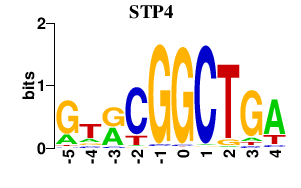
Results for STP4
Z-value: 2.90
Transcription factors associated with STP4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STP4
|
S000002206 | Protein containing a Kruppel-type zinc-finger domain |
Activity-expression correlation:
Activity profile of STP4 motif
Sorted Z-values of STP4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YMR107W | 37.42 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YOR100C | 33.56 |
CRC1
|
Mitochondrial inner membrane carnitine transporter, required for carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria during fatty acid beta-oxidation |
|
| YLR307C-A | 32.06 |
Putative protein of unknown function |
||
| YIL057C | 31.08 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YCR010C | 25.48 |
ADY2
|
Acetate transporter required for normal sporulation; phosphorylated in mitochondria |
|
| YKR097W | 24.42 |
PCK1
|
Phosphoenolpyruvate carboxykinase, key enzyme in gluconeogenesis, catalyzes early reaction in carbohydrate biosynthesis, glucose represses transcription and accelerates mRNA degradation, regulated by Mcm1p and Cat8p, located in the cytosol |
|
| YJL045W | 23.26 |
Minor succinate dehydrogenase isozyme; homologous to Sdh1p, the major isozyme reponsible for the oxidation of succinate and transfer of electrons to ubiquinone; induced during the diauxic shift in a Cat8p-dependent manner |
||
| YNR002C | 23.14 |
ATO2
|
Putative transmembrane protein involved in export of ammonia, a starvation signal that promotes cell death in aging colonies; phosphorylated in mitochondria; member of the TC 9.B.33 YaaH family; homolog of Ady2p and Y. lipolytica Gpr1p |
|
| YAL054C | 21.11 |
ACS1
|
Acetyl-coA synthetase isoform which, along with Acs2p, is the nuclear source of acetyl-coA for histone acetlyation; expressed during growth on nonfermentable carbon sources and under aerobic conditions |
|
| YOR382W | 20.97 |
FIT2
|
Mannoprotein that is incorporated into the cell wall via a glycosylphosphatidylinositol (GPI) anchor, involved in the retention of siderophore-iron in the cell wall |
|
| YAL062W | 20.21 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YPL171C | 20.07 |
OYE3
|
Widely conserved NADPH oxidoreductase containing flavin mononucleotide (FMN), homologous to Oye2p with slight differences in ligand binding and catalytic properties; may be involved in sterol metabolism |
|
| YLR327C | 20.04 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YBR296C | 19.98 |
PHO89
|
Na+/Pi cotransporter, active in early growth phase; similar to phosphate transporters of Neurospora crassa; transcription regulated by inorganic phosphate concentrations and Pho4p |
|
| YMR206W | 19.70 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YML042W | 19.00 |
CAT2
|
Carnitine acetyl-CoA transferase present in both mitochondria and peroxisomes, transfers activated acetyl groups to carnitine to form acetylcarnitine which can be shuttled across membranes |
|
| YGR236C | 18.95 |
SPG1
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YPL223C | 17.61 |
GRE1
|
Hydrophilin of unknown function; stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YPL054W | 16.26 |
LEE1
|
Zinc-finger protein of unknown function |
|
| YDR059C | 16.24 |
UBC5
|
Ubiquitin-conjugating enzyme that mediates selective degradation of short-lived and abnormal proteins, central component of the cellular stress response; expression is heat inducible |
|
| YKL217W | 15.46 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YNL194C | 15.21 |
Integral membrane protein localized to eisosomes; sporulation and plasma membrane sphingolipid content are altered in mutants; has homologs SUR7 and FMP45; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YGL146C | 15.00 |
Putative protein of unknown function, contains two putative transmembrane spans, shows no significant homology to any other known protein sequence, YGL146C is not an essential gene |
||
| YPR026W | 14.99 |
ATH1
|
Acid trehalase required for utilization of extracellular trehalose |
|
| YMR017W | 14.98 |
SPO20
|
Meiosis-specific subunit of the t-SNARE complex, required for prospore membrane formation during sporulation; similar to but not functionally redundant with Sec9p; SNAP-25 homolog |
|
| YAR053W | 13.96 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR243W | 13.86 |
FMP43
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YJR120W | 13.32 |
Protein of unknown function; essential for growth under anaerobic conditions; mutation causes decreased expression of ATP2, impaired respiration, defective sterol uptake, and altered levels/localization of ABC transporters Aus1p and Pdr11p |
||
| YOL052C-A | 11.92 |
DDR2
|
Multistress response protein, expression is activated by a variety of xenobiotic agents and environmental or physiological stresses |
|
| YDR171W | 11.90 |
HSP42
|
Small heat shock protein (sHSP) with chaperone activity; forms barrel-shaped oligomers that suppress unfolded protein aggregation; involved in cytoskeleton reorganization after heat shock |
|
| YCR005C | 11.71 |
CIT2
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate, peroxisomal isozyme involved in glyoxylate cycle; expression is controlled by Rtg1p and Rtg2p transcription factors |
|
| YGR142W | 11.03 |
BTN2
|
v-SNARE binding protein that facilitates specific protein retrieval from a late endosome to the Golgi; modulates arginine uptake, possible role in mediating pH homeostasis between the vacuole and plasma membrane H(+)-ATPase |
|
| YMR191W | 10.97 |
SPG5
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YCR021C | 10.86 |
HSP30
|
Hydrophobic plasma membrane localized, stress-responsive protein that negatively regulates the H(+)-ATPase Pma1p; induced by heat shock, ethanol treatment, weak organic acid, glucose limitation, and entry into stationary phase |
|
| YNL117W | 10.70 |
MLS1
|
Malate synthase, enzyme of the glyoxylate cycle, involved in utilization of non-fermentable carbon sources; expression is subject to carbon catabolite repression; localizes in peroxisomes during growth in oleic acid medium |
|
| YBR056W-A | 10.70 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR056C-B |
||
| YKL038W | 10.55 |
RGT1
|
Glucose-responsive transcription factor that regulates expression of several glucose transporter (HXT) genes in response to glucose; binds to promoters and acts both as a transcriptional activator and repressor |
|
| YMR306W | 10.24 |
FKS3
|
Protein involved in spore wall assembly, has similarity to 1,3-beta-D-glucan synthase catalytic subunits Fks1p and Gsc2p; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YBR105C | 10.02 |
VID24
|
Peripheral membrane protein located at Vid (vacuole import and degradation) vesicles; regulates fructose-1,6-bisphosphatase (FBPase) targeting to the vacuole; involved in proteasome-dependent catabolite degradation of FBPase |
|
| YDL210W | 9.74 |
UGA4
|
Permease that serves as a gamma-aminobutyrate (GABA) transport protein involved in the utilization of GABA as a nitrogen source; catalyzes the transport of putrescine and delta-aminolevulinic acid (ALA); localized to the vacuolar membrane |
|
| YJL185C | 9.52 |
Putative protein of unknown function; mRNA is weakly cell cycle regulated, peaking in G2 phase; YJL185C is a non-essential gene |
||
| YAR060C | 9.46 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YIL055C | 9.38 |
Putative protein of unknown function |
||
| YOL084W | 9.34 |
PHM7
|
Protein of unknown function, expression is regulated by phosphate levels; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery and vacuole |
|
| YJR146W | 9.24 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene HMS2 |
||
| YLR149C | 9.24 |
Putative protein of unknown function; YLR149C is not an essential gene |
||
| YDL085W | 9.18 |
NDE2
|
Mitochondrial external NADH dehydrogenase, catalyzes the oxidation of cytosolic NADH; Nde1p and Nde2p are involved in providing the cytosolic NADH to the mitochondrial respiratory chain |
|
| YHR212C | 8.91 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YFL052W | 8.87 |
Putative zinc cluster protein that contains a DNA binding domain; null mutant sensitive to calcofluor white, low osmolarity and heat, suggesting a role for YFL052Wp in cell wall integrity |
||
| YJL089W | 8.82 |
SIP4
|
C6 zinc cluster transcriptional activator that binds to the carbon source-responsive element (CSRE) of gluconeogenic genes; involved in the positive regulation of gluconeogenesis; regulated by Snf1p protein kinase; localized to the nucleus |
|
| YNL014W | 8.81 |
HEF3
|
Translational elongation factor EF-3; paralog of YEF3 and member of the ABC superfamily; stimulates EF-1 alpha-dependent binding of aminoacyl-tRNA by the ribosome; normally expressed in zinc deficient cells |
|
| YDR461C-A | 8.76 |
Putative protein of unknown function |
||
| YMR013C | 8.75 |
SEC59
|
Dolichol kinase, catalyzes the terminal step in dolichyl monophosphate (Dol-P) biosynthesis; required for viability and for normal rates of lipid intermediate synthesis and protein N-glycosylation |
|
| YAR070C | 8.72 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YAR069C | 8.69 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YKR102W | 8.57 |
FLO10
|
Lectin-like protein with similarity to Flo1p, thought to be involved in flocculation |
|
| YMR056C | 8.44 |
AAC1
|
Mitochondrial inner membrane ADP/ATP translocator, exchanges cytosolic ADP for mitochondrially synthesized ATP; phosphorylated; Aac1p is a minor isoform while Pet9p is the major ADP/ATP translocator |
|
| YOR348C | 8.21 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YPR001W | 8.10 |
CIT3
|
Dual specificity mitochondrial citrate and methylcitrate synthase; catalyzes the condensation of acetyl-CoA and oxaloacetate to form citrate and that of propionyl-CoA and oxaloacetate to form 2-methylcitrate |
|
| YDL199C | 7.98 |
Putative transporter, member of the sugar porter family |
||
| YHR212W-A | 7.95 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YLR438W | 7.91 |
CAR2
|
L-ornithine transaminase (OTAse), catalyzes the second step of arginine degradation, expression is dually-regulated by allophanate induction and a specific arginine induction process; not nitrogen catabolite repression sensitive |
|
| YDR343C | 7.83 |
HXT6
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt7p, expressed at high basal levels relative to other HXTs, repression of expression by high glucose requires SNF3 |
|
| YER187W | 7.83 |
Putative protein of unknown function; induced in respiratory-deficient cells |
||
| YHR217C | 7.81 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; located in the telomeric region TEL08R. |
||
| YNL305C | 7.64 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YNL305C is not an essential gene |
||
| YGL184C | 7.53 |
STR3
|
Cystathionine beta-lyase, converts cystathionine into homocysteine |
|
| YOR378W | 7.45 |
Putative protein of unknown function; belongs to the DHA2 family of drug:H+ antiporters; YOR378W is not an essential gene |
||
| YPL134C | 7.41 |
ODC1
|
Mitochondrial inner membrane transporter, exports 2-oxoadipate and 2-oxoglutarate from the mitochondrial matrix to the cytosol for lysine and glutamate biosynthesis and lysine catabolism; suppresses, in multicopy, an fmc1 null mutation |
|
| YLR356W | 7.36 |
Putative protein of unknown function with similarity to SCM4; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR356W is not an essential gene |
||
| YPL026C | 7.34 |
SKS1
|
Putative serine/threonine protein kinase; involved in the adaptation to low concentrations of glucose independent of the SNF3 regulated pathway |
|
| YPL222W | 7.23 |
FMP40
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YLR346C | 7.17 |
Putative protein of unknown function found in mitochondria; expression is regulated by transcription factors involved in pleiotropic drug resistance, Pdr1p and Yrr1p; YLR346C is not an essential gene |
||
| YPL058C | 7.17 |
PDR12
|
Plasma membrane ATP-binding cassette (ABC) transporter, weak-acid-inducible multidrug transporter required for weak organic acid resistance; induced by sorbate and benzoate and regulated by War1p; mutants exhibit sorbate hypersensitivity |
|
| YOR343C | 7.12 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YGR110W | 7.10 |
Putative protein of unknown function; transcription is increased in response to genotoxic stress; plays a role in restricting Ty1 transposition |
||
| YPL024W | 7.09 |
RMI1
|
Involved in response to DNA damage; null mutants have increased rates of recombination and delayed S phase; interacts physically and genetically with Sgs1p (RecQ family member) and Top3p (topoisomerase III) |
|
| YOL100W | 7.00 |
PKH2
|
Serine/threonine protein kinase involved in sphingolipid-mediated signaling pathway that controls endocytosis; activates Ypk1p and Ykr2p, components of signaling cascade required for maintenance of cell wall integrity; redundant with Pkh1p |
|
| YLR377C | 6.97 |
FBP1
|
Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway, required for glucose metabolism |
|
| YGR258C | 6.95 |
RAD2
|
Single-stranded DNA endonuclease, cleaves single-stranded DNA during nucleotide excision repair to excise damaged DNA; subunit of Nucleotide Excision Repair Factor 3 (NEF3); homolog of human XPG protein |
|
| YBL064C | 6.85 |
PRX1
|
Mitochondrial peroxiredoxin (1-Cys Prx) with thioredoxin peroxidase activity, has a role in reduction of hydroperoxides; induced during respiratory growth and under conditions of oxidative stress; phosphorylated |
|
| YBR051W | 6.77 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partiallly overlaps the REG2/YBR050C regulatory subunit of the Glc7p type-1 protein phosphatase |
||
| YMR096W | 6.77 |
SNZ1
|
Protein involved in vitamin B6 biosynthesis; member of a stationary phase-induced gene family; coregulated with SNO1; interacts with Sno1p and with Yhr198p, perhaps as a multiprotein complex containing other Snz and Sno proteins |
|
| YMR040W | 6.75 |
YET2
|
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; homolog of human BAP31 protein |
|
| YNR001C | 6.75 |
CIT1
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate; the rate-limiting enzyme of the TCA cycle; nuclear encoded mitochondrial protein |
|
| YAR050W | 6.71 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YDL194W | 6.68 |
SNF3
|
Plasma membrane glucose sensor that regulates glucose transport; has 12 predicted transmembrane segments; long cytoplasmic C-terminal tail is required for low glucose induction of hexose transporter genes HXT2 and HXT4 |
|
| YHL024W | 6.62 |
RIM4
|
Putative RNA-binding protein required for the expression of early and middle sporulation genes |
|
| YOL118C | 6.58 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YJR138W | 6.58 |
IML1
|
Protein of unknown function, green fluorescent protein (GFP)-fusion protein localizes to the vacuolar membrane |
|
| YGL072C | 6.57 |
Dubious open reading frame unlikely to encode a protein; partially overlaps the verified gene HSF1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YMR280C | 6.54 |
CAT8
|
Zinc cluster transcriptional activator necessary for derepression of a variety of genes under non-fermentative growth conditions, active after diauxic shift, binds carbon source responsive elements |
|
| YMR175W | 6.53 |
SIP18
|
Protein of unknown function whose expression is induced by osmotic stress |
|
| YLR437C-A | 6.50 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF CAR2/YLR438W |
||
| YGR239C | 6.49 |
PEX21
|
Peroxin required for targeting of peroxisomal matrix proteins containing PTS2; interacts with Pex7p; partially redundant with Pex18p |
|
| YBR050C | 6.40 |
REG2
|
Regulatory subunit of the Glc7p type-1 protein phosphatase; involved with Reg1p, Glc7p, and Snf1p in regulation of glucose-repressible genes, also involved in glucose-induced proteolysis of maltose permease |
|
| YLR267W | 6.32 |
BOP2
|
Protein of unknown function, overproduction suppresses a pam1 slv3 double null mutation |
|
| YBR076C-A | 6.24 |
Dubious open reading frame unlikely to encode a protein; partially overlaps verified gene ECM8; identified by fungal homology and RT-PCR |
||
| YDL214C | 6.17 |
PRR2
|
Serine/threonine protein kinase that inhibits pheromone induced signalling downstream of MAPK, possibly at the level of the Ste12p transcription factor |
|
| YLR308W | 6.16 |
CDA2
|
Chitin deacetylase, together with Cda1p involved in the biosynthesis ascospore wall component, chitosan; required for proper rigidity of the ascospore wall |
|
| YOL081W | 6.13 |
IRA2
|
GTPase-activating protein that negatively regulates RAS by converting it from the GTP- to the GDP-bound inactive form, required for reducing cAMP levels under nutrient limiting conditions, has similarity to Ira1p and human neurofibromin |
|
| YNL180C | 6.08 |
RHO5
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely involved in protein kinase C (Pkc1p)-dependent signal transduction pathway that controls cell integrity |
|
| YCR068W | 6.08 |
ATG15
|
Lipase, required for intravacuolar lysis of autophagosomes; located in the endoplasmic reticulum membrane and targeted to intravacuolar vesicles during autophagy via the multivesicular body (MVB) pathway |
|
| YER162C | 6.04 |
RAD4
|
Protein that recognizes and binds damaged DNA (with Rad23p) during nucleotide excision repair; subunit of Nuclear Excision Repair Factor 2 (NEF2); homolog of human XPC protein |
|
| YPR036W-A | 6.00 |
Protein of unknown function; transcription is regulated by Pdr1p |
||
| YGR201C | 5.92 |
Putative protein of unknown function |
||
| YAL039C | 5.92 |
CYC3
|
Cytochrome c heme lyase (holocytochrome c synthase), attaches heme to apo-cytochrome c (Cyc1p or Cyc7p) in the mitochondrial intermembrane space; human ortholog may have a role in microphthalmia with linear skin defects (MLS) |
|
| YLR094C | 5.90 |
GIS3
|
Protein of unknown function |
|
| YDR505C | 5.83 |
PSP1
|
Asn and gln rich protein of unknown function; high-copy suppressor of POL1 (DNA polymerase alpha) and partial suppressor of CDC2 (polymerase delta) and CDC6 (pre-RC loading factor) mutations; overexpression results in growth inhibition |
|
| YPR192W | 5.82 |
AQY1
|
Spore-specific water channel that mediates the transport of water across cell membranes, developmentally controlled; may play a role in spore maturation, probably by allowing water outflow, may be involved in freeze tolerance |
|
| YDR273W | 5.81 |
DON1
|
Meiosis-specific component of the spindle pole body, part of the leading edge protein (LEP) coat, forms a ring-like structure at the leading edge of the prospore membrane during meiosis II |
|
| YPL201C | 5.78 |
YIG1
|
Protein that interacts with glycerol 3-phosphatase and plays a role in anaerobic glycerol production; localizes to the nucleus and cytosol |
|
| YKL093W | 5.74 |
MBR1
|
Protein involved in mitochondrial functions and stress response; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants |
|
| YNL179C | 5.72 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YAL063C | 5.67 |
FLO9
|
Lectin-like protein with similarity to Flo1p, thought to be expressed and involved in flocculation |
|
| YOL085W-A | 5.65 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YOL085C |
||
| YGR065C | 5.61 |
VHT1
|
High-affinity plasma membrane H+-biotin (vitamin H) symporter; mutation results in fatty acid auxotrophy; 12 transmembrane domain containing major facilitator subfamily member; mRNA levels negatively regulated by iron deprivation and biotin |
|
| YFL019C | 5.58 |
Dubious open reading frame unlikely to encode a protein; YFL019C is not an essential gene |
||
| YPL018W | 5.51 |
CTF19
|
Outer kinetochore protein, required for accurate mitotic chromosome segregation; component of the kinetochore sub-complex COMA (Ctf19p, Okp1p, Mcm21p, Ame1p) that functions as a platform for kinetochore assembly |
|
| YGR067C | 5.50 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YDR536W | 5.36 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YMR014W | 5.33 |
BUD22
|
Protein involved in bud-site selection; diploid mutants display a random budding pattern instead of the wild-type bipolar pattern |
|
| YOR328W | 5.31 |
PDR10
|
ATP-binding cassette (ABC) transporter, multidrug transporter involved in the pleiotropic drug resistance network; regulated by Pdr1p and Pdr3p |
|
| YAR047C | 5.23 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDL182W | 5.21 |
LYS20
|
Homocitrate synthase isozyme, catalyzes the condensation of acetyl-CoA and alpha-ketoglutarate to form homocitrate, which is the first step in the lysine biosynthesis pathway; highly similar to the other isozyme, Lys21p |
|
| YGL163C | 5.18 |
RAD54
|
DNA-dependent ATPase, stimulates strand exchange by modifying the topology of double-stranded DNA; involved in the recombinational repair of double-strand breaks in DNA during vegetative growth and meiosis; member of the SWI/SNF family |
|
| YAR068W | 5.14 |
Fungal-specific protein of unknown function; induced in respiratory-deficient cells |
||
| YOR387C | 5.14 |
Putative protein of unknown function; regulated by the metal-responsive Aft1p transcription factor; highly inducible in zinc-depleted conditions; localizes to the soluble fraction |
||
| YLR294C | 5.09 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene ATP14 |
||
| YPL250C | 5.06 |
ICY2
|
Protein of unknown function; mobilized into polysomes upon a shift from a fermentable to nonfermentable carbon source; potential Cdc28p substrate |
|
| YPL033C | 5.01 |
Putative protein of unknown function; may be involved in DNA metabolism; expression is induced by Kar4p |
||
| YNL144C | 5.00 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YNL144C is not an essential gene |
||
| YCL025C | 4.99 |
AGP1
|
Low-affinity amino acid permease with broad substrate range, involved in uptake of asparagine, glutamine, and other amino acids; expression is regulated by the SPS plasma membrane amino acid sensor system (Ssy1p-Ptr3p-Ssy5p) |
|
| YNR064C | 4.98 |
Epoxide hydrolase, member of the alpha/beta hydrolase fold family; may have a role in detoxification of epoxides |
||
| YLR312C | 4.97 |
QNQ1
|
Protein of unknown function; null mutant quiescent and non-quiescent cells exhibit reduced reproductive capacity |
|
| YMR135C | 4.96 |
GID8
|
Protein of unknown function, involved in proteasome-dependent catabolite inactivation of fructose-1,6-bisphosphatase; contains LisH and CTLH domains, like Vid30p; dosage-dependent regulator of START |
|
| YPL271W | 4.92 |
ATP15
|
Epsilon subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; phosphorylated |
|
| YNL195C | 4.83 |
Putative protein of unknown function; shares a promoter with YNL194C; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YPL021W | 4.81 |
ECM23
|
Non-essential protein of unconfirmed function; affects pre-rRNA processing, may act as a negative regulator of the transcription of genes involved in pseudohyphal growth; homologous to Srd1p |
|
| YJR137C | 4.81 |
ECM17
|
Sulfite reductase beta subunit, involved in amino acid biosynthesis, transcription repressed by methionine |
|
| YHR218W | 4.80 |
Helicase-like protein encoded within the telomeric Y' element |
||
| YLR296W | 4.78 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YER015W | 4.72 |
FAA2
|
Long chain fatty acyl-CoA synthetase; accepts a wider range of acyl chain lengths than Faa1p, preferring C9:0-C13:0; involved in the activation of endogenous pools of fatty acids |
|
| YEL059W | 4.71 |
Dubious open reading frame unlikely to encode a functional protein |
||
| YBL015W | 4.71 |
ACH1
|
Acetyl-coA hydrolase, primarily localized to mitochondria; phosphorylated; required for acetate utilization and for diploid pseudohyphal growth |
|
| YLL039C | 4.70 |
UBI4
|
Ubiquitin, becomes conjugated to proteins, marking them for selective degradation via the ubiquitin-26S proteasome system; essential for the cellular stress response; encoded as a polyubiquitin precursor comprised of 5 head-to-tail repeats |
|
| YKR049C | 4.70 |
FMP46
|
Putative redox protein containing a thioredoxin fold; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YDR055W | 4.67 |
PST1
|
Cell wall protein that contains a putative GPI-attachment site; secreted by regenerating protoplasts; up-regulated by activation of the cell integrity pathway, as mediated by Rlm1p; upregulated by cell wall damage via disruption of FKS1 |
|
| YJL163C | 4.64 |
Putative protein of unknown function |
||
| YMR244W | 4.61 |
Putative protein of unknown function |
||
| YNL036W | 4.59 |
NCE103
|
Carbonic anhydrase; poorly transcribed under aerobic conditions and at an undetectable level under anaerobic conditions; involved in non-classical protein export pathway |
|
| YOR192C | 4.57 |
THI72
|
Transporter of thiamine or related compound; shares sequence similarity with Thi7p |
|
| YEL074W | 4.54 |
Hypothetical protein |
||
| YDR256C | 4.49 |
CTA1
|
Catalase A, breaks down hydrogen peroxide in the peroxisomal matrix formed by acyl-CoA oxidase (Pox1p) during fatty acid beta-oxidation |
|
| YEL024W | 4.41 |
RIP1
|
Ubiquinol-cytochrome-c reductase, a Rieske iron-sulfur protein of the mitochondrial cytochrome bc1 complex; transfers electrons from ubiquinol to cytochrome c1 during respiration |
|
| YNL283C | 4.41 |
WSC2
|
Partially redundant sensor-transducer of the stress-activated PKC1-MPK1 signaling pathway involved in maintenance of cell wall integrity and recovery from heat shock; secretory pathway Wsc2p is required for the arrest of secretion response |
|
| YBL043W | 4.39 |
ECM13
|
Non-essential protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YEL009C | 4.28 |
GCN4
|
Transcriptional activator of amino acid biosynthetic genes in response to amino acid starvation; expression is tightly regulated at both the transcriptional and translational levels |
|
| YOR374W | 4.27 |
ALD4
|
Mitochondrial aldehyde dehydrogenase, required for growth on ethanol and conversion of acetaldehyde to acetate; phosphorylated; activity is K+ dependent; utilizes NADP+ or NAD+ equally as coenzymes; expression is glucose repressed |
|
| YOR139C | 4.24 |
Hypothetical protein |
||
| YKL109W | 4.22 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YFL050C | 4.20 |
ALR2
|
Probable Mg(2+) transporter; overexpression confers increased tolerance to Al(3+) and Ga(3+) ions; plays a role in regulating Ty1 transposition |
|
| YBR292C | 4.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR292C is not an essential gene |
||
| YJR151C | 4.19 |
DAN4
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YKL133C | 4.18 |
Putative protein of unknown function |
||
| YPR015C | 4.15 |
Putative protein of unknown function |
||
| YBR224W | 4.11 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene TDP1 |
||
| YOR192C-C | 4.11 |
Putative protein of unknown function; identified by expression profiling and mass spectrometry |
||
| YIL146C | 4.10 |
ECM37
|
Non-essential protein of unknown function |
|
| YJR119C | 4.10 |
JHD2
|
JmjC domain family histone demethylase specific for H3-K4 (lysine at position 4 of the histone H3 protein); removes methyl groups specifically added by Set1p methyltransferase |
|
| YGL071W | 4.09 |
AFT1
|
Transcription factor involved in iron utilization and homeostasis; binds the consensus site PyPuCACCCPu and activates the expression of target genes in response to changes in iron availability |
|
| YMR095C | 4.08 |
SNO1
|
Protein of unconfirmed function, involved in pyridoxine metabolism; expression is induced during stationary phase; forms a putative glutamine amidotransferase complex with Snz1p, with Sno1p serving as the glutaminase |
|
| YDR036C | 4.06 |
EHD3
|
3-hydroxyisobutyryl-CoA hydrolase, member of a family of enoyl-CoA hydratase/isomerases; non-tagged protein is detected in highly purified mitochondria in high-throughput studies; phosphorylated; mutation affects fluid-phase endocytosis |
|
| YEL008W | 4.04 |
Hypothetical protein predicted to be involved in metabolism |
||
| YLR247C | 4.02 |
IRC20
|
Putative helicase; localizes to the mitochondrion and the nucleus; YLR247C is not an essential gene; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YNL282W | 4.02 |
POP3
|
Subunit of both RNase MRP, which cleaves pre-rRNA, and nuclear RNase P, which cleaves tRNA precursors to generate mature 5' ends |
|
| YLR402W | 4.02 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YBR117C | 3.98 |
TKL2
|
Transketolase, similar to Tkl1p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YDR231C | 3.97 |
COX20
|
Mitochondrial inner membrane protein, required for proteolytic processing of Cox2p and its assembly into cytochrome c oxidase |
|
| YER186C | 3.96 |
Putative protein of unknown function |
||
| YBR212W | 3.96 |
NGR1
|
RNA binding protein that negatively regulates growth rate; interacts with the 3' UTR of the mitochondrial porin (POR1) mRNA and enhances its degradation; overexpression impairs mitochondrial function; expressed in stationary phase |
|
| YLR299W | 3.95 |
ECM38
|
Gamma-glutamyltranspeptidase, major glutathione-degrading enzyme; involved in detoxification of electrophilic xenobiotics; expression induced mainly by nitrogen starvation |
|
| YGL180W | 3.91 |
ATG1
|
Protein serine/threonine kinase required for vesicle formation in autophagy and the cytoplasm-to-vacuole targeting (Cvt) pathway; structurally required for pre-autophagosome formation; during autophagy forms a complex with Atg13p and Atg17p |
|
| YOR034C | 3.89 |
AKR2
|
Ankyrin repeat-containing protein similar to Akr1p; member of a family of putative palmitoyltransferases containing an Asp-His-His-Cys-cysteine rich (DHHC-CRD) domain; possibly involved in constitutive endocytosis of Ste3p |
|
| YOL119C | 3.89 |
MCH4
|
Protein with similarity to mammalian monocarboxylate permeases, which are involved in transport of monocarboxylic acids across the plasma membrane; mutant is not deficient in monocarboxylate transport |
|
| YFL020C | 3.83 |
PAU5
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YLR403W | 3.83 |
SFP1
|
Transcription factor that controls expression of many ribosome biogenesis genes in response to nutrients and stress, regulates G2/M transitions during mitotic cell cycle and DNA-damage response, involved in cell size modulation |
|
| YEL075C | 3.83 |
Putative protein of unknown function |
||
| YLR164W | 3.81 |
Mitochondrial inner membrane of unknown function; similar to Tim18p and Sdh4p; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
||
| YOR140W | 3.81 |
SFL1
|
Transcriptional repressor and activator; involved in repression of flocculation-related genes, and activation of stress responsive genes; negatively regulated by cAMP-dependent protein kinase A subunit Tpk2p |
|
| YJL116C | 3.80 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YPL036W | 3.80 |
PMA2
|
Plasma membrane H+-ATPase, isoform of Pma1p, involved in pumping protons out of the cell; regulator of cytoplasmic pH and plasma membrane potential |
|
| YKL016C | 3.80 |
ATP7
|
Subunit d of the stator stalk of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YFR034C | 3.78 |
PHO4
|
Basic helix-loop-helix (bHLH) transcription factor of the myc-family; binds cooperatively with Pho2p to the PHO5 promoter; function is regulated by phosphorylation at multiple sites and by phosphate availability |
|
| YNL332W | 3.77 |
THI12
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YDR034C | 3.76 |
LYS14
|
Transcriptional activator involved in regulation of genes of the lysine biosynthesis pathway; requires 2-aminoadipate semialdehyde as co-inducer |
|
| YPR065W | 3.76 |
ROX1
|
Heme-dependent repressor of hypoxic genes; contains an HMG domain that is responsible for DNA bending activity |
|
| YBR250W | 3.75 |
SPO23
|
Protein of unknown function; associates with meiosis-specific protein Spo1p |
|
| YOR186C-A | 3.73 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YDR381C-A | 3.72 |
Protein of unknown function, localized to the mitochondrial outer membrane |
Network of associatons between targets according to the STRING database.
First level regulatory network of STP4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 22.1 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 7.3 | 29.3 | GO:0006848 | pyruvate transport(GO:0006848) |
| 6.7 | 6.7 | GO:0015755 | fructose transport(GO:0015755) |
| 5.7 | 17.0 | GO:0035948 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 5.3 | 16.0 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 5.1 | 15.3 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 4.1 | 24.4 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 3.6 | 10.9 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 3.5 | 20.9 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 3.5 | 13.8 | GO:0046323 | glucose import(GO:0046323) |
| 3.4 | 27.0 | GO:0015891 | siderophore transport(GO:0015891) |
| 3.2 | 9.6 | GO:0019541 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 3.1 | 6.1 | GO:0045980 | negative regulation of nucleotide metabolic process(GO:0045980) |
| 3.0 | 23.8 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 2.9 | 14.7 | GO:0051099 | positive regulation of binding(GO:0051099) |
| 2.9 | 8.7 | GO:0051469 | vesicle fusion with vacuole(GO:0051469) |
| 2.8 | 61.6 | GO:0019740 | nitrogen utilization(GO:0019740) |
| 2.6 | 7.7 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 2.6 | 10.2 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 2.5 | 10.2 | GO:0015886 | heme transport(GO:0015886) |
| 2.3 | 2.3 | GO:0061410 | positive regulation of transcription from RNA polymerase II promoter in response to ethanol(GO:0061410) cellular response to ethanol(GO:0071361) cellular response to alcohol(GO:0097306) |
| 2.2 | 8.9 | GO:0015847 | putrescine transport(GO:0015847) |
| 2.1 | 6.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 2.1 | 8.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 2.0 | 6.1 | GO:0036257 | multivesicular body organization(GO:0036257) |
| 2.0 | 4.0 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 2.0 | 8.0 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 1.8 | 7.4 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 1.7 | 5.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 1.7 | 8.6 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 1.7 | 5.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 1.7 | 6.9 | GO:0019346 | transsulfuration(GO:0019346) |
| 1.7 | 6.7 | GO:0043901 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 1.6 | 4.8 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 1.6 | 7.9 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 1.6 | 4.7 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 1.5 | 9.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 1.5 | 1.5 | GO:0071475 | cellular response to salt stress(GO:0071472) cellular hyperosmotic salinity response(GO:0071475) |
| 1.5 | 5.9 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 1.4 | 22.7 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 1.4 | 7.0 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) |
| 1.4 | 4.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 1.4 | 2.8 | GO:0051594 | detection of chemical stimulus(GO:0009593) detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) detection of stimulus(GO:0051606) |
| 1.4 | 23.0 | GO:0009228 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 1.3 | 6.6 | GO:0051180 | vitamin transport(GO:0051180) |
| 1.3 | 5.3 | GO:0009415 | response to water(GO:0009415) cellular response to water stimulus(GO:0071462) |
| 1.3 | 27.1 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 1.3 | 6.4 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 1.3 | 3.8 | GO:2000284 | positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 1.2 | 7.4 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 1.2 | 3.6 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 1.1 | 4.6 | GO:0015976 | carbon utilization(GO:0015976) |
| 1.1 | 3.3 | GO:0052547 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 1.1 | 3.2 | GO:0061412 | cellular response to amino acid starvation(GO:0034198) cellular alcohol catabolic process(GO:0044109) positive regulation of transcription from RNA polymerase II promoter in response to amino acid starvation(GO:0061412) |
| 1.1 | 8.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 1.1 | 2.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 1.0 | 3.1 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 1.0 | 2.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 1.0 | 5.9 | GO:0009746 | response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 1.0 | 2.9 | GO:0010039 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 1.0 | 1.0 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) regulation of endodeoxyribonuclease activity(GO:0032071) |
| 1.0 | 2.9 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.9 | 10.4 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.9 | 5.5 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.9 | 5.5 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.9 | 13.8 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 0.9 | 4.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.9 | 2.7 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.9 | 22.6 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.9 | 4.5 | GO:0032102 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.9 | 3.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.8 | 2.5 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.8 | 4.2 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.8 | 3.3 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.8 | 0.8 | GO:0051597 | response to methylmercury(GO:0051597) cellular response to methylmercury(GO:0071406) |
| 0.8 | 5.7 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.8 | 3.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.8 | 0.8 | GO:0015688 | iron chelate transport(GO:0015688) |
| 0.8 | 11.0 | GO:0006885 | regulation of pH(GO:0006885) |
| 0.8 | 10.5 | GO:0034087 | establishment of sister chromatid cohesion(GO:0034085) establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.7 | 18.7 | GO:0098660 | inorganic ion transmembrane transport(GO:0098660) |
| 0.7 | 2.2 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.7 | 4.5 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.7 | 2.9 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) |
| 0.7 | 5.9 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.7 | 2.9 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.7 | 3.6 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.7 | 4.2 | GO:0000338 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.7 | 1.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.7 | 3.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.7 | 3.3 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.6 | 3.0 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.6 | 5.5 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.6 | 3.6 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.6 | 1.8 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.6 | 1.8 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.6 | 6.4 | GO:0000436 | carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.6 | 0.6 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) |
| 0.6 | 1.7 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.6 | 4.4 | GO:0006078 | (1->6)-beta-D-glucan biosynthetic process(GO:0006078) beta-glucan biosynthetic process(GO:0051274) |
| 0.6 | 5.5 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.5 | 12.0 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.5 | 0.5 | GO:0071243 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) positive regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061395) cellular response to arsenic-containing substance(GO:0071243) |
| 0.5 | 0.5 | GO:0071941 | urea catabolic process(GO:0043419) nitrogen cycle metabolic process(GO:0071941) |
| 0.5 | 12.3 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.5 | 2.1 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.5 | 20.3 | GO:0071940 | ascospore wall assembly(GO:0030476) spore wall assembly(GO:0042244) spore wall biogenesis(GO:0070590) ascospore wall biogenesis(GO:0070591) fungal-type cell wall assembly(GO:0071940) |
| 0.5 | 4.8 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.5 | 2.0 | GO:0015893 | drug transport(GO:0015893) |
| 0.5 | 9.7 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.5 | 2.4 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.5 | 2.8 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.5 | 7.4 | GO:0007129 | synapsis(GO:0007129) |
| 0.5 | 9.2 | GO:0012501 | apoptotic process(GO:0006915) cell death(GO:0008219) programmed cell death(GO:0012501) |
| 0.5 | 1.4 | GO:0043954 | cellular component maintenance(GO:0043954) |
| 0.4 | 1.3 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of metal ion(GO:0051238) sequestering of iron ion(GO:0097577) |
| 0.4 | 0.4 | GO:0000196 | MAPK cascade involved in cell wall organization or biogenesis(GO:0000196) |
| 0.4 | 5.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.4 | 10.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.4 | 1.6 | GO:0019563 | alditol catabolic process(GO:0019405) glycerol catabolic process(GO:0019563) |
| 0.4 | 1.6 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.4 | 1.2 | GO:0042843 | D-xylose catabolic process(GO:0042843) |
| 0.4 | 0.8 | GO:0009190 | cAMP biosynthetic process(GO:0006171) cyclic nucleotide biosynthetic process(GO:0009190) cyclic purine nucleotide metabolic process(GO:0052652) |
| 0.4 | 1.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.4 | 1.1 | GO:1903138 | inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.4 | 1.8 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.4 | 2.2 | GO:0015851 | nucleobase transport(GO:0015851) |
| 0.4 | 0.4 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.3 | 2.1 | GO:0019932 | calcium-mediated signaling(GO:0019722) second-messenger-mediated signaling(GO:0019932) |
| 0.3 | 1.6 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.3 | 5.7 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.3 | 0.9 | GO:1903727 | positive regulation of phospholipid biosynthetic process(GO:0071073) positive regulation of phospholipid metabolic process(GO:1903727) |
| 0.3 | 1.8 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.3 | 1.5 | GO:0034503 | protein localization to nucleolar rDNA repeats(GO:0034503) |
| 0.3 | 0.9 | GO:0055078 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.3 | 1.2 | GO:0051383 | centromere complex assembly(GO:0034508) kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.3 | 0.9 | GO:0019933 | cAMP-mediated signaling(GO:0019933) cyclic-nucleotide-mediated signaling(GO:0019935) |
| 0.3 | 0.9 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.3 | 3.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.3 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 0.9 | GO:0071050 | snoRNA polyadenylation(GO:0071050) |
| 0.3 | 2.8 | GO:0000753 | cell morphogenesis involved in conjugation with cellular fusion(GO:0000753) |
| 0.3 | 0.8 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.3 | 0.6 | GO:0007534 | gene conversion at mating-type locus(GO:0007534) |
| 0.3 | 1.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.3 | 1.4 | GO:0005977 | glycogen metabolic process(GO:0005977) energy reserve metabolic process(GO:0006112) |
| 0.3 | 2.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.3 | 0.5 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) negative regulation of developmental process(GO:0051093) |
| 0.3 | 3.1 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.3 | 0.8 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.2 | 1.0 | GO:0030835 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) barbed-end actin filament capping(GO:0051016) actin filament capping(GO:0051693) |
| 0.2 | 0.7 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 10.5 | GO:0007124 | pseudohyphal growth(GO:0007124) |
| 0.2 | 1.0 | GO:0009065 | glutamine family amino acid catabolic process(GO:0009065) |
| 0.2 | 0.5 | GO:0061392 | regulation of transcription from RNA polymerase II promoter in response to osmotic stress(GO:0061392) |
| 0.2 | 0.5 | GO:0000709 | meiotic joint molecule formation(GO:0000709) |
| 0.2 | 1.6 | GO:1902532 | negative regulation of intracellular signal transduction(GO:1902532) |
| 0.2 | 1.2 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.2 | 0.7 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.2 | 2.0 | GO:0043487 | regulation of RNA stability(GO:0043487) regulation of mRNA stability(GO:0043488) |
| 0.2 | 0.6 | GO:0006784 | heme a biosynthetic process(GO:0006784) heme a metabolic process(GO:0046160) |
| 0.2 | 0.9 | GO:0043200 | response to amino acid(GO:0043200) |
| 0.2 | 1.7 | GO:0031578 | mitotic spindle orientation checkpoint(GO:0031578) |
| 0.2 | 2.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.2 | 0.4 | GO:1902932 | positive regulation of alcohol biosynthetic process(GO:1902932) |
| 0.2 | 0.6 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.2 | 3.1 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.2 | 5.7 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.2 | 10.1 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.2 | 0.8 | GO:0034486 | amino acid export(GO:0032973) amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 0.2 | 6.2 | GO:0034293 | sexual sporulation(GO:0034293) sexual sporulation resulting in formation of a cellular spore(GO:0043935) |
| 0.2 | 0.6 | GO:0051259 | protein oligomerization(GO:0051259) |
| 0.2 | 0.8 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.2 | 1.0 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.2 | 0.6 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.2 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.6 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.2 | 1.7 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.2 | 2.2 | GO:0046854 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.2 | 1.6 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.2 | 0.4 | GO:0051304 | chromosome separation(GO:0051304) |
| 0.2 | 0.9 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 0.2 | GO:0001178 | regulation of transcriptional start site selection at RNA polymerase II promoter(GO:0001178) |
| 0.2 | 0.5 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.2 | 0.7 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.2 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.2 | 0.5 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.2 | 1.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 1.1 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) manganese ion homeostasis(GO:0055071) |
| 0.2 | 0.3 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.1 | 2.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.9 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) proton-transporting ATP synthase complex assembly(GO:0043461) |
| 0.1 | 3.9 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 2.0 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 0.4 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.1 | 0.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 1.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.6 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.1 | 0.5 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.4 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.1 | 1.4 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 0.2 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 | 0.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 1.2 | GO:0015918 | sterol transport(GO:0015918) |
| 0.1 | 1.1 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.8 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.1 | 0.3 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 0.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.2 | GO:0055067 | monovalent inorganic cation homeostasis(GO:0055067) |
| 0.1 | 1.6 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.3 | GO:0000114 | obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.1 | 0.6 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 1.3 | GO:0043633 | polyadenylation-dependent RNA catabolic process(GO:0043633) |
| 0.1 | 1.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.1 | GO:0016320 | endoplasmic reticulum membrane fusion(GO:0016320) |
| 0.1 | 3.5 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.1 | 0.3 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.1 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.3 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of vesicle targeting, to, from or within Golgi(GO:0048209) regulation of ER to Golgi vesicle-mediated transport(GO:0060628) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 0.9 | GO:0016571 | histone methylation(GO:0016571) |
| 0.1 | 0.2 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 1.2 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.1 | 0.3 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.1 | 0.7 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.2 | GO:0033875 | coenzyme A metabolic process(GO:0015936) nucleoside bisphosphate metabolic process(GO:0033865) ribonucleoside bisphosphate metabolic process(GO:0033875) purine nucleoside bisphosphate metabolic process(GO:0034032) |
| 0.1 | 0.2 | GO:0097033 | mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.2 | GO:0035690 | cellular response to drug(GO:0035690) regulation of response to drug(GO:2001023) positive regulation of response to drug(GO:2001025) regulation of cellular response to drug(GO:2001038) positive regulation of cellular response to drug(GO:2001040) |
| 0.1 | 1.8 | GO:0048646 | anatomical structure formation involved in morphogenesis(GO:0048646) |
| 0.0 | 0.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.0 | GO:0032845 | negative regulation of telomere maintenance(GO:0032205) negative regulation of homeostatic process(GO:0032845) |
| 0.0 | 0.4 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) copper ion homeostasis(GO:0055070) |
| 0.0 | 2.2 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.1 | GO:0051126 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.1 | GO:0070550 | rDNA condensation(GO:0070550) |
| 0.0 | 0.3 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.1 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:0045861 | negative regulation of proteolysis(GO:0045861) |
| 0.0 | 0.1 | GO:0072511 | zinc II ion transport(GO:0006829) divalent metal ion transport(GO:0070838) divalent inorganic cation transport(GO:0072511) |
| 0.0 | 0.0 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0031081 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.0 | 0.1 | GO:0009209 | CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) |
| 0.0 | 0.1 | GO:0000147 | actin cortical patch assembly(GO:0000147) actin cortical patch organization(GO:0044396) |
| 0.0 | 0.1 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 | 0.1 | GO:0051666 | actin cortical patch localization(GO:0051666) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0051181 | cofactor transport(GO:0051181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 15.2 | GO:0030287 | cell wall-bounded periplasmic space(GO:0030287) |
| 3.5 | 7.0 | GO:0032126 | eisosome(GO:0032126) |
| 3.2 | 9.5 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 2.6 | 7.8 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 2.6 | 10.2 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 2.1 | 15.0 | GO:0034657 | GID complex(GO:0034657) |
| 1.7 | 5.2 | GO:0000111 | nucleotide-excision repair factor 2 complex(GO:0000111) |
| 1.7 | 6.7 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 1.6 | 4.9 | GO:0045269 | mitochondrial proton-transporting ATP synthase, central stalk(GO:0005756) proton-transporting ATP synthase, central stalk(GO:0045269) |
| 1.6 | 4.8 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial oxoglutarate dehydrogenase complex(GO:0009353) dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 1.5 | 4.6 | GO:0000274 | mitochondrial proton-transporting ATP synthase, stator stalk(GO:0000274) proton-transporting ATP synthase, stator stalk(GO:0045265) |
| 1.4 | 5.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 1.4 | 2.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 1.4 | 4.2 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 1.4 | 16.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 1.3 | 8.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 1.3 | 8.0 | GO:0045263 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 1.2 | 2.5 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 1.2 | 4.6 | GO:0000817 | COMA complex(GO:0000817) |
| 1.1 | 7.7 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 1.1 | 5.3 | GO:0044426 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 1.1 | 4.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 1.0 | 11.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 1.0 | 4.1 | GO:0042597 | periplasmic space(GO:0042597) |
| 1.0 | 2.0 | GO:0070985 | TFIIK complex(GO:0070985) |
| 1.0 | 53.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 1.0 | 8.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.9 | 2.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.9 | 22.4 | GO:0042764 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.9 | 6.0 | GO:0005619 | ascospore wall(GO:0005619) spore wall(GO:0031160) |
| 0.8 | 35.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.8 | 3.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.7 | 3.6 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) mitochondrial inner membrane protein insertion complex(GO:0042721) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.7 | 4.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.7 | 6.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.6 | 1.3 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.6 | 1.3 | GO:0001400 | mating projection base(GO:0001400) |
| 0.6 | 1.8 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.6 | 5.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.5 | 2.1 | GO:0033309 | SBF transcription complex(GO:0033309) |
| 0.5 | 6.4 | GO:0090544 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.5 | 15.9 | GO:0000322 | storage vacuole(GO:0000322) lytic vacuole(GO:0000323) fungal-type vacuole(GO:0000324) |
| 0.4 | 74.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.4 | 2.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.4 | 1.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.4 | 1.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.4 | 1.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.4 | 1.5 | GO:0033551 | monopolin complex(GO:0033551) |
| 0.4 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.4 | 10.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.4 | 3.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.3 | 12.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.3 | 1.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.3 | 1.6 | GO:0070772 | PAS complex(GO:0070772) |
| 0.3 | 1.6 | GO:0035361 | Cul8-RING ubiquitin ligase complex(GO:0035361) |
| 0.3 | 15.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.2 | 1.0 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.2 | 1.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 7.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.2 | 0.7 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.2 | 3.1 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 3.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.2 | 0.2 | GO:0038201 | TOR complex(GO:0038201) |
| 0.2 | 1.0 | GO:0000133 | polarisome(GO:0000133) |
| 0.2 | 0.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 29.3 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.2 | 0.4 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) catalytic step 2 spliceosome(GO:0071013) |
| 0.2 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.7 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) Sec62/Sec63 complex(GO:0031207) |
| 0.2 | 2.2 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 2.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.3 | GO:0030907 | MBF transcription complex(GO:0030907) |
| 0.1 | 1.6 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.6 | GO:0044232 | ERMES complex(GO:0032865) organelle membrane contact site(GO:0044232) ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.3 | GO:0000417 | HIR complex(GO:0000417) |
| 0.1 | 0.9 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 1.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.7 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 0.5 | GO:0000144 | cellular bud neck septin ring(GO:0000144) |
| 0.1 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.7 | GO:0098797 | plasma membrane protein complex(GO:0098797) |
| 0.1 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 45.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 2.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0031499 | TRAMP complex(GO:0031499) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 0.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0000818 | MIS12/MIND type complex(GO:0000444) nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.1 | GO:0016587 | Isw1 complex(GO:0016587) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0000799 | condensin complex(GO:0000796) nuclear condensin complex(GO:0000799) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 26.6 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 7.4 | 22.1 | GO:0016406 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 7.0 | 21.1 | GO:0016208 | AMP binding(GO:0016208) |
| 6.1 | 24.4 | GO:0005537 | mannose binding(GO:0005537) |
| 6.0 | 30.1 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 5.3 | 16.0 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 4.6 | 13.9 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 4.4 | 13.1 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 4.2 | 16.8 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 3.5 | 13.9 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 3.4 | 10.2 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 3.2 | 9.5 | GO:0005536 | glucose binding(GO:0005536) |
| 3.1 | 34.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 3.1 | 9.3 | GO:0008137 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 2.9 | 8.8 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 2.7 | 24.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 2.4 | 4.8 | GO:0015116 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 1.8 | 7.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 1.7 | 6.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 1.7 | 13.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.6 | 11.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 1.5 | 16.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 1.5 | 5.9 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 1.4 | 5.7 | GO:0042927 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 1.4 | 7.0 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 1.4 | 6.9 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 1.3 | 20.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 1.2 | 11.0 | GO:0000149 | SNARE binding(GO:0000149) |
| 1.2 | 1.2 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 1.2 | 6.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 1.2 | 3.6 | GO:0048038 | quinone binding(GO:0048038) |
| 1.1 | 3.3 | GO:0030414 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 1.1 | 6.6 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 1.1 | 3.2 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 1.0 | 7.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.0 | 23.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 1.0 | 46.7 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.9 | 13.9 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.9 | 5.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.8 | 6.7 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.8 | 5.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.8 | 3.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.8 | 0.8 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.7 | 4.4 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.7 | 2.9 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.7 | 2.9 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.7 | 2.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.6 | 3.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.6 | 5.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.6 | 1.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.6 | 1.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.6 | 2.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.6 | 1.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.6 | 1.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.6 | 1.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.6 | 1.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.6 | 1.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.6 | 7.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.6 | 1.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.6 | 1.7 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.5 | 4.8 | GO:0016723 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.5 | 1.0 | GO:0019903 | phosphatase binding(GO:0019902) protein phosphatase binding(GO:0019903) |
| 0.5 | 2.5 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.5 | 1.5 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) |
| 0.5 | 2.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.5 | 3.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.4 | 2.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.4 | 1.3 | GO:0015103 | inorganic anion transmembrane transporter activity(GO:0015103) |
| 0.4 | 1.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.4 | 0.8 | GO:0015151 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.4 | 4.5 | GO:0004872 | receptor activity(GO:0004872) transmembrane signaling receptor activity(GO:0004888) signaling receptor activity(GO:0038023) molecular transducer activity(GO:0060089) transmembrane receptor activity(GO:0099600) |
| 0.4 | 5.9 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.4 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.4 | 8.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.4 | 2.2 | GO:0005102 | receptor binding(GO:0005102) |
| 0.4 | 6.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.4 | 1.8 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.4 | 1.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.4 | 11.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.3 | 1.0 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.3 | 2.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.3 | 3.3 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.3 | 1.7 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.3 | 1.0 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.3 | 1.5 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.3 | 3.3 | GO:0022838 | substrate-specific channel activity(GO:0022838) |
| 0.3 | 1.5 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.3 | 0.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 0.9 | GO:0008972 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.3 | 0.6 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) TFIIH-class transcription factor binding(GO:0001097) |
| 0.3 | 3.3 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.2 | 3.2 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.2 | 1.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 2.1 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 7.2 | GO:0001159 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.2 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 2.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 1.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 0.7 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.2 | 1.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.2 | 4.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 1.7 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.2 | 1.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 2.0 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.2 | 1.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.2 | 1.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 11.3 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.2 | 1.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 3.0 | GO:0016769 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.2 | 0.7 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.2 | 0.9 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.2 | 3.4 | GO:0005342 | organic acid transmembrane transporter activity(GO:0005342) |
| 0.2 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 0.9 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 0.5 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.2 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 0.5 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.2 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 2.2 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.2 | 0.3 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 1.2 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 8.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 8.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.1 | 0.4 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.1 | 1.3 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.1 | 1.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.7 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.1 | 0.4 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.1 | 1.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.7 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 1.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.7 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.8 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.1 | 1.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 2.6 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.1 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 7.3 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.1 | 0.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 0.2 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.1 | 0.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) flap endonuclease activity(GO:0048256) |
| 0.1 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.3 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 0.1 | 0.2 | GO:0043177 | amino acid binding(GO:0016597) carboxylic acid binding(GO:0031406) organic acid binding(GO:0043177) |
| 0.1 | 0.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 0.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 5.7 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.4 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 1.1 | GO:0008233 | peptidase activity(GO:0008233) |
| 0.0 | 0.1 | GO:0034595 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) phosphatidylinositol phosphate phosphatase activity(GO:0052866) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.6 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.1 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 24.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 1.2 | 7.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.9 | 2.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.9 | 8.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.9 | 2.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.4 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.4 | 1.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.3 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 368.6 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 0.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 0.3 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 23.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 1.2 | 4.7 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.8 | 2.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.5 | 1.6 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.5 | 2.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.5 | 3.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.5 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 0.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 368.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | Genes involved in Transmembrane transport of small molecules |
| 0.0 | 0.3 | REACTOME POST TRANSLATIONAL PROTEIN MODIFICATION | Genes involved in Post-translational protein modification |
| 0.0 | 0.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |