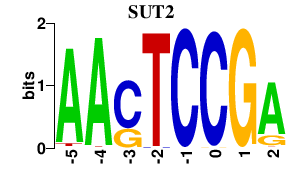
Results for SUT2
Z-value: 0.52
Transcription factors associated with SUT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SUT2
|
S000006213 | Putative transcription factor |
Activity-expression correlation:
Activity profile of SUT2 motif
Sorted Z-values of SUT2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YGL258W-A | 2.07 |
Putative protein of unknown function |
||
| YLR174W | 1.96 |
IDP2
|
Cytosolic NADP-specific isocitrate dehydrogenase, catalyzes oxidation of isocitrate to alpha-ketoglutarate; levels are elevated during growth on non-fermentable carbon sources and reduced during growth on glucose |
|
| YJR095W | 1.84 |
SFC1
|
Mitochondrial succinate-fumarate transporter, transports succinate into and fumarate out of the mitochondrion; required for ethanol and acetate utilization |
|
| YLR377C | 1.84 |
FBP1
|
Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway, required for glucose metabolism |
|
| YER065C | 1.68 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YFR053C | 1.67 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YNR002C | 1.50 |
ATO2
|
Putative transmembrane protein involved in export of ammonia, a starvation signal that promotes cell death in aging colonies; phosphorylated in mitochondria; member of the TC 9.B.33 YaaH family; homolog of Ady2p and Y. lipolytica Gpr1p |
|
| YBR072W | 1.36 |
HSP26
|
Small heat shock protein (sHSP) with chaperone activity; forms hollow, sphere-shaped oligomers that suppress unfolded proteins aggregation; oligomer activation requires a heat-induced conformational change; not expressed in unstressed cells |
|
| YER066W | 1.33 |
Putative protein of unknown function; YER066W is not an essential gene |
||
| YOR049C | 1.32 |
RSB1
|
Suppressor of sphingoid long chain base (LCB) sensitivity of an LCB-lyase mutation; putative integral membrane transporter or flippase that may transport LCBs from the cytoplasmic side toward the extracytoplasmic side of the membrane |
|
| YIR039C | 1.27 |
YPS6
|
Putative GPI-anchored aspartic protease |
|
| YPL223C | 1.23 |
GRE1
|
Hydrophilin of unknown function; stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YFL030W | 1.20 |
AGX1
|
Alanine:glyoxylate aminotransferase (AGT), catalyzes the synthesis of glycine from glyoxylate, which is one of three pathways for glycine biosynthesis in yeast; has similarity to mammalian and plant alanine:glyoxylate aminotransferases |
|
| YMR206W | 1.16 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YDL215C | 1.07 |
GDH2
|
NAD(+)-dependent glutamate dehydrogenase, degrades glutamate to ammonia and alpha-ketoglutarate; expression sensitive to nitrogen catabolite repression and intracellular ammonia levels |
|
| YBL015W | 1.05 |
ACH1
|
Acetyl-coA hydrolase, primarily localized to mitochondria; phosphorylated; required for acetate utilization and for diploid pseudohyphal growth |
|
| YGL162W | 1.04 |
SUT1
|
Transcription factor of the Zn[II]2Cys6 family involved in sterol uptake; involved in induction of hypoxic gene expression |
|
| YLR356W | 1.00 |
Putative protein of unknown function with similarity to SCM4; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR356W is not an essential gene |
||
| YJL161W | 0.96 |
FMP33
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YML042W | 0.94 |
CAT2
|
Carnitine acetyl-CoA transferase present in both mitochondria and peroxisomes, transfers activated acetyl groups to carnitine to form acetylcarnitine which can be shuttled across membranes |
|
| YEL008W | 0.93 |
Hypothetical protein predicted to be involved in metabolism |
||
| YMR272C | 0.91 |
SCS7
|
Sphingolipid alpha-hydroxylase, functions in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids, has both cytochrome b5-like and hydroxylase/desaturase domains, not essential for growth |
|
| YNL055C | 0.90 |
POR1
|
Mitochondrial porin (voltage-dependent anion channel), outer membrane protein required for the maintenance of mitochondrial osmotic stability and mitochondrial membrane permeability; phosphorylated |
|
| YNL160W | 0.89 |
YGP1
|
Cell wall-related secretory glycoprotein; induced by nutrient deprivation-associated growth arrest and upon entry into stationary phase; may be involved in adaptation prior to stationary phase entry; has similarity to Sps100p |
|
| YFL052W | 0.89 |
Putative zinc cluster protein that contains a DNA binding domain; null mutant sensitive to calcofluor white, low osmolarity and heat, suggesting a role for YFL052Wp in cell wall integrity |
||
| YJL116C | 0.88 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YGL053W | 0.86 |
PRM8
|
Pheromone-regulated protein with 2 predicted transmembrane segments and an FF sequence, a motif involved in COPII binding; forms a complex with Prp9p in the ER; member of DUP240 gene family |
|
| YOR120W | 0.85 |
GCY1
|
Putative NADP(+) coupled glycerol dehydrogenase, proposed to be involved in an alternative pathway for glycerol catabolism; member of the aldo-keto reductase (AKR) family |
|
| YKR097W | 0.84 |
PCK1
|
Phosphoenolpyruvate carboxykinase, key enzyme in gluconeogenesis, catalyzes early reaction in carbohydrate biosynthesis, glucose represses transcription and accelerates mRNA degradation, regulated by Mcm1p and Cat8p, located in the cytosol |
|
| YJL045W | 0.83 |
Minor succinate dehydrogenase isozyme; homologous to Sdh1p, the major isozyme reponsible for the oxidation of succinate and transfer of electrons to ubiquinone; induced during the diauxic shift in a Cat8p-dependent manner |
||
| YKL217W | 0.81 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YJL166W | 0.81 |
QCR8
|
Subunit 8 of ubiquinol cytochrome-c reductase complex, which is a component of the mitochondrial inner membrane electron transport chain; oriented facing the intermembrane space; expression is regulated by Abf1p and Cpf1p |
|
| YGR045C | 0.80 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR036C | 0.79 |
EHD3
|
3-hydroxyisobutyryl-CoA hydrolase, member of a family of enoyl-CoA hydratase/isomerases; non-tagged protein is detected in highly purified mitochondria in high-throughput studies; phosphorylated; mutation affects fluid-phase endocytosis |
|
| YDR277C | 0.75 |
MTH1
|
Negative regulator of the glucose-sensing signal transduction pathway, required for repression of transcription by Rgt1p; interacts with Rgt1p and the Snf3p and Rgt2p glucose sensors; phosphorylated by Yck1p, triggering Mth1p degradation |
|
| YDR343C | 0.75 |
HXT6
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt7p, expressed at high basal levels relative to other HXTs, repression of expression by high glucose requires SNF3 |
|
| YNL200C | 0.74 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YIL125W | 0.70 |
KGD1
|
Component of the mitochondrial alpha-ketoglutarate dehydrogenase complex, which catalyzes a key step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation of alpha-ketoglutarate to form succinyl-CoA |
|
| YPL017C | 0.70 |
IRC15
|
Putative S-adenosylmethionine-dependent methyltransferase of the seven beta-strand family, required for accurate meiotic chromosome segregation; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YMR210W | 0.68 |
Putative acyltransferase with similarity to Eeb1p and Eht1p, has a minor role in medium-chain fatty acid ethyl ester biosynthesis; may be involved in lipid metabolism and detoxification |
||
| YMR008C | 0.67 |
PLB1
|
Phospholipase B (lysophospholipase) involved in lipid metabolism, required for deacylation of phosphatidylcholine and phosphatidylethanolamine but not phosphatidylinositol |
|
| YEL009C | 0.66 |
GCN4
|
Transcriptional activator of amino acid biosynthetic genes in response to amino acid starvation; expression is tightly regulated at both the transcriptional and translational levels |
|
| YHR211W | 0.65 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YAR060C | 0.64 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPL200W | 0.64 |
CSM4
|
Protein required for accurate chromosome segregation during meiosis |
|
| YGR288W | 0.62 |
MAL13
|
MAL-activator protein, part of complex locus MAL1; nonfunctional in genomic reference strain S288C |
|
| YML100W | 0.62 |
TSL1
|
Large subunit of trehalose 6-phosphate synthase (Tps1p)/phosphatase (Tps2p) complex, which converts uridine-5'-diphosphoglucose and glucose 6-phosphate to trehalose, homologous to Tps3p and may share function |
|
| YPL201C | 0.62 |
YIG1
|
Protein that interacts with glycerol 3-phosphatase and plays a role in anaerobic glycerol production; localizes to the nucleus and cytosol |
|
| YKR040C | 0.61 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YKR041W |
||
| YNL274C | 0.60 |
GOR1
|
Glyoxylate reductase; null mutation results in increased biomass after diauxic shift; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YDR031W | 0.59 |
MIC14
|
Mitochondrial intermembrane space cysteine motif protein of 14 kDa |
|
| YPL014W | 0.59 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and to the nucleus |
||
| YNL018C | 0.59 |
Putative protein of unknown function |
||
| YJL127C-B | 0.58 |
Putative protein of unknown function; identified based on homology to the filamentous fungus, Ashbya gossypii |
||
| YPR151C | 0.58 |
SUE1
|
Mitochondrial protein required for degradation of unstable forms of cytochrome c |
|
| YKR102W | 0.57 |
FLO10
|
Lectin-like protein with similarity to Flo1p, thought to be involved in flocculation |
|
| YGR028W | 0.57 |
MSP1
|
Mitochondrial protein involved in sorting of proteins in the mitochondria; putative membrane-spanning ATPase |
|
| YJL093C | 0.56 |
TOK1
|
Outward-rectifier potassium channel of the plasma membrane with two pore domains in tandem, each of which forms a functional channel permeable to potassium; carboxy tail functions to prevent inner gate closures; target of K1 toxin |
|
| YPL222W | 0.56 |
FMP40
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YCR005C | 0.56 |
CIT2
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate, peroxisomal isozyme involved in glyoxylate cycle; expression is controlled by Rtg1p and Rtg2p transcription factors |
|
| YFL024C | 0.55 |
EPL1
|
Component of NuA4, which is an essential histone H4/H2A acetyltransferase complex; homologous to Drosophila Enhancer of Polycomb |
|
| YPR196W | 0.55 |
Putative maltose activator |
||
| YDL015C | 0.54 |
TSC13
|
Enoyl reductase that catalyzes the last step in each cycle of very long chain fatty acid elongation, localizes to the ER, highly enriched in a structure marking nuclear-vacuolar junctions, coimmunoprecipitates with elongases Fen1p and Sur4p |
|
| YKL223W | 0.54 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOL059W | 0.54 |
GPD2
|
NAD-dependent glycerol 3-phosphate dehydrogenase, homolog of Gpd1p, expression is controlled by an oxygen-independent signaling pathway required to regulate metabolism under anoxic conditions; located in cytosol and mitochondria |
|
| YDR216W | 0.52 |
ADR1
|
Carbon source-responsive zinc-finger transcription factor, required for transcription of the glucose-repressed gene ADH2, of peroxisomal protein genes, and of genes required for ethanol, glycerol, and fatty acid utilization |
|
| YLR149C | 0.52 |
Putative protein of unknown function; YLR149C is not an essential gene |
||
| YKL224C | 0.52 |
PAU16
|
Putative protein of unknown function |
|
| YOL126C | 0.51 |
MDH2
|
Cytoplasmic malate dehydrogenase, one of three isozymes that catalyze interconversion of malate and oxaloacetate; involved in the glyoxylate cycle and gluconeogenesis during growth on two-carbon compounds; interacts with Pck1p and Fbp1 |
|
| YPL185W | 0.50 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene UIP4/YPL186C |
||
| YHR212C | 0.50 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPL015C | 0.49 |
HST2
|
Cytoplasmic member of the silencing information regulator 2 (Sir2) family of NAD(+)-dependent protein deacetylases; modulates nucleolar (rDNA) and telomeric silencing; possesses NAD(+)-dependent histone deacetylase activity in vitro |
|
| YNL237W | 0.49 |
YTP1
|
Probable type-III integral membrane protein of unknown function, has regions of similarity to mitochondrial electron transport proteins |
|
| YLR295C | 0.49 |
ATP14
|
Subunit h of the F0 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YJR048W | 0.47 |
CYC1
|
Cytochrome c, isoform 1; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YPL274W | 0.46 |
SAM3
|
High-affinity S-adenosylmethionine permease, required for utilization of S-adenosylmethionine as a sulfur source; has similarity to S-methylmethionine permease Mmp1p |
|
| YGL055W | 0.46 |
OLE1
|
Delta(9) fatty acid desaturase, required for monounsaturated fatty acid synthesis and for normal distribution of mitochondria |
|
| YAR053W | 0.46 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOL001W | 0.46 |
PHO80
|
Cyclin, negatively regulates phosphate metabolism; Pho80p-Pho85p (cyclin-CDK complex) phosphorylates Pho4p and Swi5p; deletion of PHO80 leads to aminoglycoside supersensitivity; truncated form of PHO80 affects vacuole inheritance |
|
| YGR088W | 0.45 |
CTT1
|
Cytosolic catalase T, has a role in protection from oxidative damage by hydrogen peroxide |
|
| YOL060C | 0.45 |
MAM3
|
Protein required for normal mitochondrial morphology, has similarity to hemolysins |
|
| YHR212W-A | 0.45 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YOR343C | 0.45 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YHR092C | 0.44 |
HXT4
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YHR210C | 0.44 |
Putative protein of unknown function; non-essential gene; highly expressed under anaeorbic conditions; sequence similarity to aldose 1-epimerases such as GAL10 |
||
| YPL186C | 0.43 |
UIP4
|
Protein that interacts with Ulp1p, a Ubl (ubiquitin-like protein)-specific protease for Smt3p protein conjugates; detected in a phosphorylated state in the mitochondrial outer membrane; also detected in ER and nuclear envelope |
|
| YML081C-A | 0.43 |
ATP18
|
Subunit of the mitochondrial F1F0 ATP synthase, which is a large enzyme complex required for ATP synthesis; termed subunit I or subunit j; does not correspond to known ATP synthase subunits in other organisms |
|
| YGR043C | 0.43 |
NQM1
|
Protein of unknown function; transcription is repressed by Mot1p and induced by alpha-factor and during diauxic shift; null mutant non-quiescent cells exhibit reduced reproductive capacity |
|
| YBL074C | 0.42 |
AAR2
|
Component of the U5 snRNP, required for splicing of U3 precursors; originally described as a splicing factor specifically required for splicing pre-mRNA of the MATa1 cistron |
|
| YKL004W | 0.42 |
AUR1
|
Phosphatidylinositol:ceramide phosphoinositol transferase (IPC synthase), required for sphingolipid synthesis; can mutate to confer aureobasidin A resistance |
|
| YLR296W | 0.41 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YJL213W | 0.41 |
Protein of unknown function that may interact with ribosomes; periodically expressed during the yeast metabolic cycle; phosphorylated in vitro by the mitotic exit network (MEN) kinase complex, Dbf2p/Mob1p |
||
| YGR087C | 0.41 |
PDC6
|
Minor isoform of pyruvate decarboxylase, decarboxylates pyruvate to acetaldehyde, involved in amino acid catabolism; transcription is glucose- and ethanol-dependent, and is strongly induced during sulfur limitation |
|
| YDR032C | 0.41 |
PST2
|
Protein with similarity to members of a family of flavodoxin-like proteins; induced by oxidative stress in a Yap1p dependent manner; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YPR023C | 0.41 |
EAF3
|
Esa1p-associated factor, nonessential component of the NuA4 acetyltransferase complex, homologous to Drosophila dosage compensation protein MSL3; plays a role in regulating Ty1 transposition |
|
| YKL216W | 0.40 |
URA1
|
Dihydroorotate dehydrogenase, catalyzes the fourth enzymatic step in the de novo biosynthesis of pyrimidines, converting dihydroorotic acid into orotic acid |
|
| YOL151W | 0.39 |
GRE2
|
3-methylbutanal reductase and NADPH-dependent methylglyoxal reductase (D-lactaldehyde dehydrogenase); stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YMR110C | 0.39 |
HFD1
|
Putative fatty aldehyde dehydrogenase, located in the mitochondrial outer membrane and also in lipid particles; has similarity to human fatty aldehyde dehydrogenase (FALDH) which is implicated in Sjogren-Larsson syndrome |
|
| YBL073W | 0.39 |
Hypothetical protein; open reading frame overlaps 5' end of essential AAR2 gene encoding a component of the U5 snRNP required for splicing of U3 precursors |
||
| YPR030W | 0.38 |
CSR2
|
Nuclear protein with a potential regulatory role in utilization of galactose and nonfermentable carbon sources; overproduction suppresses the lethality at high temperature of a chs5 spa2 double null mutation; potential Cdc28p substrate |
|
| YDR022C | 0.38 |
CIS1
|
Autophagy-specific protein required for autophagosome formation; may form a complex with Atg17p and Atg29p that localizes other proteins to the pre-autophagosomal structure; high-copy suppressor of CIK1 deletion |
|
| YLR350W | 0.38 |
ORM2
|
Evolutionarily conserved protein with similarity to Orm1p, required for resistance to agents that induce the unfolded protein response; human ortholog is located in the endoplasmic reticulum |
|
| YGR250C | 0.38 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YFL051C | 0.38 |
Putative protein of unknown function; YFL051C is not an essential gene |
||
| YMR107W | 0.38 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YMR081C | 0.37 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YOR378W | 0.37 |
Putative protein of unknown function; belongs to the DHA2 family of drug:H+ antiporters; YOR378W is not an essential gene |
||
| YDR030C | 0.37 |
RAD28
|
Protein involved in DNA repair, related to the human CSA protein that is involved in transcription-coupled repair nucleotide excision repair |
|
| YJL152W | 0.36 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPL171C | 0.36 |
OYE3
|
Widely conserved NADPH oxidoreductase containing flavin mononucleotide (FMN), homologous to Oye2p with slight differences in ligand binding and catalytic properties; may be involved in sterol metabolism |
|
| YNR071C | 0.36 |
Putative protein of unknown function |
||
| YOR152C | 0.35 |
Putative protein of unknown function; has no similarity to any known protein; YOR152C is not an essential gene |
||
| YOR228C | 0.35 |
Protein of unknown function, localized to the mitochondrial outer membrane |
||
| YHR139C-A | 0.35 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBR223C | 0.35 |
TDP1
|
Tyrosyl-DNA Phosphodiesterase I, hydrolyzes 3'-phosphotyrosyl bonds to generate 3'-phosphate DNA and tyrosine, involved in the repair of DNA lesions created by topoisomerase I |
|
| YJR151C | 0.35 |
DAN4
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YDL222C | 0.34 |
FMP45
|
Integral membrane protein localized to mitochondria (untagged protein) and eisosomes, immobile patches at the cortex associated with endocytosis; sporulation and sphingolipid content are altered in mutants; has homologs SUR7 and YNL194C |
|
| YIL102C | 0.34 |
Putative protein of unknown function |
||
| YPR024W | 0.34 |
YME1
|
Subunit, with Mgr1p, of the mitochondrial inner membrane i-AAA protease complex, which is responsible for degradation of unfolded or misfolded mitochondrial gene products; mutation causes an elevated rate of mitochondrial turnover |
|
| YMR148W | 0.33 |
Putative protein of unknown function; predicted to contain a transmembrane domain; YMR148W is not an essential gene |
||
| YBR116C | 0.33 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene TKL2 |
||
| YHR139C | 0.33 |
SPS100
|
Protein required for spore wall maturation; expressed during sporulation; may be a component of the spore wall |
|
| YOR387C | 0.32 |
Putative protein of unknown function; regulated by the metal-responsive Aft1p transcription factor; highly inducible in zinc-depleted conditions; localizes to the soluble fraction |
||
| YKL038W | 0.32 |
RGT1
|
Glucose-responsive transcription factor that regulates expression of several glucose transporter (HXT) genes in response to glucose; binds to promoters and acts both as a transcriptional activator and repressor |
|
| YPR007C | 0.32 |
REC8
|
Meiosis-specific component of sister chromatid cohesion complex; maintains cohesion between sister chromatids during meiosis I; maintains cohesion between centromeres of sister chromatids until meiosis II; homolog of S. pombe Rec8p |
|
| YLR331C | 0.31 |
JIP3
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 98% of ORF overlaps the verified gene MID2 |
|
| YML004C | 0.31 |
GLO1
|
Monomeric glyoxalase I, catalyzes the detoxification of methylglyoxal (a by-product of glycolysis) via condensation with glutathione to produce S-D-lactoylglutathione; expression regulated by methylglyoxal levels and osmotic stress |
|
| YDL234C | 0.30 |
GYP7
|
GTPase-activating protein for yeast Rab family members including: Ypt7p (most effective), Ypt1p, Ypt31p, and Ypt32p (in vitro); involved in vesicle mediated protein trafficking |
|
| YOL084W | 0.30 |
PHM7
|
Protein of unknown function, expression is regulated by phosphate levels; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery and vacuole |
|
| YPR001W | 0.30 |
CIT3
|
Dual specificity mitochondrial citrate and methylcitrate synthase; catalyzes the condensation of acetyl-CoA and oxaloacetate to form citrate and that of propionyl-CoA and oxaloacetate to form 2-methylcitrate |
|
| YJR020W | 0.30 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YIL162W | 0.30 |
SUC2
|
Invertase, sucrose hydrolyzing enzyme; a secreted, glycosylated form is regulated by glucose repression, and an intracellular, nonglycosylated enzyme is produced constitutively |
|
| YHL024W | 0.30 |
RIM4
|
Putative RNA-binding protein required for the expression of early and middle sporulation genes |
|
| YMR250W | 0.30 |
GAD1
|
Glutamate decarboxylase, converts glutamate into gamma-aminobutyric acid (GABA) during glutamate catabolism; involved in response to oxidative stress |
|
| YIL120W | 0.29 |
QDR1
|
Multidrug transporter of the major facilitator superfamily, required for resistance to quinidine, ketoconazole, fluconazole, and barban |
|
| YGR144W | 0.29 |
THI4
|
Thiazole synthase, catalyzes formation of the thiazole moiety of thiamin pyrophosphate; required for thiamine biosynthesis and for mitochondrial genome stability |
|
| YMR082C | 0.29 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNL034W | 0.29 |
Putative protein of unknown function; YNL034W is not an essential gene |
||
| YBL017C | 0.29 |
PEP1
|
Type I transmembrane sorting receptor for multiple vacuolar hydrolases; cycles between the late-Golgi and prevacuolar endosome-like compartments |
|
| YEL052W | 0.29 |
AFG1
|
Conserved protein that may act as a chaperone in the degradation of misfolded or unassembled cytochrome c oxidase subunits; localized to matrix face of the mitochondrial inner membrane; member of the AAA family but lacks a protease domain |
|
| YBR203W | 0.29 |
COS111
|
Protein required for resistance to the antifungal drug ciclopirox olamine; not related to the subtelomerically-encoded COS family; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YLR332W | 0.29 |
MID2
|
O-glycosylated plasma membrane protein that acts as a sensor for cell wall integrity signaling and activates the pathway; interacts with Rom2p, a guanine nucleotide exchange factor for Rho1p, and with cell integrity pathway protein Zeo1p |
|
| YOR317W | 0.29 |
FAA1
|
Long chain fatty acyl-CoA synthetase with a preference for C12:0-C16:0 fatty acids; involved in the activation of imported fatty acids; localized to both lipid particles and mitochondrial outer membrane; essential for stationary phase |
|
| YLR297W | 0.29 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YLR297W is not an essential gene; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YNL202W | 0.28 |
SPS19
|
Peroxisomal 2,4-dienoyl-CoA reductase, auxiliary enzyme of fatty acid beta-oxidation; homodimeric enzyme required for growth and sporulation on petroselineate medium; expression induced during late sporulation and in the presence of oleate |
|
| YPR015C | 0.28 |
Putative protein of unknown function |
||
| YCR007C | 0.28 |
Putative integral membrane protein, member of DUP240 gene family; YCR007C is not an essential gene |
||
| YEL024W | 0.28 |
RIP1
|
Ubiquinol-cytochrome-c reductase, a Rieske iron-sulfur protein of the mitochondrial cytochrome bc1 complex; transfers electrons from ubiquinol to cytochrome c1 during respiration |
|
| YHR140W | 0.28 |
Putative integral membrane protein of unknown function |
||
| YPL271W | 0.27 |
ATP15
|
Epsilon subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; phosphorylated |
|
| YOL106W | 0.27 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YML058W-A | 0.27 |
HUG1
|
Protein involved in the Mec1p-mediated checkpoint pathway that responds to DNA damage or replication arrest, transcription is induced by DNA damage |
|
| YLR279W | 0.27 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER033C | 0.27 |
ZRG8
|
Protein of unknown function; authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; GFP-fusion protein is localized to the cytoplasm; transcription induced under conditions of zinc deficiency |
|
| YJL089W | 0.27 |
SIP4
|
C6 zinc cluster transcriptional activator that binds to the carbon source-responsive element (CSRE) of gluconeogenic genes; involved in the positive regulation of gluconeogenesis; regulated by Snf1p protein kinase; localized to the nucleus |
|
| YPR006C | 0.27 |
ICL2
|
2-methylisocitrate lyase of the mitochondrial matrix, functions in the methylcitrate cycle to catalyze the conversion of 2-methylisocitrate to succinate and pyruvate; ICL2 transcription is repressed by glucose and induced by ethanol |
|
| YNL017C | 0.26 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps the tRNA ORF tI(AAU)N2 |
||
| YLR366W | 0.26 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR364C-A |
||
| YDL244W | 0.26 |
THI13
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YJR019C | 0.26 |
TES1
|
Peroxisomal acyl-CoA thioesterase likely to be involved in fatty acid oxidation rather than fatty acid synthesis; conserved protein also found in human peroxisomes; TES1 mRNA levels increase during growth on fatty acids |
|
| YIL014C-A | 0.26 |
Putative protein of unknown function |
||
| YOL002C | 0.25 |
IZH2
|
Plasma membrane protein involved in zinc metabolism and osmotin-induced apoptosis; transcription regulated by Zap1p, zinc and fatty acid levels; similar to mammalian adiponectins; deletion increases sensitivity to elevated zinc |
|
| YGL205W | 0.25 |
POX1
|
Fatty-acyl coenzyme A oxidase, involved in the fatty acid beta-oxidation pathway; localized to the peroxisomal matrix |
|
| YJR078W | 0.25 |
BNA2
|
Putative tryptophan 2,3-dioxygenase or indoleamine 2,3-dioxygenase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p and Aft2p |
|
| YJL217W | 0.25 |
Cytoplasmic protein of unknown function; expression induced by calcium shortage and via the copper sensing transciption factor Mac1p during conditons of copper deficiency; mRNA is cell cycle regulated, peaking in G1 phase |
||
| YGL033W | 0.25 |
HOP2
|
Meiosis-specific protein that localizes to chromosomes, preventing synapsis between nonhomologous chromosomes and ensuring synapsis between homologs; complexes with Mnd1p to promote homolog pairing and meiotic double-strand break repair |
|
| YKR098C | 0.25 |
UBP11
|
Ubiquitin-specific protease that cleaves ubiquitin from ubiquitinated proteins |
|
| YLL019C | 0.25 |
KNS1
|
Nonessential putative protein kinase of unknown cellular role; member of the LAMMER family of protein kinases, which are serine/threonine kinases also capable of phosphorylating tyrosine residues |
|
| YCL021W-A | 0.25 |
Putative protein of unknown function |
||
| YOR383C | 0.24 |
FIT3
|
Mannoprotein that is incorporated into the cell wall via a glycosylphosphatidylinositol (GPI) anchor, involved in the retention of siderophore-iron in the cell wall |
|
| YEL012W | 0.24 |
UBC8
|
Ubiquitin-conjugating enzyme that negatively regulates gluconeogenesis by mediating the glucose-induced ubiquitination of fructose-1,6-bisphosphatase (FBPase); cytoplasmic enzyme that catalyzes the ubiquitination of histones in vitro |
|
| YMR034C | 0.24 |
Putative transporter, member of the SLC10 carrier family; identified in a transposon mutagenesis screen as a gene involved in azole resistance; YMR034C is not an essential gene |
||
| YCL001W-B | 0.24 |
Putative protein of unknown function; identified by homology |
||
| YOL030W | 0.24 |
GAS5
|
1,3-beta-glucanosyltransferase, has similarity to Gas1p; localizes to the cell wall |
|
| YJL130C | 0.24 |
URA2
|
Bifunctional carbamoylphosphate synthetase (CPSase)-aspartate transcarbamylase (ATCase), catalyzes the first two enzymatic steps in the de novo biosynthesis of pyrimidines; both activities are subject to feedback inhibition by UTP |
|
| YBR224W | 0.24 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene TDP1 |
||
| YLL053C | 0.24 |
Putative protein; in the Sigma 1278B strain background YLL053C is contiguous with AQY2 which encodes an aquaporin |
||
| YDR058C | 0.23 |
TGL2
|
Protein with lipolytic activity towards triacylglycerols and diacylglycerols when expressed in E. coli; role in yeast lipid degradation is unclear |
|
| YKL150W | 0.23 |
MCR1
|
Mitochondrial NADH-cytochrome b5 reductase, involved in ergosterol biosynthesis |
|
| YAL062W | 0.23 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YML089C | 0.23 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression induced by calcium shortage |
||
| YLR135W | 0.23 |
SLX4
|
Subunit of a complex, with Slx1p, that hydrolyzes 5' branches from duplex DNA in response to stalled or converging replication forks; function overlaps with that of Sgs1p-Top3p |
|
| YDL037C | 0.23 |
BSC1
|
Protein of unconfirmed function, similar to cell surface flocculin Muc1p; ORF exhibits genomic organization compatible with a translational readthrough-dependent mode of expression |
|
| YJR077C | 0.22 |
MIR1
|
Mitochondrial phosphate carrier, imports inorganic phosphate into mitochondria; functionally redundant with Pic2p but more abundant than Pic2p under normal conditions; phosphorylated |
|
| YDR287W | 0.22 |
INM2
|
Inositol monophosphatase, involved in biosynthesis of inositol; enzymatic activity requires magnesium ions and is inhibited by lithium and sodium ions; inm1 inm2 double mutant lacks inositol auxotrophy |
|
| YIR040C | 0.22 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YLL052C | 0.22 |
AQY2
|
Water channel that mediates the transport of water across cell membranes, only expressed in proliferating cells, controlled by osmotic signals, may be involved in freeze tolerance; disrupted by a stop codon in many S. cerevisiae strains |
|
| YIR041W | 0.22 |
PAU15
|
Hypothetical protein |
|
| YIL082W | 0.22 |
Retrotransposon TYA Gag gene co-transcribed with TYB Pol; translated as TYA or TYA-TYB polyprotein; Gag is a nucleocapsid protein that is the structural constituent of virus-like particles (VLPs); similar to retroviral Gag |
||
| YMR170C | 0.22 |
ALD2
|
Cytoplasmic aldehyde dehydrogenase, involved in ethanol oxidation and beta-alanine biosynthesis; uses NAD+ as the preferred coenzyme; expression is stress induced and glucose repressed; very similar to Ald3p |
|
| YGR137W | 0.21 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR204W | 0.21 |
COQ4
|
Protein with a role in ubiquinone (Coenzyme Q) biosynthesis, possibly functioning in stabilization of Coq7p; located on the matrix face of the mitochondrial inner membrane; component of a mitochondrial ubiquinone-synthesizing complex |
|
| YDR234W | 0.21 |
LYS4
|
Homoaconitase, catalyzes the conversion of homocitrate to homoisocitrate, which is a step in the lysine biosynthesis pathway |
|
| YOR011W-A | 0.21 |
Putative protein of unknown function |
||
| YGR213C | 0.21 |
RTA1
|
Protein involved in 7-aminocholesterol resistance; has seven potential membrane-spanning regions; expression is induced under both low-heme and low-oxygen conditions |
|
| YEL074W | 0.21 |
Hypothetical protein |
||
| YDL130W-A | 0.21 |
STF1
|
Protein involved in regulation of the mitochondrial F1F0-ATP synthase; Stf1p and Stf2p may act as stabilizing factors that enhance inhibitory action of the Inh1p protein |
|
| YBR208C | 0.21 |
DUR1,2
|
Urea amidolyase, contains both urea carboxylase and allophanate hydrolase activities, degrades urea to CO2 and NH3; expression sensitive to nitrogen catabolite repression and induced by allophanate, an intermediate in allantoin degradation |
|
| YGR067C | 0.20 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YLL041C | 0.20 |
SDH2
|
Iron-sulfur protein subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
Network of associatons between targets according to the STRING database.
First level regulatory network of SUT2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.6 | 1.7 | GO:0015755 | fructose transport(GO:0015755) |
| 0.5 | 2.0 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.5 | 4.8 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.4 | 1.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.3 | 0.9 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.3 | 0.8 | GO:0042843 | D-xylose catabolic process(GO:0042843) |
| 0.3 | 1.3 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.2 | 1.6 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.2 | 0.9 | GO:2000911 | positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) positive regulation of sterol transport(GO:0032373) positive regulation of transmembrane transport(GO:0034764) regulation of sterol import by regulation of transcription from RNA polymerase II promoter(GO:0035968) positive regulation of sterol import by positive regulation of transcription from RNA polymerase II promoter(GO:0035969) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.2 | 0.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.2 | 0.8 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.2 | 0.6 | GO:0019626 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 0.2 | 0.6 | GO:0034762 | regulation of transmembrane transport(GO:0034762) |
| 0.2 | 1.1 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.2 | 0.9 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.2 | 0.5 | GO:0046321 | regulation of fatty acid beta-oxidation(GO:0031998) positive regulation of fatty acid beta-oxidation(GO:0032000) regulation of fatty acid oxidation(GO:0046320) positive regulation of fatty acid oxidation(GO:0046321) |
| 0.2 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.7 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.2 | 0.5 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.2 | 1.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 0.6 | GO:0051303 | meiotic telomere clustering(GO:0045141) establishment of chromosome localization(GO:0051303) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.2 | 0.3 | GO:0046323 | glucose import(GO:0046323) |
| 0.2 | 0.5 | GO:0033559 | unsaturated fatty acid biosynthetic process(GO:0006636) unsaturated fatty acid metabolic process(GO:0033559) |
| 0.2 | 1.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 2.2 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.1 | 1.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.3 | GO:0035948 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 0.1 | 0.4 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) regulation of membrane lipid metabolic process(GO:1905038) |
| 0.1 | 2.0 | GO:0072593 | reactive oxygen species metabolic process(GO:0072593) |
| 0.1 | 0.4 | GO:0019627 | urea metabolic process(GO:0019627) |
| 0.1 | 0.1 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.6 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.3 | GO:0051596 | methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.4 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.1 | 0.4 | GO:0060194 | antisense RNA transcription(GO:0009300) regulation of antisense RNA transcription(GO:0060194) negative regulation of antisense RNA transcription(GO:0060195) |
| 0.1 | 1.1 | GO:0016241 | regulation of macroautophagy(GO:0016241) |
| 0.1 | 1.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.2 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 | 0.4 | GO:0045980 | negative regulation of nucleotide metabolic process(GO:0045980) |
| 0.1 | 0.3 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.1 | 0.7 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.2 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.8 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 0.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.2 | GO:0051469 | vesicle fusion with vacuole(GO:0051469) |
| 0.1 | 0.3 | GO:0070054 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) IRE1-mediated unfolded protein response(GO:0036498) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) |
| 0.1 | 0.4 | GO:0034354 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.1 | 0.9 | GO:0045144 | meiosis II(GO:0007135) meiotic sister chromatid segregation(GO:0045144) |
| 0.1 | 0.2 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.1 | 0.2 | GO:0034059 | response to anoxia(GO:0034059) cellular response to anoxia(GO:0071454) |
| 0.1 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.1 | 0.6 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.3 | GO:0000256 | allantoin metabolic process(GO:0000255) allantoin catabolic process(GO:0000256) cellular amide catabolic process(GO:0043605) |
| 0.1 | 0.3 | GO:0046503 | glycerolipid catabolic process(GO:0046503) |
| 0.1 | 0.2 | GO:0000736 | double-strand break repair via single-strand annealing, removal of nonhomologous ends(GO:0000736) |
| 0.1 | 0.4 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.2 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.1 | 0.1 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.1 | 0.5 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.1 | 1.3 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.1 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.2 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.1 | 0.2 | GO:0019405 | alditol catabolic process(GO:0019405) glycerol catabolic process(GO:0019563) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0009107 | lipoate metabolic process(GO:0009106) lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0006074 | (1->3)-beta-D-glucan metabolic process(GO:0006074) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.3 | GO:0005992 | trehalose biosynthetic process(GO:0005992) oligosaccharide biosynthetic process(GO:0009312) disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.4 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.3 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.4 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.0 | 0.1 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.0 | 0.3 | GO:0042773 | respiratory electron transport chain(GO:0022904) ATP synthesis coupled electron transport(GO:0042773) mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.1 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.2 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.0 | 0.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.7 | GO:0042724 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.0 | 0.1 | GO:0006827 | high-affinity iron ion transmembrane transport(GO:0006827) |
| 0.0 | 0.1 | GO:0031135 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.0 | 0.2 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of vesicle targeting, to, from or within Golgi(GO:0048209) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.3 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.6 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:0001881 | receptor recycling(GO:0001881) protein import into peroxisome matrix, receptor recycling(GO:0016562) receptor metabolic process(GO:0043112) |
| 0.0 | 0.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:1901661 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) ketone biosynthetic process(GO:0042181) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0048017 | inositol lipid-mediated signaling(GO:0048017) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.0 | 0.1 | GO:2000219 | positive regulation of invasive growth in response to glucose limitation by positive regulation of transcription from RNA polymerase II promoter(GO:0036095) positive regulation of invasive growth in response to glucose limitation(GO:2000219) |
| 0.0 | 0.2 | GO:0000409 | regulation of transcription by galactose(GO:0000409) positive regulation of transcription by galactose(GO:0000411) |
| 0.0 | 0.1 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.1 | GO:0051099 | positive regulation of binding(GO:0051099) |
| 0.0 | 0.1 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) pantothenate biosynthetic process(GO:0015940) |
| 0.0 | 0.1 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.0 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.0 | GO:1902932 | positive regulation of alcohol biosynthetic process(GO:1902932) positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.8 | GO:0070726 | cell wall assembly(GO:0070726) |
| 0.0 | 0.1 | GO:0035384 | acetyl-CoA biosynthetic process(GO:0006085) acetyl-CoA biosynthetic process from pyruvate(GO:0006086) thioester biosynthetic process(GO:0035384) acyl-CoA biosynthetic process(GO:0071616) |
| 0.0 | 0.1 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) |
| 0.0 | 0.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.1 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) ribonucleoside catabolic process(GO:0042454) |
| 0.0 | 0.1 | GO:0009231 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.0 | 0.0 | GO:1903138 | inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.0 | 0.7 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) DNA recombinase assembly(GO:0000730) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.2 | GO:0044088 | regulation of vacuole fusion, non-autophagic(GO:0032889) regulation of vacuole organization(GO:0044088) |
| 0.0 | 0.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.3 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.0 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.1 | GO:0060238 | regulation of pheromone-dependent signal transduction involved in conjugation with cellular fusion(GO:0010969) regulation of signal transduction involved in conjugation with cellular fusion(GO:0060238) |
| 0.0 | 0.1 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0043954 | cellular component maintenance(GO:0043954) |
| 0.0 | 0.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.0 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.0 | GO:0034033 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.0 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.0 | GO:0071825 | lipid tube assembly(GO:0060988) protein-lipid complex assembly(GO:0065005) protein-lipid complex subunit organization(GO:0071825) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.3 | 2.4 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.3 | 0.9 | GO:0045240 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial oxoglutarate dehydrogenase complex(GO:0009353) dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0005756 | mitochondrial proton-transporting ATP synthase, central stalk(GO:0005756) proton-transporting ATP synthase, central stalk(GO:0045269) |
| 0.1 | 0.3 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.1 | 0.4 | GO:0032221 | Rpd3S complex(GO:0032221) |
| 0.1 | 0.2 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.1 | 1.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0031307 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0032126 | eisosome(GO:0032126) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 1.7 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 2.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.2 | GO:0035339 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.3 | GO:0031160 | spore wall(GO:0031160) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 14.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0031226 | integral component of plasma membrane(GO:0005887) intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.0 | GO:0032116 | SMC loading complex(GO:0032116) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.7 | 2.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.5 | 1.8 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.4 | 1.8 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.4 | 1.9 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.3 | 1.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.3 | 1.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 0.9 | GO:0016406 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.3 | 0.9 | GO:0022829 | anion channel activity(GO:0005253) voltage-gated anion channel activity(GO:0008308) porin activity(GO:0015288) wide pore channel activity(GO:0022829) |
| 0.3 | 0.9 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.3 | 1.9 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 0.5 | GO:0015489 | putrescine transmembrane transporter activity(GO:0015489) |
| 0.2 | 0.8 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 1.0 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.2 | 0.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 0.8 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.2 | 0.6 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) |
| 0.2 | 1.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 0.5 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 0.5 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 1.2 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.5 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 1.9 | GO:0046906 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.1 | 0.4 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.3 | GO:0022832 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.1 | 0.4 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 0.1 | 0.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.3 | GO:0004564 | beta-fructofuranosidase activity(GO:0004564) sucrose alpha-glucosidase activity(GO:0004575) |
| 0.1 | 0.6 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.8 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 0.6 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.2 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.1 | 0.2 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 1.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.4 | GO:0008483 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.4 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.7 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 0.1 | 0.3 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.2 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.1 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 0.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.7 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0042124 | glucanosyltransferase activity(GO:0042123) 1,3-beta-glucanosyltransferase activity(GO:0042124) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.1 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.5 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0008902 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.0 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.1 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 0.4 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0097027 | cyclin binding(GO:0030332) ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.4 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.2 | GO:0016298 | lipase activity(GO:0016298) |
| 0.0 | 0.4 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0015151 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.0 | 0.2 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.1 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.0 | GO:0016642 | glycine dehydrogenase (decarboxylating) activity(GO:0004375) oxidoreductase activity, acting on the CH-NH2 group of donors, disulfide as acceptor(GO:0016642) |
| 0.0 | 0.1 | GO:0052742 | phosphatidylinositol kinase activity(GO:0052742) |
| 0.0 | 0.3 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0016830 | carbon-carbon lyase activity(GO:0016830) |
| 0.0 | 0.1 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 0.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 0.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.3 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | Genes involved in Transmembrane transport of small molecules |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |