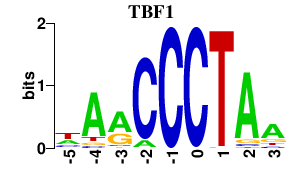
Results for TBF1
Z-value: 0.67
Transcription factors associated with TBF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBF1
|
S000006049 | Telobox-containing general regulatory factor |
Activity-expression correlation:
Activity profile of TBF1 motif
Sorted Z-values of TBF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YLR312C | 2.19 |
QNQ1
|
Protein of unknown function; null mutant quiescent and non-quiescent cells exhibit reduced reproductive capacity |
|
| YHR212W-A | 1.52 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YMR107W | 1.51 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YAR053W | 1.41 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YJL133C-A | 1.31 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YCR068W | 1.26 |
ATG15
|
Lipase, required for intravacuolar lysis of autophagosomes; located in the endoplasmic reticulum membrane and targeted to intravacuolar vesicles during autophagy via the multivesicular body (MVB) pathway |
|
| YDR490C | 1.09 |
PKH1
|
Serine/threonine protein kinase involved in sphingolipid-mediated signaling pathway that controls endocytosis; activates Ypk1p and Ykr2p, components of signaling cascade required for maintenance of cell wall integrity; redundant with Pkh2p |
|
| YFL033C | 1.06 |
RIM15
|
Glucose-repressible protein kinase involved in signal transduction during cell proliferation in response to nutrients, specifically the establishment of stationary phase; identified as a regulator of IME2; substrate of Pho80p-Pho85p kinase |
|
| YGL146C | 1.04 |
Putative protein of unknown function, contains two putative transmembrane spans, shows no significant homology to any other known protein sequence, YGL146C is not an essential gene |
||
| YKL171W | 1.04 |
Putative protein of unknown function; predicted protein kinase; implicated in proteasome function; epitope-tagged protein localizes to the cytoplasm |
||
| YNL117W | 1.03 |
MLS1
|
Malate synthase, enzyme of the glyoxylate cycle, involved in utilization of non-fermentable carbon sources; expression is subject to carbon catabolite repression; localizes in peroxisomes during growth in oleic acid medium |
|
| YAR060C | 1.01 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR212C | 0.98 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJR120W | 0.92 |
Protein of unknown function; essential for growth under anaerobic conditions; mutation causes decreased expression of ATP2, impaired respiration, defective sterol uptake, and altered levels/localization of ABC transporters Aus1p and Pdr11p |
||
| YGL219C | 0.92 |
MDM34
|
Mitochondrial outer membrane protein, required for normal mitochondrial morphology and inheritance; localizes to dots on the mitochondrial surface near mtDNA nucleoids; interacts genetically with MDM31 and MDM32 |
|
| YMR302C | 0.91 |
YME2
|
Integral inner mitochondrial membrane protein with a role in maintaining mitochondrial nucleoid structure and number; mutants exhibit an increased rate of mitochondrial DNA escape; shows some sequence similarity to exonucleases |
|
| YER169W | 0.90 |
RPH1
|
JmjC domain-containing histone demethylase which can specifically demethylate H3K36 tri- and dimethyl modification states; transcriptional repressor of PHR1; Rph1p phosphorylation during DNA damage is under control of the MEC1-RAD53 pathway |
|
| YMR172W | 0.88 |
HOT1
|
Transcription factor required for the transient induction of glycerol biosynthetic genes GPD1 and GPP2 in response to high osmolarity; targets Hog1p to osmostress responsive promoters; has similarity to Msn1p and Gcr1p |
|
| YOL060C | 0.87 |
MAM3
|
Protein required for normal mitochondrial morphology, has similarity to hemolysins |
|
| YML017W | 0.86 |
PSP2
|
Asn rich cytoplasmic protein that contains RGG motifs; high-copy suppressor of group II intron-splicing defects of a mutation in MRS2 and of a conditional mutation in POL1 (DNA polymerase alpha); possible role in mitochondrial mRNA splicing |
|
| YCR073W-A | 0.84 |
SOL2
|
Protein with a possible role in tRNA export; shows similarity to 6-phosphogluconolactonase non-catalytic domains but does not exhibit this enzymatic activity; homologous to Sol1p, Sol3p, and Sol4p |
|
| YLR094C | 0.84 |
GIS3
|
Protein of unknown function |
|
| YKR089C | 0.83 |
TGL4
|
Triacylglycerol lipase involved in triacylglycerol mobilization and degradation; found in lipid particles; potential Cdc28p substrate |
|
| YJR119C | 0.82 |
JHD2
|
JmjC domain family histone demethylase specific for H3-K4 (lysine at position 4 of the histone H3 protein); removes methyl groups specifically added by Set1p methyltransferase |
|
| YPL116W | 0.80 |
HOS3
|
Trichostatin A-insensitive homodimeric histone deacetylase (HDAC) with specificity in vitro for histones H3, H4, H2A, and H2B; similar to Hda1p, Rpd3p, Hos1p, and Hos2p; deletion results in increased histone acetylation at rDNA repeats |
|
| YFR040W | 0.79 |
SAP155
|
Protein that forms a complex with the Sit4p protein phosphatase and is required for its function; member of a family of similar proteins including Sap4p, Sap185p, and Sap190p |
|
| YOL051W | 0.78 |
GAL11
|
Subunit of the RNA polymerase II mediator complex; associates with core polymerase subunits to form the RNA polymerase II holoenzyme; affects transcription by acting as target of activators and repressors |
|
| YKR025W | 0.77 |
RPC37
|
RNA polymerase III subunit C37 |
|
| YJR109C | 0.75 |
CPA2
|
Large subunit of carbamoyl phosphate synthetase, which catalyzes a step in the synthesis of citrulline, an arginine precursor |
|
| YMR171C | 0.73 |
EAR1
|
Specificity factor required for Rsp5p-dependent ubiquitination and sorting of specific cargo proteins at the multivesicular body; mRNA is targeted to the bud via the mRNA transport system involving She2p: YMR171C is not an essential gene |
|
| YLL013C | 0.71 |
PUF3
|
Protein of the mitochondrial outer surface, links the Arp2/3 complex with the mitochore during anterograde mitochondrial movement; also binds to and promotes degradation of mRNAs for select nuclear-encoded mitochondrial proteins |
|
| YHR206W | 0.71 |
SKN7
|
Nuclear response regulator and transcription factor, part of a branched two-component signaling system; required for optimal induction of heat-shock genes in response to oxidative stress; involved in osmoregulation |
|
| YFL051C | 0.71 |
Putative protein of unknown function; YFL051C is not an essential gene |
||
| YBR072W | 0.68 |
HSP26
|
Small heat shock protein (sHSP) with chaperone activity; forms hollow, sphere-shaped oligomers that suppress unfolded proteins aggregation; oligomer activation requires a heat-induced conformational change; not expressed in unstressed cells |
|
| YOL084W | 0.68 |
PHM7
|
Protein of unknown function, expression is regulated by phosphate levels; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery and vacuole |
|
| YGL217C | 0.64 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF KIP3/YGL216W |
||
| YDL170W | 0.64 |
UGA3
|
Transcriptional activator necessary for gamma-aminobutyrate (GABA)-dependent induction of GABA genes (such as UGA1, UGA2, UGA4); zinc-finger transcription factor of the Zn(2)-Cys(6) binuclear cluster domain type; localized to the nucleus |
|
| YBL033C | 0.64 |
RIB1
|
GTP cyclohydrolase II; catalyzes the first step of the riboflavin biosynthesis pathway |
|
| YPR184W | 0.63 |
GDB1
|
Glycogen debranching enzyme containing glucanotranferase and alpha-1,6-amyloglucosidase activities, required for glycogen degradation; phosphorylated in mitochondria |
|
| YGL205W | 0.63 |
POX1
|
Fatty-acyl coenzyme A oxidase, involved in the fatty acid beta-oxidation pathway; localized to the peroxisomal matrix |
|
| YAL028W | 0.63 |
FRT2
|
Tail-anchored endoplasmic reticulum membrane protein, interacts with homolog Frt1p but is not a substrate of calcineurin (unlike Frt1p), promotes growth in conditions of high Na+, alkaline pH, or cell wall stress; potential Cdc28p substrate |
|
| YGL216W | 0.62 |
KIP3
|
Kinesin-related motor protein involved in mitotic spindle positioning |
|
| YMR017W | 0.61 |
SPO20
|
Meiosis-specific subunit of the t-SNARE complex, required for prospore membrane formation during sporulation; similar to but not functionally redundant with Sec9p; SNAP-25 homolog |
|
| YLR189C | 0.61 |
ATG26
|
UDP-glucose:sterol glucosyltransferase, conserved enzyme involved in synthesis of sterol glucoside membrane lipids; in contrast to ATG26 from P. pastoris, S. cerevisiae ATG26 is not involved in autophagy |
|
| YGR230W | 0.60 |
BNS1
|
Protein with some similarity to Spo12p; overexpression bypasses need for Spo12p, but not required for meiosis |
|
| YBR037C | 0.59 |
SCO1
|
Copper-binding protein of the mitochondrial inner membrane, required for cytochrome c oxidase activity and respiration; may function to deliver copper to cytochrome c oxidase; has similarity to thioredoxins |
|
| YOR235W | 0.59 |
IRC13
|
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YCR086W | 0.59 |
CSM1
|
Nucleolar protein that forms a complex with Lrs4p which binds Mam1p at kinetochores during meiosis I to mediate accurate chromosome segregation, may be involved in premeiotic DNA replication; possible role in telomere maintenance |
|
| YDR536W | 0.59 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YGR256W | 0.57 |
GND2
|
6-phosphogluconate dehydrogenase (decarboxylating), catalyzes an NADPH regenerating reaction in the pentose phosphate pathway; required for growth on D-glucono-delta-lactone |
|
| YPR030W | 0.57 |
CSR2
|
Nuclear protein with a potential regulatory role in utilization of galactose and nonfermentable carbon sources; overproduction suppresses the lethality at high temperature of a chs5 spa2 double null mutation; potential Cdc28p substrate |
|
| YKL217W | 0.57 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YJR128W | 0.57 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF RSF2 |
||
| YLR055C | 0.56 |
SPT8
|
Subunit of the SAGA transcriptional regulatory complex but not present in SAGA-like complex SLIK/SALSA, required for SAGA-mediated inhibition at some promoters |
|
| YHR150W | 0.55 |
PEX28
|
Peroxisomal integral membrane peroxin, involved in the regulation of peroxisomal size, number and distribution; genetic interactions suggest that Pex28p and Pex29p act at steps upstream of those mediated by Pex30p, Pex31p, and Pex32p |
|
| YJL020C | 0.55 |
BBC1
|
Protein possibly involved in assembly of actin patches; interacts with an actin assembly factor Las17p and with the SH3 domains of Type I myosins Myo3p and Myo5p; localized predominantly to cortical actin patches |
|
| YCL016C | 0.55 |
DCC1
|
Subunit of a complex with Ctf8p and Ctf18p that shares some components with Replication Factor C, required for sister chromatid cohesion and telomere length maintenance |
|
| YKL105C | 0.55 |
Putative protein of unknown function |
||
| YDL230W | 0.55 |
PTP1
|
Phosphotyrosine-specific protein phosphatase that dephosphorylates a broad range of substrates in vivo, including Fpr3p; localized to the cytoplasm and the mitochondria |
|
| YEL060C | 0.54 |
PRB1
|
Vacuolar proteinase B (yscB), a serine protease of the subtilisin family; involved in protein degradation in the vacuole and required for full protein degradation during sporulation |
|
| YMR175W | 0.53 |
SIP18
|
Protein of unknown function whose expression is induced by osmotic stress |
|
| YLR402W | 0.53 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR446W | 0.53 |
ECM11
|
Non-essential protein apparently involved in meiosis, GFP fusion protein is present in discrete clusters in the nucleus throughout mitosis; may be involved in maintaining chromatin structure |
|
| YGR243W | 0.53 |
FMP43
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YKL103C | 0.52 |
LAP4
|
Vacuolar aminopeptidase, often used as a marker protein in studies of autophagy and cytosol to vacuole targeting (CVT) pathway |
|
| YEL009C-A | 0.51 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR013C | 0.51 |
SEC59
|
Dolichol kinase, catalyzes the terminal step in dolichyl monophosphate (Dol-P) biosynthesis; required for viability and for normal rates of lipid intermediate synthesis and protein N-glycosylation |
|
| YLL010C | 0.50 |
PSR1
|
Plasma membrane associated protein phosphatase involved in the general stress response; required along with binding partner Whi2p for full activation of STRE-mediated gene expression, possibly through dephosphorylation of Msn2p |
|
| YJR094C | 0.50 |
IME1
|
Master regulator of meiosis that is active only during meiotic events, activates transcription of early meiotic genes through interaction with Ume6p, degraded by the 26S proteasome following phosphorylation by Ime2p |
|
| YML007W | 0.50 |
YAP1
|
Basic leucine zipper (bZIP) transcription factor required for oxidative stress tolerance; activated by H2O2 through the multistep formation of disulfide bonds and transit from the cytoplasm to the nucleus; mediates resistance to cadmium |
|
| YOR147W | 0.50 |
MDM32
|
Mitochondrial inner membrane protein with similarity to Mdm31p, required for normal mitochondrial morphology and inheritance; interacts genetically with MMM1, MDM10, MDM12, and MDM34 |
|
| YDR277C | 0.50 |
MTH1
|
Negative regulator of the glucose-sensing signal transduction pathway, required for repression of transcription by Rgt1p; interacts with Rgt1p and the Snf3p and Rgt2p glucose sensors; phosphorylated by Yck1p, triggering Mth1p degradation |
|
| YBL038W | 0.49 |
MRPL16
|
Mitochondrial ribosomal protein of the large subunit |
|
| YIL055C | 0.49 |
Putative protein of unknown function |
||
| YCR007C | 0.47 |
Putative integral membrane protein, member of DUP240 gene family; YCR007C is not an essential gene |
||
| YOL100W | 0.46 |
PKH2
|
Serine/threonine protein kinase involved in sphingolipid-mediated signaling pathway that controls endocytosis; activates Ypk1p and Ykr2p, components of signaling cascade required for maintenance of cell wall integrity; redundant with Pkh1p |
|
| YPL223C | 0.46 |
GRE1
|
Hydrophilin of unknown function; stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YDR542W | 0.45 |
PAU10
|
Hypothetical protein |
|
| YLR403W | 0.45 |
SFP1
|
Transcription factor that controls expression of many ribosome biogenesis genes in response to nutrients and stress, regulates G2/M transitions during mitotic cell cycle and DNA-damage response, involved in cell size modulation |
|
| YER067C-A | 0.44 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YER067W |
||
| YLR149C | 0.43 |
Putative protein of unknown function; YLR149C is not an essential gene |
||
| YKL139W | 0.43 |
CTK1
|
Catalytic (alpha) subunit of C-terminal domain kinase I (CTDK-I), which phosphorylates the C-terminal repeated domain of the RNA polymerase II large subunit (Rpo21p) to affect both transcription and pre-mRNA 3' end processing |
|
| YGL228W | 0.43 |
SHE10
|
Putative glycosylphosphatidylinositol (GPI)-anchored protein of unknown function; overexpression causes growth arrest |
|
| YNL172W | 0.43 |
APC1
|
Largest subunit of the Anaphase-Promoting Complex/Cyclosome (APC/C), which is a ubiquitin-protein ligase required for degradation of anaphase inhibitors, including mitotic cyclins, during the metaphase/anaphase transition |
|
| YKL102C | 0.43 |
Dubious open reading frame unlikely to encode a functional protein; deletion confers sensitivity to citric acid; predicted protein would include a thiol-disulfide oxidoreductase active site |
||
| YDR342C | 0.43 |
HXT7
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels |
|
| YHR211W | 0.42 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YLR366W | 0.42 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR364C-A |
||
| YER014C-A | 0.42 |
BUD25
|
Protein involved in bud-site selection; diploid mutants display a random budding pattern instead of the wild-type bipolar pattern |
|
| YOR324C | 0.42 |
FRT1
|
Tail-anchored endoplasmic reticulum membrane protein that is a substrate of the phosphatase calcineurin, interacts with homolog Frt2p, promotes cell growth in conditions of high Na+, alkaline pH, and cell wall stress |
|
| YCL014W | 0.41 |
BUD3
|
Protein involved in bud-site selection and required for axial budding pattern; localizes with septins to bud neck in mitosis and may constitute an axial landmark for next round of budding |
|
| YDR102C | 0.40 |
Dubious open reading frame; homozygous diploid deletion strain exhibits high budding index |
||
| YGL089C | 0.40 |
MF(ALPHA)2
|
Mating pheromone alpha-factor, made by alpha cells; interacts with mating type a cells to induce cell cycle arrest and other responses leading to mating; also encoded by MF(ALPHA)1, which is more highly expressed than MF(ALPHA)2 |
|
| YDR103W | 0.40 |
STE5
|
Pheromone-response scaffold protein; binds kinases Ste11p, Ste7p, and Fus3p to form a MAPK cascade complex that interacts with the plasma membrane, via a PH (pleckstrin homology) and PM/NLS domain, and with Ste4p-Ste18p, during signaling |
|
| YOL035C | 0.40 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER065C | 0.40 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YLR174W | 0.39 |
IDP2
|
Cytosolic NADP-specific isocitrate dehydrogenase, catalyzes oxidation of isocitrate to alpha-ketoglutarate; levels are elevated during growth on non-fermentable carbon sources and reduced during growth on glucose |
|
| YNL093W | 0.39 |
YPT53
|
GTPase, similar to Ypt51p and Ypt52p and to mammalian rab5; required for vacuolar protein sorting and endocytosis |
|
| YLR019W | 0.39 |
PSR2
|
Functionally redundant Psr1p homolog, a plasma membrane phosphatase involved in the general stress response; required with Psr1p and Whi2p for full activation of STRE-mediated gene expression, possibly through dephosphorylation of Msn2p |
|
| YEL010W | 0.39 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR325W | 0.38 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps the verified ORF FRT1 |
||
| YPR010C-A | 0.38 |
Putative protein of unknown function; conserved among Saccharomyces sensu stricto species |
||
| YPR151C | 0.38 |
SUE1
|
Mitochondrial protein required for degradation of unstable forms of cytochrome c |
|
| YLR356W | 0.38 |
Putative protein of unknown function with similarity to SCM4; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR356W is not an essential gene |
||
| YCR005C | 0.38 |
CIT2
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate, peroxisomal isozyme involved in glyoxylate cycle; expression is controlled by Rtg1p and Rtg2p transcription factors |
|
| YPL026C | 0.37 |
SKS1
|
Putative serine/threonine protein kinase; involved in the adaptation to low concentrations of glucose independent of the SNF3 regulated pathway |
|
| YOR211C | 0.37 |
MGM1
|
Mitochondrial GTPase related to dynamin, present in a complex containing Ugo1p and Fzo1p; required for normal morphology of cristae and for stability of Tim11p; homolog of human OPA1 involved in autosomal dominant optic atrophy |
|
| YPL024W | 0.37 |
RMI1
|
Involved in response to DNA damage; null mutants have increased rates of recombination and delayed S phase; interacts physically and genetically with Sgs1p (RecQ family member) and Top3p (topoisomerase III) |
|
| YJR115W | 0.37 |
Putative protein of unknown function |
||
| YMR118C | 0.37 |
Protein of unknown function with similarity to succinate dehydrogenase cytochrome b subunit; YMR118C is not an essential gene |
||
| YOL131W | 0.37 |
Putative protein of unknown function |
||
| YML034W | 0.36 |
SRC1
|
Inner nuclear membrane (INM) protein with a putative role in sister chromatid segregation, potentially phosphorylated by Cdc28p; contains helix-extension-helix (HEH) motif, nuclear localization signal sequence |
|
| YJR061W | 0.36 |
Putative protein of unknown function; non-essential gene with similarity to Mnn4, a putative membrane protein involved in glycosylation; transcription repressed by Rm101p |
||
| YNL339C | 0.36 |
YRF1-6
|
Helicase encoded by the Y' element of subtelomeric regions, highly expressed in the mutants lacking the telomerase component TLC1; potentially phosphorylated by Cdc28p |
|
| YOR382W | 0.35 |
FIT2
|
Mannoprotein that is incorporated into the cell wall via a glycosylphosphatidylinositol (GPI) anchor, involved in the retention of siderophore-iron in the cell wall |
|
| YPL230W | 0.35 |
USV1
|
Putative transcription factor containing a C2H2 zinc finger; mutation affects transcriptional regulation of genes involved in protein folding, ATP binding, and cell wall biosynthesis |
|
| YGL177W | 0.35 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR301W | 0.34 |
CFT1
|
RNA-binding subunit of the mRNA cleavage and polyadenylation factor; involved in poly(A) site recognition and required for both pre-mRNA cleavage and polyadenylation, 51% sequence similarity with mammalian AAUAA-binding subunit of CPSF |
|
| YHR193C-A | 0.34 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps verified ORF MDM31 |
||
| YAL039C | 0.34 |
CYC3
|
Cytochrome c heme lyase (holocytochrome c synthase), attaches heme to apo-cytochrome c (Cyc1p or Cyc7p) in the mitochondrial intermembrane space; human ortholog may have a role in microphthalmia with linear skin defects (MLS) |
|
| YBL073W | 0.34 |
Hypothetical protein; open reading frame overlaps 5' end of essential AAR2 gene encoding a component of the U5 snRNP required for splicing of U3 precursors |
||
| YLR425W | 0.33 |
TUS1
|
Guanine nucleotide exchange factor (GEF) that functions to modulate Rho1p activity as part of the cell integrity signaling pathway; multicopy suppressor of tor2 mutation and ypk1 ypk2 double mutation; potential Cdc28p substrate |
|
| YPL058C | 0.33 |
PDR12
|
Plasma membrane ATP-binding cassette (ABC) transporter, weak-acid-inducible multidrug transporter required for weak organic acid resistance; induced by sorbate and benzoate and regulated by War1p; mutants exhibit sorbate hypersensitivity |
|
| YMR313C | 0.33 |
TGL3
|
Triacylglycerol lipase of the lipid particle, responsible for all the TAG lipase activity of the lipid particle; contains the consensus sequence motif GXSXG, which is found in lipolytic enzymes |
|
| YAL062W | 0.33 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YFR036W | 0.33 |
CDC26
|
Subunit of the Anaphase-Promoting Complex/Cyclosome (APC/C), which is a ubiquitin-protein ligase required for degradation of anaphase inhibitors, including mitotic cyclins, during the metaphase/anaphase transition |
|
| YCL001W-B | 0.32 |
Putative protein of unknown function; identified by homology |
||
| YDL231C | 0.32 |
BRE4
|
Zinc finger protein containing five transmembrane domains; null mutant exhibits strongly fragmented vacuoles and sensitivity to brefeldin A, a drug which is known to affect intracellular transport |
|
| YHR194W | 0.32 |
MDM31
|
Mitochondrial inner membrane protein with similarity to Mdm32p, required for normal mitochondrial morphology and inheritance; interacts genetically with MMM1, MDM10, MDM12, and MDM34 |
|
| YKL023W | 0.32 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YOR098C | 0.32 |
NUP1
|
Nuclear pore complex (NPC) subunit, involved in protein import/export and in export of RNAs, possible karyopherin release factor that accelerates release of karyopherin-cargo complexes after transport across NPC; potential Cdc28p substrate |
|
| YDR059C | 0.32 |
UBC5
|
Ubiquitin-conjugating enzyme that mediates selective degradation of short-lived and abnormal proteins, central component of the cellular stress response; expression is heat inducible |
|
| YPL127C | 0.31 |
HHO1
|
Histone H1, a linker histone required for nucleosome packaging at restricted sites; suppresses DNA repair involving homologous recombination; not required for telomeric silencing, basal transcriptional repression, or efficient sporulation |
|
| YLR431C | 0.31 |
ATG23
|
Peripheral membrane protein required for the cytoplasm-to-vacuole targeting (Cvt) pathway; cycles between the pre-autophagosome (PAS) and non-PAS locations; forms a complex with Atg9p and Atg27p |
|
| YDR358W | 0.31 |
GGA1
|
Golgi-localized protein with homology to gamma-adaptin, interacts with and regulates Arf1p and Arf2p in a GTP-dependent manner in order to facilitate traffic through the late Golgi |
|
| YHR151C | 0.31 |
MTC6
|
Putative protein of unknown function; YHR151C is synthetically sick with cdc13-1 |
|
| YMR068W | 0.31 |
AVO2
|
Component of a complex containing the Tor2p kinase and other proteins, which may have a role in regulation of cell growth |
|
| YJL163C | 0.31 |
Putative protein of unknown function |
||
| YOR140W | 0.30 |
SFL1
|
Transcriptional repressor and activator; involved in repression of flocculation-related genes, and activation of stress responsive genes; negatively regulated by cAMP-dependent protein kinase A subunit Tpk2p |
|
| YBL074C | 0.30 |
AAR2
|
Component of the U5 snRNP, required for splicing of U3 precursors; originally described as a splicing factor specifically required for splicing pre-mRNA of the MATa1 cistron |
|
| YGL013C | 0.30 |
PDR1
|
Zinc cluster protein that is a master regulator involved in recruiting other zinc cluster proteins to pleiotropic drug response elements (PDREs) to fine tune the regulation of multidrug resistance genes |
|
| YJL019W | 0.30 |
MPS3
|
Essential integral membrane protein required for spindle pole body duplication and for nuclear fusion, localizes to the spindle pole body half bridge, interacts with DnaJ-like chaperone Jem1p and with centrin homolog Cdc31p |
|
| YOR192C | 0.30 |
THI72
|
Transporter of thiamine or related compound; shares sequence similarity with Thi7p |
|
| YFL024C | 0.30 |
EPL1
|
Component of NuA4, which is an essential histone H4/H2A acetyltransferase complex; homologous to Drosophila Enhancer of Polycomb |
|
| YMR014W | 0.30 |
BUD22
|
Protein involved in bud-site selection; diploid mutants display a random budding pattern instead of the wild-type bipolar pattern |
|
| YGR286C | 0.30 |
BIO2
|
Biotin synthase, catalyzes the conversion of dethiobiotin to biotin, which is the last step of the biotin biosynthesis pathway; complements E. coli bioB mutant |
|
| YJL037W | 0.30 |
IRC18
|
Putative protein of unknown function; expression induced in respiratory-deficient cells and in carbon-limited chemostat cultures; similar to adjacent ORF, YJL038C; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YKR100C | 0.29 |
SKG1
|
Transmembrane protein with a role in cell wall polymer composition; localizes on the inner surface of the plasma membrane at the bud and in the daughter cell |
|
| YMR016C | 0.29 |
SOK2
|
Nuclear protein that plays a regulatory role in the cyclic AMP (cAMP)-dependent protein kinase (PKA) signal transduction pathway; negatively regulates pseudohyphal differentiation; homologous to several transcription factors |
|
| YMR240C | 0.29 |
CUS1
|
Protein required for assembly of U2 snRNP into the spliceosome, forms a complex with Hsh49p and Hsh155p |
|
| YOL034W | 0.29 |
SMC5
|
Structural maintenance of chromosomes (SMC) protein; essential subunit of the Mms21-Smc5-Smc6 complex; required for growth and DNA repair; S. pombe homolog forms a heterodimer with S. pombe Rad18p that is involved in DNA repair |
|
| YHR096C | 0.29 |
HXT5
|
Hexose transporter with moderate affinity for glucose, induced in the presence of non-fermentable carbon sources, induced by a decrease in growth rate, contains an extended N-terminal domain relative to other HXTs |
|
| YPL213W | 0.29 |
LEA1
|
Component of U2 snRNP; disruption causes reduced U2 snRNP levels; physically interacts with Msl1p; invovled in telomere maintenance; putative homolog of human U2A' snRNP protein |
|
| YBR059C | 0.29 |
AKL1
|
Ser-Thr protein kinase, member (with Ark1p and Prk1p) of the Ark kinase family; involved in endocytosis and actin cytoskeleton organization |
|
| YLR007W | 0.29 |
NSE1
|
Essential subunit of the Mms21-Smc5-Smc6 complex; nuclear protein required for DNA repair and growth; has a nonstructural role in the maintenance of chromosomes |
|
| YIL119C | 0.29 |
RPI1
|
Putative transcriptional regulator; overexpression suppresses the heat shock sensitivity of wild-type RAS2 overexpression and also suppresses the cell lysis defect of an mpk1 mutation |
|
| YMR174C | 0.28 |
PAI3
|
Cytoplasmic proteinase A (Pep4p) inhibitor, dependent on Pbs2p and Hog1p protein kinases for osmotic induction; intrinsically unstructured, N-terminal half becomes ordered in the active site of proteinase A upon contact |
|
| YKR011C | 0.28 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus |
||
| YDR113C | 0.28 |
PDS1
|
Securin that inhibits anaphase by binding separin Esp1p, also blocks cyclin destruction and mitotic exit, essential for cell cycle arrest in mitosis in the presence of DNA damage or aberrant mitotic spindles; also present in meiotic nuclei |
|
| YCR019W | 0.28 |
MAK32
|
Protein necessary for structural stability of L-A double-stranded RNA-containing particles |
|
| YOR139C | 0.28 |
Hypothetical protein |
||
| YDR043C | 0.28 |
NRG1
|
Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters; mediates glucose repression and negatively regulates a variety of processes including filamentous growth and alkaline pH response |
|
| YHR075C | 0.27 |
PPE1
|
Protein with carboxyl methyl esterase activity that may have a role in demethylation of the phosphoprotein phosphatase catalytic subunit; also identified as a small subunit mitochondrial ribosomal protein |
|
| YKL163W | 0.27 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YLR136C | 0.27 |
TIS11
|
mRNA-binding protein expressed during iron starvation; binds to a sequence element in the 3'-untranslated regions of specific mRNAs to mediate their degradation; involved in iron homeostasis |
|
| YKL038W | 0.27 |
RGT1
|
Glucose-responsive transcription factor that regulates expression of several glucose transporter (HXT) genes in response to glucose; binds to promoters and acts both as a transcriptional activator and repressor |
|
| YJL038C | 0.27 |
LOH1
|
Putative protein of unknown function; expression induced during sporulation and repressed during vegetative growth by Sum1p and Hst1p; similar to adjacent open reading frame, YJL037W |
|
| YCR021C | 0.27 |
HSP30
|
Hydrophobic plasma membrane localized, stress-responsive protein that negatively regulates the H(+)-ATPase Pma1p; induced by heat shock, ethanol treatment, weak organic acid, glucose limitation, and entry into stationary phase |
|
| YDR255C | 0.26 |
RMD5
|
Cytosolic protein required for sporulation; also required for the ubiquitination of the gluconeogenetic enzyme fructose-1,6-bisphosphatase, which is degraded rapidly after the switch from gluconeogenesis to glycolysis |
|
| YML035C | 0.26 |
AMD1
|
AMP deaminase, tetrameric enzyme that catalyzes the deamination of AMP to form IMP and ammonia; may be involved in regulation of intracellular adenine nucleotide pools |
|
| YDR441C | 0.25 |
APT2
|
Apparent pseudogene, not transcribed or translated under normal conditions; encodes a protein with similarity to adenine phosphoribosyltransferase, but artificially expressed protein exhibits no enzymatic activity |
|
| YBL101C | 0.25 |
ECM21
|
Non-essential protein of unknown function; promoter contains several Gcn4p binding elements |
|
| YJR038C | 0.25 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER096W | 0.24 |
SHC1
|
Sporulation-specific activator of Chs3p (chitin synthase III), required for the synthesis of the chitosan layer of ascospores; has similarity to Skt5p, which activates Chs3p during vegetative growth; transcriptionally induced at alkaline pH |
|
| YKL101W | 0.24 |
HSL1
|
Nim1p-related protein kinase that regulates the morphogenesis and septin checkpoints; associates with the assembled septin filament; required along with Hsl7p for bud neck recruitment, phosphorylation, and degradation of Swe1p |
|
| YKL097C | 0.24 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species |
||
| YDR231C | 0.24 |
COX20
|
Mitochondrial inner membrane protein, required for proteolytic processing of Cox2p and its assembly into cytochrome c oxidase |
|
| YIL102C-A | 0.24 |
Putative protein of unknown function, identified based on comparisons of the genome sequences of six Saccharomyces species |
||
| YHR071W | 0.24 |
PCL5
|
Cyclin, interacts with Pho85p cyclin-dependent kinase (Cdk), induced by Gcn4p at level of transcription, specifically required for Gcn4p degradation, may be sensor of cellular protein biosynthetic capacity |
|
| YHR080C | 0.24 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YOR192C-C | 0.24 |
Putative protein of unknown function; identified by expression profiling and mass spectrometry |
||
| YER166W | 0.24 |
DNF1
|
Aminophospholipid translocase (flippase) that localizes primarily to the plasma membrane; contributes to endocytosis, protein transport and cell polarity; type 4 P-type ATPase |
|
| YKR102W | 0.24 |
FLO10
|
Lectin-like protein with similarity to Flo1p, thought to be involved in flocculation |
|
| YMR306W | 0.24 |
FKS3
|
Protein involved in spore wall assembly, has similarity to 1,3-beta-D-glucan synthase catalytic subunits Fks1p and Gsc2p; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YLR164W | 0.24 |
Mitochondrial inner membrane of unknown function; similar to Tim18p and Sdh4p; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
||
| YPR169W-A | 0.24 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps two other dubious ORFs: YPR170C and YPR170W-B |
||
| YJL220W | 0.23 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YJL221C/FSP2 |
||
| YPL189W | 0.23 |
GUP2
|
Probable membrane protein with a possible role in proton symport of glycerol; member of the MBOAT family of putative membrane-bound O-acyltransferases; Gup1p homolog |
|
| YEL049W | 0.23 |
PAU2
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YNL025C | 0.23 |
SSN8
|
Cyclin-like component of the RNA polymerase II holoenzyme, involved in phosphorylation of the RNA polymerase II C-terminal domain; involved in glucose repression and telomere maintenance |
|
| YDR362C | 0.23 |
TFC6
|
One of six subunits of RNA polymerase III transcription initiation factor complex (TFIIIC); part of TFIIIC TauB domain that binds BoxB promoter sites of tRNA and other genes; cooperates with Tfc3p in DNA binding; human homolog is TFIIIC-110 |
|
| YFR020W | 0.23 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR296W | 0.22 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YNL076W | 0.22 |
MKS1
|
Pleiotropic negative transcriptional regulator involved in Ras-CAMP and lysine biosynthetic pathways and nitrogen regulation; involved in retrograde (RTG) mitochondria-to-nucleus signaling |
|
| YKL065W-A | 0.22 |
Putative protein of unknown function |
||
| YER016W | 0.22 |
BIM1
|
Microtubule-binding protein that together with Kar9p makes up the cortical microtubule capture site and delays the exit from mitosis when the spindle is oriented abnormally |
|
| YBR208C | 0.22 |
DUR1,2
|
Urea amidolyase, contains both urea carboxylase and allophanate hydrolase activities, degrades urea to CO2 and NH3; expression sensitive to nitrogen catabolite repression and induced by allophanate, an intermediate in allantoin degradation |
|
| YFL023W | 0.22 |
BUD27
|
Protein involved in bud-site selection, nutrient signaling, and gene expression controlled by TOR kinase; diploid mutants show a random budding pattern rather than the wild-type bipolar pattern; plays a role in regulating Ty1 transposition |
|
| YDR406W | 0.22 |
PDR15
|
Plasma membrane ATP binding cassette (ABC) transporter, multidrug transporter and general stress response factor implicated in cellular detoxification; regulated by Pdr1p, Pdr3p and Pdr8p; promoter contains a PDR responsive element |
|
| YFL064C | 0.22 |
Putative protein of unknown function |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 0.4 | 1.3 | GO:0036257 | multivesicular body organization(GO:0036257) |
| 0.4 | 1.2 | GO:0000304 | response to singlet oxygen(GO:0000304) |
| 0.3 | 1.5 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) |
| 0.3 | 0.3 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.3 | 1.2 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.2 | 0.5 | GO:0009415 | response to water(GO:0009415) cellular response to water stimulus(GO:0071462) |
| 0.2 | 2.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 1.2 | GO:0046464 | triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.2 | 1.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 1.0 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.2 | 1.1 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.2 | 0.8 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.2 | 1.3 | GO:0071472 | cellular response to salt stress(GO:0071472) |
| 0.2 | 0.8 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.2 | 0.6 | GO:0009450 | gamma-aminobutyric acid metabolic process(GO:0009448) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.2 | 1.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 0.6 | GO:0034503 | protein localization to nucleolar rDNA repeats(GO:0034503) |
| 0.2 | 0.2 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) |
| 0.2 | 0.3 | GO:2001023 | cellular response to drug(GO:0035690) regulation of response to drug(GO:2001023) positive regulation of response to drug(GO:2001025) regulation of cellular response to drug(GO:2001038) positive regulation of cellular response to drug(GO:2001040) |
| 0.2 | 0.6 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 0.4 | GO:0018209 | peptidyl-serine phosphorylation(GO:0018105) peptidyl-serine modification(GO:0018209) |
| 0.1 | 0.6 | GO:0035753 | maintenance of DNA trinucleotide repeats(GO:0035753) |
| 0.1 | 1.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) regulation of transcription from RNA polymerase II promoter in response to zinc ion starvation(GO:0034225) |
| 0.1 | 0.6 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.5 | GO:0042357 | thiamine diphosphate biosynthetic process(GO:0009229) thiamine diphosphate metabolic process(GO:0042357) |
| 0.1 | 0.3 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 0.3 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.2 | GO:0043419 | urea catabolic process(GO:0043419) |
| 0.1 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.7 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) manganese ion homeostasis(GO:0055071) |
| 0.1 | 1.6 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.6 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 0.3 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.3 | GO:0009730 | detection of chemical stimulus(GO:0009593) detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) detection of stimulus(GO:0051606) |
| 0.1 | 0.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 0.1 | 0.2 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.1 | 1.0 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.1 | 0.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.4 | GO:0043388 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.1 | 0.4 | GO:0046323 | glucose import(GO:0046323) |
| 0.1 | 0.4 | GO:0032105 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.1 | 0.6 | GO:0006771 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.1 | 1.3 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 0.1 | 0.4 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.3 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.1 | 0.3 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.5 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.1 | 0.4 | GO:0043007 | maintenance of rDNA(GO:0043007) |
| 0.1 | 0.6 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.1 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.3 | GO:0045141 | meiotic telomere clustering(GO:0045141) establishment of chromosome localization(GO:0051303) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.1 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.2 | GO:0090527 | actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.1 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 0.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 0.4 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) |
| 0.1 | 0.8 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.2 | GO:0006343 | establishment of chromatin silencing(GO:0006343) |
| 0.1 | 0.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.3 | GO:0000078 | obsolete cytokinesis after mitosis checkpoint(GO:0000078) |
| 0.1 | 0.1 | GO:0042710 | biofilm formation(GO:0042710) |
| 0.1 | 0.4 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.1 | 0.3 | GO:0032048 | cardiolipin metabolic process(GO:0032048) |
| 0.1 | 0.2 | GO:0010994 | free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 0.2 | GO:0055070 | cellular copper ion homeostasis(GO:0006878) copper ion homeostasis(GO:0055070) |
| 0.1 | 0.2 | GO:0043901 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.1 | 0.1 | GO:0061414 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 0.1 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.2 | GO:0006003 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.1 | 0.1 | GO:2000058 | regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.1 | 0.2 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.1 | 0.2 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.1 | 0.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.3 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 0.4 | GO:0009749 | response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.1 | 0.2 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.1 | 0.1 | GO:0051099 | positive regulation of binding(GO:0051099) |
| 0.0 | 0.3 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.0 | 0.0 | GO:0019627 | urea metabolic process(GO:0019627) |
| 0.0 | 1.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.0 | 0.2 | GO:0071041 | antisense RNA transcript catabolic process(GO:0071041) |
| 0.0 | 0.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.2 | GO:0031930 | mitochondria-nucleus signaling pathway(GO:0031930) |
| 0.0 | 0.2 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.0 | 0.3 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.2 | GO:0032845 | negative regulation of telomere maintenance(GO:0032205) negative regulation of homeostatic process(GO:0032845) |
| 0.0 | 0.2 | GO:0048209 | regulation of COPII vesicle coating(GO:0003400) regulation of vesicle targeting, to, from or within Golgi(GO:0048209) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.3 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.5 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.0 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.0 | 0.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0009065 | glutamine family amino acid catabolic process(GO:0009065) |
| 0.0 | 0.3 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.0 | GO:1903313 | positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.0 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0000092 | mitotic anaphase B(GO:0000092) |
| 0.0 | 0.1 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.0 | 0.2 | GO:0000411 | regulation of transcription by galactose(GO:0000409) positive regulation of transcription by galactose(GO:0000411) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) |
| 0.0 | 0.5 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.0 | 0.1 | GO:0018216 | peptidyl-arginine modification(GO:0018195) peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.4 | GO:0043086 | negative regulation of catalytic activity(GO:0043086) |
| 0.0 | 0.2 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.3 | GO:0045910 | negative regulation of DNA recombination(GO:0045910) |
| 0.0 | 0.1 | GO:0043970 | regulation of transcriptional start site selection at RNA polymerase II promoter(GO:0001178) histone H3 acetylation(GO:0043966) histone H3-K9 acetylation(GO:0043970) histone H3-K14 acetylation(GO:0044154) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.5 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.1 | GO:0070542 | response to lipid(GO:0033993) response to oleic acid(GO:0034201) response to fatty acid(GO:0070542) cellular response to lipid(GO:0071396) cellular response to fatty acid(GO:0071398) cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0009967 | positive regulation of signal transduction(GO:0009967) positive regulation of cell communication(GO:0010647) positive regulation of signaling(GO:0023056) |
| 0.0 | 0.1 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0001009 | transcription from RNA polymerase III type 2 promoter(GO:0001009) transcription from a RNA polymerase III hybrid type promoter(GO:0001041) |
| 0.0 | 0.0 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 0.0 | GO:0001324 | age-dependent general metabolic decline involved in chronological cell aging(GO:0001323) age-dependent response to oxidative stress involved in chronological cell aging(GO:0001324) |
| 0.0 | 0.1 | GO:0031134 | sister chromatid biorientation(GO:0031134) |
| 0.0 | 0.3 | GO:0046854 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0006529 | asparagine metabolic process(GO:0006528) asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.1 | GO:2000284 | positive regulation of cellular amino acid metabolic process(GO:0045764) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.3 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.0 | 0.0 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.0 | 0.4 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:0007534 | gene conversion at mating-type locus(GO:0007534) |
| 0.0 | 0.0 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.3 | GO:0000767 | cell morphogenesis involved in conjugation(GO:0000767) |
| 0.0 | 0.1 | GO:0046503 | glycerolipid catabolic process(GO:0046503) |
| 0.0 | 0.1 | GO:0032106 | positive regulation of response to external stimulus(GO:0032103) positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.0 | 0.1 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.2 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 0.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.6 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.0 | GO:1902932 | positive regulation of alcohol biosynthetic process(GO:1902932) |
| 0.0 | 0.1 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:0070058 | tRNA gene clustering(GO:0070058) |
| 0.0 | 0.2 | GO:0045991 | carbon catabolite activation of transcription(GO:0045991) |
| 0.0 | 0.2 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.0 | GO:0055075 | cellular potassium ion homeostasis(GO:0030007) potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.1 | GO:0019935 | cAMP-mediated signaling(GO:0019933) cyclic-nucleotide-mediated signaling(GO:0019935) |
| 0.0 | 0.2 | GO:0009228 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.0 | 0.4 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.2 | GO:0006109 | regulation of carbohydrate metabolic process(GO:0006109) |
| 0.0 | 0.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.0 | GO:0061392 | regulation of transcription from RNA polymerase II promoter in response to osmotic stress(GO:0061392) |
| 0.0 | 0.1 | GO:0006078 | (1->6)-beta-D-glucan biosynthetic process(GO:0006078) beta-glucan biosynthetic process(GO:0051274) |
| 0.0 | 0.1 | GO:0032365 | intracellular lipid transport(GO:0032365) |
| 0.0 | 0.1 | GO:0051597 | response to methylmercury(GO:0051597) cellular response to methylmercury(GO:0071406) |
| 0.0 | 0.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.2 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) re-entry into mitotic cell cycle after pheromone arrest(GO:0000321) |
| 0.0 | 0.2 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.4 | GO:0007124 | pseudohyphal growth(GO:0007124) |
| 0.0 | 0.6 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 0.0 | GO:0000161 | MAPK cascade involved in osmosensory signaling pathway(GO:0000161) |
| 0.0 | 0.0 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.4 | GO:0000725 | recombinational repair(GO:0000725) |
| 0.0 | 0.7 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0016562 | receptor recycling(GO:0001881) protein import into peroxisome matrix, receptor recycling(GO:0016562) receptor metabolic process(GO:0043112) |
| 0.0 | 0.0 | GO:0072367 | positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) positive regulation of sterol transport(GO:0032373) positive regulation of transmembrane transport(GO:0034764) regulation of sterol import by regulation of transcription from RNA polymerase II promoter(GO:0035968) positive regulation of sterol import by positive regulation of transcription from RNA polymerase II promoter(GO:0035969) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.0 | 0.0 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.0 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.0 | 0.2 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.0 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0031070 | intronic snoRNA processing(GO:0031070) intronic box C/D snoRNA processing(GO:0034965) |
| 0.0 | 0.1 | GO:0001101 | response to acid chemical(GO:0001101) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 1.5 | GO:0032126 | eisosome(GO:0032126) |
| 0.2 | 0.9 | GO:0032865 | ERMES complex(GO:0032865) ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 1.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 0.6 | GO:0033551 | monopolin complex(GO:0033551) |
| 0.1 | 0.4 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.1 | 0.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.3 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.1 | 0.7 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.6 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.3 | GO:0005825 | half bridge of spindle pole body(GO:0005825) |
| 0.1 | 0.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.3 | GO:0001400 | mating projection base(GO:0001400) |
| 0.1 | 0.2 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.1 | 1.0 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.6 | GO:0005619 | ascospore wall(GO:0005619) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0032806 | carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 0.1 | GO:0009353 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial oxoglutarate dehydrogenase complex(GO:0009353) dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0033309 | SBF transcription complex(GO:0033309) |
| 0.0 | 0.9 | GO:0042764 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.0 | 0.3 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.0 | 0.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.6 | GO:0044438 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.1 | GO:0031499 | TRAMP complex(GO:0031499) |
| 0.0 | 0.2 | GO:0005940 | septin ring(GO:0005940) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.0 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0045263 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.0 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.0 | 0.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.0 | GO:0032161 | cellular bud neck septin structure(GO:0000399) cleavage apparatus septin structure(GO:0032161) split septin rings(GO:0032176) cellular bud neck split septin rings(GO:0032177) |
| 0.0 | 0.0 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.1 | GO:0000446 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) nucleoplasmic THO complex(GO:0000446) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.0 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.0 | 0.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.0 | GO:0000417 | HIR complex(GO:0000417) |
| 0.0 | 0.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0050253 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.5 | 1.8 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.4 | 1.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.3 | 0.9 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.2 | 1.2 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.2 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 0.9 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 0.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 0.8 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.2 | 0.2 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.2 | 0.5 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.2 | 0.8 | GO:0097372 | NAD-dependent histone deacetylase activity (H3-K18 specific)(GO:0097372) |
| 0.1 | 0.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.1 | 0.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 1.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.4 | GO:0005186 | mating pheromone activity(GO:0000772) pheromone activity(GO:0005186) |
| 0.1 | 0.3 | GO:0005536 | glucose binding(GO:0005536) |
| 0.1 | 0.3 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 0.1 | 0.3 | GO:0008972 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.1 | 0.5 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 0.1 | 0.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.6 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.2 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.1 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 0.8 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.1 | 0.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.2 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.1 | 0.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 0.2 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 0.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.9 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.1 | GO:0042736 | NAD+ kinase activity(GO:0003951) NADH kinase activity(GO:0042736) |
| 0.0 | 0.4 | GO:0004601 | peroxidase activity(GO:0004601) oxidoreductase activity, acting on peroxide as acceptor(GO:0016684) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 0.0 | 0.0 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 1.1 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.3 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.4 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 2.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 3.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 0.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0001067 | regulatory region nucleic acid binding(GO:0001067) |
| 0.0 | 0.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.2 | GO:0016723 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.5 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.1 | GO:0052742 | phosphatidylinositol kinase activity(GO:0052742) |
| 0.0 | 0.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.7 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.1 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0004872 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
| 0.0 | 0.7 | GO:0000988 | transcription factor activity, protein binding(GO:0000988) |
| 0.0 | 0.3 | GO:0061733 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.1 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 0.0 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 0.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.5 | GO:0019899 | enzyme binding(GO:0019899) |
| 0.0 | 0.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.1 | REACTOME DNA REPAIR | Genes involved in DNA Repair |