047W | 1.18 |
HEM12
|
Uroporphyrinogen decarboxylase, catalyzes the fifth step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus; activity inhibited by Cu2+, Zn2+, Fe2+, Fe3+ and sulfhydryl-specific reagents |
|
| YFL014W | 1.17 |
HSP12
|
Plasma membrane localized protein that protects membranes from desiccation; induced by heat shock, oxidative stress, osmostress, stationary phase entry, glucose depletion, oleate and alcohol; regulated by the HOG and Ras-Pka pathways |
|
| YFL015C | 1.17 |
Dubious open reading frame unlikely to encode a protein; partially overlaps dubious ORF YFL015W-A; YFL015C is not an essential gene |
||
| YDR033W | 1.14 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YLR154W-A | 1.12 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YGR234W | 1.10 |
YHB1
|
Nitric oxide oxidoreductase, flavohemoglobin involved in nitric oxide detoxification; plays a role in the oxidative and nitrosative stress responses |
|
| YLR154W-B | 1.09 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YGL009C | 1.09 |
LEU1
|
Isopropylmalate isomerase, catalyzes the second step in the leucine biosynthesis pathway |
|
| YCR102C | 1.08 |
Putative protein of unknown function; involved in copper metabolism; similar to C.carbonum toxD gene; YCR102C is not an essential gene |
||
| YMR318C | 1.07 |
ADH6
|
NADPH-dependent medium chain alcohol dehydrogenase with broad substrate specificity; member of the cinnamyl family of alcohol dehydrogenases; may be involved in fusel alcohol synthesis or in aldehyde tolerance |
|
| YBL100C | 1.06 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; almost completely overlaps the 5' end of ATP1 |
||
| YGR055W | 1.05 |
MUP1
|
High affinity methionine permease, integral membrane protein with 13 putative membrane-spanning regions; also involved in cysteine uptake |
|
| YCL064C | 1.00 |
CHA1
|
Catabolic L-serine (L-threonine) deaminase, catalyzes the degradation of both L-serine and L-threonine; required to use serine or threonine as the sole nitrogen source, transcriptionally induced by serine and threonine |
|
| YGR279C | 0.98 |
SCW4
|
Cell wall protein with similarity to glucanases; scw4 scw10 double mutants exhibit defects in mating |
|
| YCR102W-A | 0.98 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YMR251W-A | 0.98 |
HOR7
|
Protein of unknown function; overexpression suppresses Ca2+ sensitivity of mutants lacking inositol phosphorylceramide mannosyltransferases Csg1p and Csh1p; transcription is induced under hyperosmotic stress and repressed by alpha factor |
|
| YBR178W | 0.96 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YBR177C |
||
| YDR309C | 0.95 |
GIC2
|
Redundant rho-like GTPase Cdc42p effector; homolog of Gic1p; involved in initiation of budding and cellular polarization; interacts with Cdc42p via the Cdc42/Rac-interactive binding (CRIB) domain and with PI(4,5)P2 via a polybasic region |
|
| YHL003C | 0.92 |
LAG1
|
Ceramide synthase component, involved in synthesis of ceramide from C26(acyl)-coenzyme A and dihydrosphingosine or phytosphingosine, functionally equivalent to Lac1p |
|
| YMR243C | 0.87 |
ZRC1
|
Vacuolar membrane zinc transporter, transports zinc from the cytosol into the vacuole for storage; also has a role in resistance to zinc shock resulting from a sudden influx of zinc into the cytoplasm |
|
| YER001W | 0.87 |
MNN1
|
Alpha-1,3-mannosyltransferase, integral membrane glycoprotein of the Golgi complex, required for addition of alpha1,3-mannose linkages to N-linked and O-linked oligosaccharides, one of five S. cerevisiae proteins of the MNN1 family |
|
| YHR208W | 0.87 |
BAT1
|
Mitochondrial branched-chain amino acid aminotransferase, homolog of murine ECA39; highly expressed during logarithmic phase and repressed during stationary phase |
|
| YCR024C-B | 0.84 |
Putative protein of unknown function; identified by expression profiling and mass spectrometry |
||
| YNL327W | 0.83 |
EGT2
|
Glycosylphosphatidylinositol (GPI)-anchored cell wall endoglucanase required for proper cell separation after cytokinesis, expression is activated by Swi5p and tightly regulated in a cell cycle-dependent manner |
|
| YDL125C | 0.82 |
HNT1
|
Adenosine 5'-monophosphoramidase; interacts physically and genetically with Kin28p, a CDK and TFIIK subunit, and genetically with CAK1; member of the histidine triad (HIT) superfamily of nucleotide-binding proteins and similar to Hint |
|
| YIL123W | 0.81 |
SIM1
|
Protein of the SUN family (Sim1p, Uth1p, Nca3p, Sun4p) that may participate in DNA replication, promoter contains SCB regulation box at -300 bp indicating that expression may be cell cycle-regulated |
|
| YPR149W | 0.81 |
NCE102
|
Protein of unknown function; contains transmembrane domains; involved in secretion of proteins that lack classical secretory signal sequences; component of the detergent-insoluble glycolipid-enriched complexes (DIGs) |
|
| YKL219W | 0.81 |
COS9
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YFL022C | 0.80 |
FRS2
|
Alpha subunit of cytoplasmic phenylalanyl-tRNA synthetase, forms a tetramer with Frs1p to form active enzyme; evolutionarily distant from mitochondrial phenylalanyl-tRNA synthetase based on protein sequence, but substrate binding is similar |
|
| YOR029W | 0.79 |
Dubious open reading frame unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YER134C | 0.79 |
Putative protein of unknown function; non-essential gene |
||
| YBR032W | 0.78 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YLR212C | 0.78 |
TUB4
|
Gamma-tubulin, involved in nucleating microtubules from both the cytoplasmic and nuclear faces of the spindle pole body |
|
| YLR411W | 0.75 |
CTR3
|
High-affinity copper transporter of the plasma membrane, acts as a trimer; gene is disrupted by a Ty2 transposon insertion in many laboratory strains of S. cerevisiae |
|
| YPR063C | 0.75 |
ER-localized protein of unknown function |
||
| YOR376W-A | 0.75 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YBL006C | 0.75 |
LDB7
|
Component of the RSC chromatin remodeling complex; interacts with Rsc3p, Rsc30p, Npl6p, and Htl1p to form a module important for a broad range of RSC functions |
|
| YOR272W | 0.74 |
YTM1
|
Constituent of 66S pre-ribosomal particles, required for maturation of the large ribosomal subunit |
|
| YBL099W | 0.72 |
ATP1
|
Alpha subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; phosphorylated |
|
| YOR247W | 0.72 |
SRL1
|
Mannoprotein that exhibits a tight association with the cell wall, required for cell wall stability in the absence of GPI-anchored mannoproteins; has a high serine-threonine content; expression is induced in cell wall mutants |
|
| YER117W | 0.72 |
RPL23B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl23Ap and has similarity to E. coli L14 and rat L23 ribosomal proteins |
|
| YFR056C | 0.69 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YKL209C | 0.64 |
STE6
|
Plasma membrane ATP-binding cassette (ABC) transporter required for the export of a-factor, catalyzes ATP hydrolysis coupled to a-factor transport; contains 12 transmembrane domains and two ATP binding domains; expressed only in MATa cells |
|
| YML127W | 0.64 |
RSC9
|
Component of the RSC chromatin remodeling complex; DNA-binding protein involved in the synthesis of rRNA and in transcriptional repression and activation of genes regulated by the Target of Rapamycin (TOR) pathway |
|
| YMR246W | 0.63 |
FAA4
|
Long chain fatty acyl-CoA synthetase, regulates protein modification during growth in the presence of ethanol, functions to incorporate palmitic acid into phospholipids and neutral lipids |
|
| YGR254W | 0.61 |
ENO1
|
Enolase I, a phosphopyruvate hydratase that catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate during glycolysis and the reverse reaction during gluconeogenesis; expression is repressed in response to glucose |
|
| YDL174C | 0.58 |
DLD1
|
D-lactate dehydrogenase, oxidizes D-lactate to pyruvate, transcription is heme-dependent, repressed by glucose, and derepressed in ethanol or lactate; located in the mitochondrial inner membrane |
|
| YNL104C | 0.56 |
LEU4
|
Alpha-isopropylmalate synthase (2-isopropylmalate synthase); the main isozyme responsible for the first step in the leucine biosynthesis pathway |
|
| YGL202W | 0.56 |
ARO8
|
Aromatic aminotransferase I, expression is regulated by general control of amino acid biosynthesis |
|
| YOR012W | 0.55 |
Putative protein of unknown function |
||
| YFL016C | 0.55 |
MDJ1
|
Co-chaperone that stimulates the ATPase activity of the HSP70 protein Ssc1p; involved in protein folding/refolding in the mitochodrial matrix; required for proteolysis of misfolded proteins; member of the HSP40 (DnaJ) family of chaperones |
|
| YLR258W | 0.54 |
GSY2
|
Glycogen synthase, similar to Gsy1p; expression induced by glucose limitation, nitrogen starvation, heat shock, and stationary phase; activity regulated by cAMP-dependent, Snf1p and Pho85p kinases as well as by the Gac1p-Glc7p phosphatase |
|
| YGR251W | 0.54 |
Putative protein of unknown function; deletion mutant has defects in pre-rRNA processing; green fluorescent protein (GFP)-fusion protein localizes to both the nucleus and the nucleolus; YGR251W is an essential gene |
||
| YER010C | 0.53 |
Protein of unknown function, forms a ring-shaped homotrimer; has similarity to members of the prokaryotic rraA family; possibly involved in a phosphotransfer reaction |
||
| YNL055C | 0.53 |
POR1
|
Mitochondrial porin (voltage-dependent anion channel), outer membrane protein required for the maintenance of mitochondrial osmotic stability and mitochondrial membrane permeability; phosphorylated |
|
| YHR140W | 0.52 |
Putative integral membrane protein of unknown function |
||
| YDR385W | 0.52 |
EFT2
|
Elongation factor 2 (EF-2), also encoded by EFT1; catalyzes ribosomal translocation during protein synthesis; contains diphthamide, the unique posttranslationally modified histidine residue specifically ADP-ribosylated by diphtheria toxin |
|
| YKR075C | 0.52 |
Protein of unknown function; similar to YOR062Cp and Reg1p; expression regulated by glucose and Rgt1p; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YIL051C | 0.51 |
MMF1
|
Mitochondrial protein involved in maintenance of the mitochondrial genome |
|
| YOR092W | 0.51 |
ECM3
|
Non-essential protein of unknown function; involved in signal transduction and the genotoxic response; induced rapidly in response to treatment with 8-methoxypsoralen and UVA irradiation |
|
| YOR271C | 0.51 |
FSF1
|
Putative protein, predicted to be an alpha-isopropylmalate carrier; belongs to the sideroblastic-associated protein family; non-tagged protein is detected in purified mitochondria; likely to play a role in iron homeostasis |
|
| YMR304C-A | 0.51 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene SCW10 |
||
| YOL052C | 0.50 |
SPE2
|
S-adenosylmethionine decarboxylase, required for the biosynthesis of spermidine and spermine; cells lacking Spe2p require spermine or spermidine for growth in the presence of oxygen but not when grown anaerobically |
|
| YPR156C | 0.49 |
TPO3
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; member of the major facilitator superfamily |
|
| YLR312W-A | 0.49 |
MRPL15
|
Mitochondrial ribosomal protein of the large subunit |
|
| YKR041W | 0.48 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YJL104W | 0.47 |
PAM16
|
Constituent of the mitochondrial import motor associated with the presequence translocase, along with Ssc1p, Tim44p, Mge1p, and Pam18p; forms a 1:1 subcomplex with Pam18p and inhibits its cochaperone activity; contains J-like domain |
|
| YDR010C | 0.47 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOL012C | 0.47 |
HTZ1
|
Histone variant H2AZ, exchanged for histone H2A in nucleosomes by the SWR1 complex; involved in transcriptional regulation through prevention of the spread of silent heterochromatin |
|
| YOR375C | 0.46 |
GDH1
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh3p; expression regulated by nitrogen and carbon sources |
|
| YOL039W | 0.46 |
RPP2A
|
Ribosomal protein P2 alpha, a component of the ribosomal stalk, which is involved in the interaction between translational elongation factors and the ribosome; regulates the accumulation of P1 (Rpp1Ap and Rpp1Bp) in the cytoplasm |
|
| YOL154W | 0.46 |
ZPS1
|
Putative GPI-anchored protein; transcription is induced under low-zinc conditions, as mediated by the Zap1p transcription factor, and at alkaline pH |
|
| YNR013C | 0.46 |
PHO91
|
Low-affinity phosphate transporter of the vacuolar membrane; deletion of pho84, pho87, pho89, pho90, and pho91 causes synthetic lethality; transcription independent of Pi and Pho4p activity; overexpression results in vigorous growth |
|
| YDR041W | 0.46 |
RSM10
|
Mitochondrial ribosomal protein of the small subunit, has similarity to E. coli S10 ribosomal protein; essential for viability, unlike most other mitoribosomal proteins |
|
| YGL147C | 0.45 |
RPL9A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl9Bp and has similarity to E. coli L6 and rat L9 ribosomal proteins |
|
| YFL015W-A | 0.45 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBR069C | 0.44 |
TAT1
|
Amino acid transport protein for valine, leucine, isoleucine, and tyrosine, low-affinity tryptophan and histidine transporter; overexpression confers FK506 and FTY720 resistance |
|
| YDR533C | 0.44 |
HSP31
|
Possible chaperone and cysteine protease with similarity to E. coli Hsp31; member of the DJ-1/ThiJ/PfpI superfamily, which includes human DJ-1 involved in Parkinson's disease; exists as a dimer and contains a putative metal-binding site |
|
| YDR094W | 0.43 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified ORF DNF2 |
||
| YDR040C | 0.43 |
ENA1
|
P-type ATPase sodium pump, involved in Na+ and Li+ efflux to allow salt tolerance |
|
| YER146W | 0.42 |
LSM5
|
Lsm (Like Sm) protein; part of heteroheptameric complexes (Lsm2p-7p and either Lsm1p or 8p): cytoplasmic Lsm1p complex involved in mRNA decay; nuclear Lsm8p complex part of U6 snRNP and possibly involved in processing tRNA, snoRNA, and rRNA |
|
| YGR124W | 0.42 |
ASN2
|
Asparagine synthetase, isozyme of Asn1p; catalyzes the synthesis of L-asparagine from L-aspartate in the asparagine biosynthetic pathway |
|
| YDR215C | 0.42 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; null mutant displays elevated sensitivity to expression of a mutant huntingtin fragment or of alpha-synuclein |
||
| YDR032C | 0.42 |
PST2
|
Protein with similarity to members of a family of flavodoxin-like proteins; induced by oxidative stress in a Yap1p dependent manner; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YCR018C | 0.41 |
SRD1
|
Protein involved in the processing of pre-rRNA to mature rRNA; contains a C2/C2 zinc finger motif; srd1 mutation suppresses defects caused by the rrp1-1 mutation |
|
| YJR114W | 0.41 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF RSM7/YJR113C |
||
| YGR052W | 0.41 |
FMP48
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YPL170W | 0.41 |
DAP1
|
Heme-binding protein involved in regulation of cytochrome P450 protein Erg11p; damage response protein, related to mammalian membrane progesterone receptors; mutations lead to defects in telomeres, mitochondria, and sterol synthesis |
|
| YLL045C | 0.41 |
RPL8B
|
Ribosomal protein L4 of the large (60S) ribosomal subunit, nearly identical to Rpl8Ap and has similarity to rat L7a ribosomal protein; mutation results in decreased amounts of free 60S subunits |
|
| YER182W | 0.40 |
FMP10
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YPL265W | 0.40 |
DIP5
|
Dicarboxylic amino acid permease, mediates high-affinity and high-capacity transport of L-glutamate and L-aspartate; also a transporter for Gln, Asn, Ser, Ala, and Gly |
|
| YDL173W | 0.39 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YDL173W is not an essential gene |
||
| YER011W | 0.38 |
TIR1
|
Cell wall mannoprotein of the Srp1p/Tip1p family of serine-alanine-rich proteins; expression is downregulated at acidic pH and induced by cold shock and anaerobiosis; abundance is increased in cells cultured without shaking |
|
| YER136W | 0.38 |
GDI1
|
GDP dissociation inhibitor, regulates vesicle traffic in secretory pathways by regulating the dissociation of GDP from the Sec4/Ypt/rab family of GTP binding proteins |
|
| YPL014W | 0.38 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and to the nucleus |
||
| YPL250W-A | 0.37 |
Identified by fungal homology and RT-PCR |
||
| YBR068C | 0.37 |
BAP2
|
High-affinity leucine permease, functions as a branched-chain amino acid permease involved in the uptake of leucine, isoleucine and valine; contains 12 predicted transmembrane domains |
|
| YMR263W | 0.37 |
SAP30
|
Subunit of a histone deacetylase complex, along with Rpd3p and Sin3p, that is involved in silencing at telomeres, rDNA, and silent mating-type loci; involved in telomere maintenance |
|
| YCL042W | 0.37 |
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm |
||
| YGR146C-A | 0.37 |
Putative protein of unknown function |
||
| YNL024C | 0.37 |
Putative protein of unknown function with seven beta-strand methyltransferase motif; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YNL024C is not an essential gene |
||
| YKL043W | 0.36 |
PHD1
|
Transcriptional activator that enhances pseudohyphal growth; regulates expression of FLO11, an adhesin required for pseudohyphal filament formation; similar to StuA, an A. nidulans developmental regulator; potential Cdc28p substrate |
|
| YHR181W | 0.36 |
SVP26
|
Integral membrane protein of the early Golgi apparatus and endoplasmic reticulum, involved in COP II vesicle transport; may also function to promote retention of proteins in the early Golgi compartment |
|
| YEL033W | 0.36 |
MTC7
|
Predicted metabolic role based on network analysis derived from ChIP experiments, a large-scale deletion study and localization of transcription factor binding sites; null mutant is sensitive to temperature oscillation in a cdc13-1 mutant |
|
| YDR091C | 0.36 |
RLI1
|
Essential iron-sulfur protein required for ribosome biogenesis and translation initiation; facilitates binding of a multifactor complex (MFC) of translation initiation factors to the small ribosomal subunit; predicted ABC family ATPase |
|
| YPR170W-B | 0.35 |
Putative protein of unknown function, conserved in fungi; partially overlaps the dubious genes YPR169W-A, YPR170W-A and YRP170C |
||
| YMR108W | 0.35 |
ILV2
|
Acetolactate synthase, catalyses the first common step in isoleucine and valine biosynthesis and is the target of several classes of inhibitors, localizes to the mitochondria; expression of the gene is under general amino acid control |
|
| YJR113C | 0.35 |
RSM7
|
Mitochondrial ribosomal protein of the small subunit, has similarity to E. coli S7 ribosomal protein |
|
| YHL021C | 0.35 |
AIM17
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) |
|
| YLR447C | 0.35 |
VMA6
|
Subunit d of the five-subunit V0 integral membrane domain of vacuolar H+-ATPase (V-ATPase), an electrogenic proton pump found in the endomembrane system; stabilizes VO subunits; required for V1 domain assembly on the vacuolar membrane |
|
| YBL044W | 0.35 |
Putative protein of unknown function; YBL044W is not an essential protein |
||
| YPR017C | 0.34 |
DSS4
|
Guanine nucleotide dissociation stimulator for Sec4p, functions in the post-Golgi secretory pathway; binds zinc, found both on membranes and in the cytosol |
|
| YNL035C | 0.34 |
Putative protein of unknown function with similarity to proteins containing WD-40 domains; green fluorescent protein (GFP)-fusion protein localizes to the nucleus; YNL035C is not an essential gene |
||
| YDR344C | 0.34 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR154C | 0.34 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YPL256C | 0.34 |
CLN2
|
G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) |
|
| YNL160W | 0.34 |
YGP1
|
Cell wall-related secretory glycoprotein; induced by nutrient deprivation-associated growth arrest and upon entry into stationary phase; may be involved in adaptation prior to stationary phase entry; has similarity to Sps100p |
|
| YER153C | 0.34 |
PET122
|
Mitochondrial translational activator specific for the COX3 mRNA, acts together with Pet54p and Pet494p; located in the mitochondrial inner membrane |
|
| YDR384C | 0.33 |
ATO3
|
Plasma membrane protein, regulation pattern suggests a possible role in export of ammonia from the cell; phosphorylated in mitochondria; member of the TC 9.B.33 YaaH family of putative transporters |
|
| YBR126W-A | 0.33 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR126W-B; identified by gene-trapping, microarray analysis, and genome-wide homology searches |
||
| YJR147W | 0.33 |
HMS2
|
Protein with similarity to heat shock transcription factors; overexpression suppresses the pseudohyphal filamentation defect of a diploid mep1 mep2 homozygous null mutant |
|
| YHR021C | 0.33 |
RPS27B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps27Ap and has similarity to rat S27 ribosomal protein |
|
| YDR044W | 0.33 |
HEM13
|
Coproporphyrinogen III oxidase, an oxygen requiring enzyme that catalyzes the sixth step in the heme biosynthetic pathway; localizes to the mitochondrial inner membrane; transcription is repressed by oxygen and heme (via Rox1p and Hap1p) |
|
| YOL120C | 0.32 |
RPL18A
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl18Bp and has similarity to rat L18 ribosomal protein; intron of RPL18A pre-mRNA forms stem-loop structures that are a target for Rnt1p cleavage leading to degradation |
|
| YJR015W | 0.32 |
Putative protein of unknown function; localizes to the endoplasmic reticulum and cytoplasm; predicted to encode a membrane transporter based on phylogenetic analysis; YJR015W is a non-essential gene |
||
| YMR199W | 0.32 |
CLN1
|
G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) |
|
| YBR183W | 0.31 |
YPC1
|
Alkaline ceramidase that also has reverse (CoA-independent) ceramide synthase activity, catalyzes both breakdown and synthesis of phytoceramide; overexpression confers fumonisin B1 resistance |
|
| YMR272C | 0.31 |
SCS7
|
Sphingolipid alpha-hydroxylase, functions in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids, has both cytochrome b5-like and hydroxylase/desaturase domains, not essential for growth |
|
| YBR093C | 0.31 |
PHO5
|
Repressible acid phosphatase (1 of 3) that also mediates extracellular nucleotide-derived phosphate hydrolysis; secretory pathway derived cell surface glycoprotein; induced by phosphate starvation and coordinately regulated by PHO4 and PHO2 |
|
| YIL011W | 0.31 |
TIR3
|
Cell wall mannoprotein of the Srp1p/Tip1p family of serine-alanine-rich proteins; expressed under anaerobic conditions and required for anaerobic growth |
|
| YPR198W | 0.30 |
SGE1
|
Plasma membrane multidrug transporter of the major facilitator superfamily, acts as an extrusion permease; partial multicopy suppressor of gal11 mutations |
|
| YML123C | 0.30 |
PHO84
|
High-affinity inorganic phosphate (Pi) transporter and low-affinity manganese transporter; regulated by Pho4p and Spt7p; mutation confers resistance to arsenate; exit from the ER during maturation requires Pho86p |
|
| YHR092C | 0.30 |
HXT4
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YOR107W | 0.30 |
RGS2
|
Negative regulator of glucose-induced cAMP signaling; directly activates the GTPase activity of the heterotrimeric G protein alpha subunit Gpa2p |
|
| YLR178C | 0.29 |
TFS1
|
Carboxypeptidase Y inhibitor, function requires acetylation by the NatB N-terminal acetyltransferase; phosphatidylethanolamine-binding protein involved in protein kinase A signaling pathway |
|
| YLR415C | 0.29 |
Putative protein of unknown function; YLR415C is not an essential gene |
||
| YOR226C | 0.29 |
ISU2
|
Conserved protein of the mitochondrial matrix, required for synthesis of mitochondrial and cytosolic iron-sulfur proteins, performs a scaffolding function in mitochondria during Fe/S cluster assembly; isu1 isu2 double mutant is inviable |
|
| YKL060C | 0.29 |
FBA1
|
Fructose 1,6-bisphosphate aldolase, required for glycolysis and gluconeogenesis; catalyzes conversion of fructose 1,6 bisphosphate to glyceraldehyde-3-P and dihydroxyacetone-P; locates to mitochondrial outer surface upon oxidative stress |
|
| YBR052C | 0.29 |
RFS1
|
Protein of unknown function; member of a flavodoxin-like fold protein family that includes Pst2p and Ycp4p; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YOR072W-B | 0.29 |
Putative protein of unknown function; identified by expression profiling and mass spectrometry |
||
| YGR108W | 0.29 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YIL096C | 0.28 |
Putative protein of unknown function; associates with precursors of the 60S ribosomal subunit |
||
| YCL063W | 0.28 |
VAC17
|
Protein involved in vacuole inheritance; acts as a vacuole-specific receptor for myosin Myo2p |
|
| YPR197C | 0.28 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOL011W | 0.27 |
PLB3
|
Phospholipase B (lysophospholipase) involved in phospholipid metabolism; hydrolyzes phosphatidylinositol and phosphatidylserine and displays transacylase activity in vitro |
|
| YBR126C | 0.27 |
TPS1
|
Synthase subunit of trehalose-6-phosphate synthase/phosphatase complex, which synthesizes the storage carbohydrate trehalose; also found in a monomeric form; expression is induced by the stress response and repressed by the Ras-cAMP pathway |
|
| YDR276C | 0.27 |
PMP3
|
Small plasma membrane protein related to a family of plant polypeptides that are overexpressed under high salt concentration or low temperature, not essential for viability, deletion causes hyperpolarization of the plasma membrane potential |
|
| YER026C | 0.27 |
CHO1
|
Phosphatidylserine synthase, functions in phospholipid biosynthesis; catalyzes the reaction CDP-diaclyglycerol + L-serine = CMP + L-1-phosphatidylserine, transcriptionally repressed by myo-inositol and choline |
|
| YPL031C | 0.27 |
PHO85
|
Cyclin-dependent kinase, with ten cyclin partners; involved in regulating the cellular response to nutrient levels and environmental conditions and progression through the cell cycle |
|
| YGL255W | 0.27 |
ZRT1
|
High-affinity zinc transporter of the plasma membrane, responsible for the majority of zinc uptake; transcription is induced under low-zinc conditions by the Zap1p transcription factor |
|
| YJL136C | 0.27 |
RPS21B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps21Ap and has similarity to rat S21 ribosomal protein |
|
| YLR355C | 0.26 |
ILV5
|
Acetohydroxyacid reductoisomerase, mitochondrial protein involved in branched-chain amino acid biosynthesis, also required for maintenance of wild-type mitochondrial DNA and found in mitochondrial nucleoids |
|
| YBR158W | 0.26 |
AMN1
|
Protein required for daughter cell separation, multiple mitotic checkpoints, and chromosome stability; contains 12 degenerate leucine-rich repeat motifs; expression is induced by the Mitotic Exit Network (MEN) |
|
| YNL178W | 0.26 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YCL048W-A | 0.26 |
Putative protein of unknown function |
||
| YJR116W | 0.26 |
Putative protein of unknown function |
||
| YBR293W | 0.26 |
VBA2
|
Permease of basic amino acids in the vacuolar membrane |
|
| YCL043C | 0.26 |
PDI1
|
Protein disulfide isomerase, multifunctional protein resident in the endoplasmic reticulum lumen, essential for the formation of disulfide bonds in secretory and cell-surface proteins, unscrambles non-native disulfide bonds |
|
| YGL080W | 0.25 |
FMP37
|
Putative protein of unknown function; highly conserved across species and orthologous to human gene BRP44L; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YGL225W | 0.24 |
VRG4
|
Golgi GDP-mannose transporter; regulates Golgi function and glycosylation in Golgi |
|
| YOL058W | 0.24 |
ARG1
|
Arginosuccinate synthetase, catalyzes the formation of L-argininosuccinate from citrulline and L-aspartate in the arginine biosynthesis pathway; potential Cdc28p substrate |
|
| YNL231C | 0.24 |
PDR16
|
Phosphatidylinositol transfer protein (PITP) controlled by the multiple drug resistance regulator Pdr1p, localizes to lipid particles and microsomes, controls levels of various lipids, may regulate lipid synthesis, homologous to Pdr17p |
|
| YDR346C | 0.24 |
SVF1
|
Protein with a potential role in cell survival pathways, required for the diauxic growth shift; expression in mammalian cells increases survival under conditions inducing apoptosis |
|
| YCL040W | 0.24 |
GLK1
|
Glucokinase, catalyzes the phosphorylation of glucose at C6 in the first irreversible step of glucose metabolism; one of three glucose phosphorylating enzymes; expression regulated by non-fermentable carbon sources |
|
| YEL017W | 0.24 |
GTT3
|
Protein of unknown function with a possible role in glutathione metabolism, as suggested by computational analysis of large-scale protein-protein interaction data; GFP-fusion protein localizes to the nuclear periphery |
|
| YBR085W | 0.24 |
AAC3
|
Mitochondrial inner membrane ADP/ATP translocator, exchanges cytosolic ADP for mitochondrially synthesized ATP; expressed under anaerobic conditions; similar to Pet9p and Aac1p; has roles in maintenance of viability and in respiration |
|
| YML028W | 0.24 |
TSA1
|
Thioredoxin peroxidase, acts as both a ribosome-associated and free cytoplasmic antioxidant; self-associates to form a high-molecular weight chaperone complex under oxidative stress; deletion results in mutator phenotype |
|
| YBR082C | 0.23 |
UBC4
|
Ubiquitin-conjugating enzyme (E2), mediates degradation of short-lived and abnormal proteins; interacts with E3-CaM in ubiquitinating calmodulin; interacts with many SCF ubiquitin protein ligases; component of the cellular stress response |
|
| YOR103C | 0.23 |
OST2
|
Epsilon subunit of the oligosaccharyltransferase complex of the ER lumen, which catalyzes asparagine-linked glycosylation of newly synthesized proteins |
|
| YCL049C | 0.23 |
Protein of unknown function; localizes to membrane fraction; YCL049C is not an essential gene; |
||
| YHL006C | 0.23 |
SHU1
|
Protein of unassigned function involved in mutation suppression, important for error-free repair of spontaneous and induced DNA lesions to protect the genome from mutation; associates with Shu2p, Psy3p, and Csm2p |
|
| YPL059W | 0.23 |
GRX5
|
Hydroperoxide and superoxide-radical responsive glutathione-dependent oxidoreductase; mitochondrial matrix protein involved in the synthesis/assembly of iron-sulfur centers; monothiol glutaredoxin subfamily member along with Grx3p and Grx4p |
|
| YIR027C | 0.23 |
DAL1
|
Allantoinase, converts allantoin to allantoate in the first step of allantoin degradation; expression sensitive to nitrogen catabolite repression |
|
| YER074W | 0.23 |
RPS24A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps24Bp and has similarity to rat S24 ribosomal protein |
|
| YMR143W | 0.23 |
RPS16A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps16Bp and has similarity to E. coli S9 and rat S16 ribosomal proteins |
|
| YGL209W | 0.22 |
MIG2
|
Protein containing zinc fingers, involved in repression, along with Mig1p, of SUC2 (invertase) expression by high levels of glucose; binds to Mig1p-binding sites in SUC2 promoter |
|
| YOL155C | 0.22 |
HPF1
|
Haze-protective mannoprotein that reduces the particle size of aggregated proteins in white wines |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBS1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0000296 | spermine transport(GO:0000296) |
| 0.6 | 4.4 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.6 | 1.7 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) regulation of membrane lipid metabolic process(GO:1905038) |
| 0.5 | 2.0 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.5 | 1.5 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.4 | 2.5 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.3 | 1.0 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.3 | 1.4 | GO:0098609 | agglutination involved in conjugation with cellular fusion(GO:0000752) agglutination involved in conjugation(GO:0000771) heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) cell-cell adhesion(GO:0098609) adhesion between unicellular organisms(GO:0098610) multi organism cell adhesion(GO:0098740) cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.3 | 1.0 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 0.3 | 0.9 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.3 | 0.8 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 0.7 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.2 | 1.6 | GO:0019655 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.2 | 1.5 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 | 1.2 | GO:0006672 | ceramide metabolic process(GO:0006672) ceramide biosynthetic process(GO:0046513) |
| 0.2 | 1.2 | GO:0007155 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.2 | 0.6 | GO:0009094 | tyrosine biosynthetic process(GO:0006571) L-phenylalanine biosynthetic process(GO:0009094) erythrose 4-phosphate/phosphoenolpyruvate family amino acid biosynthetic process(GO:1902223) |
| 0.2 | 0.5 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) isoleucine biosynthetic process(GO:0009097) |
| 0.2 | 0.5 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.2 | 0.6 | GO:0035337 | long-chain fatty acid transport(GO:0015909) fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.2 | 0.5 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.2 | 0.8 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.3 | GO:0044091 | ascospore-type prospore membrane assembly(GO:0032120) membrane biogenesis(GO:0044091) membrane assembly(GO:0071709) |
| 0.1 | 0.8 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.1 | 0.5 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.1 | 0.2 | GO:0072367 | positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) positive regulation of sterol transport(GO:0032373) positive regulation of transmembrane transport(GO:0034764) regulation of sterol import by regulation of transcription from RNA polymerase II promoter(GO:0035968) positive regulation of sterol import by positive regulation of transcription from RNA polymerase II promoter(GO:0035969) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.1 | 0.3 | GO:0023021 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 1.1 | GO:0090662 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 0.7 | GO:0031506 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) |
| 0.1 | 0.4 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.5 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 2.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.8 | GO:0006797 | polyphosphate metabolic process(GO:0006797) |
| 0.1 | 0.3 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) |
| 0.1 | 1.1 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.1 | 0.4 | GO:0032443 | regulation of steroid metabolic process(GO:0019218) regulation of ergosterol biosynthetic process(GO:0032443) regulation of steroid biosynthetic process(GO:0050810) |
| 0.1 | 0.4 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.1 | 0.3 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) |
| 0.1 | 0.1 | GO:0033239 | negative regulation of cellular amine metabolic process(GO:0033239) |
| 0.1 | 1.0 | GO:0007009 | plasma membrane organization(GO:0007009) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.2 | GO:0036170 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.1 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.5 | GO:0009312 | trehalose biosynthetic process(GO:0005992) oligosaccharide biosynthetic process(GO:0009312) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.2 | GO:0052547 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 0.1 | 1.9 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.1 | 0.3 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.3 | GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000461) |
| 0.1 | 0.8 | GO:0015833 | peptide transport(GO:0015833) |
| 0.1 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.2 | GO:0030048 | actin filament-based movement(GO:0030048) actin filament-based transport(GO:0099515) |
| 0.1 | 1.0 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 0.2 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.1 | 0.4 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.3 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 0.2 | GO:0019413 | acetate biosynthetic process(GO:0019413) |
| 0.0 | 0.5 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.2 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 0.1 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) regulation of vacuole organization(GO:0044088) |
| 0.0 | 1.0 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.0 | 0.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.2 | GO:0043007 | maintenance of rDNA(GO:0043007) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0000947 | amino acid catabolic process to alcohol via Ehrlich pathway(GO:0000947) |
| 0.0 | 0.1 | GO:0048313 | Golgi inheritance(GO:0048313) |
| 0.0 | 0.4 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.3 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.4 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) |
| 0.0 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of vesicle targeting, to, from or within Golgi(GO:0048209) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.0 | 0.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.5 | GO:0006407 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.2 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.0 | 0.1 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.2 | GO:0043487 | regulation of RNA stability(GO:0043487) regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.1 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 3.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.3 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.5 | GO:0070481 | nuclear-transcribed mRNA catabolic process, non-stop decay(GO:0070481) |
| 0.0 | 0.5 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.3 | GO:0000011 | vacuole inheritance(GO:0000011) |
| 0.0 | 0.3 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.0 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) |
| 0.0 | 0.1 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 1.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.3 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.7 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.3 | 0.8 | GO:0030428 | cell septum(GO:0030428) |
| 0.3 | 2.2 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.3 | 1.1 | GO:0044462 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 0.2 | 0.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 0.7 | GO:0000928 | gamma-tubulin small complex, spindle pole body(GO:0000928) gamma-tubulin complex(GO:0000930) gamma-tubulin small complex(GO:0008275) |
| 0.2 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 0.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.7 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.5 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.1 | 2.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 0.3 | GO:0033179 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 1.4 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.2 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.2 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 3.2 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 3.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.0 | 1.1 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.6 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) R2TP complex(GO:0097255) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.7 | GO:0044445 | cytosolic part(GO:0044445) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 2.7 | GO:0000329 | fungal-type vacuole membrane(GO:0000329) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 2.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 3.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.2 | GO:0033698 | Rpd3L complex(GO:0033698) |
| 0.0 | 3.3 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 1.3 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.1 | GO:0032045 | eukaryotic translation initiation factor 2B complex(GO:0005851) guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.5 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.1 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 2.6 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.3 | 1.7 | GO:0034318 | alcohol O-acetyltransferase activity(GO:0004026) alcohol O-acyltransferase activity(GO:0034318) |
| 0.3 | 1.9 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.3 | 1.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 1.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.3 | 2.5 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.3 | 0.8 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 0.8 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.2 | 1.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 0.7 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) |
| 0.2 | 0.2 | GO:0004805 | trehalose-phosphatase activity(GO:0004805) |
| 0.2 | 0.6 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.2 | 1.0 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 0.8 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.2 | 0.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.2 | 0.5 | GO:0022829 | anion channel activity(GO:0005253) voltage-gated anion channel activity(GO:0008308) porin activity(GO:0015288) wide pore channel activity(GO:0022829) |
| 0.2 | 0.9 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 0.5 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.2 | 1.1 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.2 | 0.5 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.6 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.6 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.5 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 0.1 | 2.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 0.7 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.3 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) |
| 0.1 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.5 | GO:0008106 | aldo-keto reductase (NADP) activity(GO:0004033) alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 0.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 1.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 2.0 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 0.9 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.1 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.3 | GO:0005384 | inorganic phosphate transmembrane transporter activity(GO:0005315) manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.5 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.2 | GO:0030414 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 0.1 | 1.0 | GO:0051540 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.1 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.9 | GO:0016769 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.4 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.4 | GO:0015079 | potassium ion transmembrane transporter activity(GO:0015079) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 1.7 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 1.1 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.1 | GO:0043141 | 5'-3' DNA helicase activity(GO:0043139) ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.5 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.4 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0051880 | RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 0.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 6.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.0 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.1 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.5 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 0.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 0.2 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.2 | REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | Genes involved in Insulin receptor signalling cascade |
| 0.1 | 0.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME SIGNALING BY GPCR | Genes involved in Signaling by GPCR |
| 0.0 | 0.8 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.1 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.0 | 0.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |