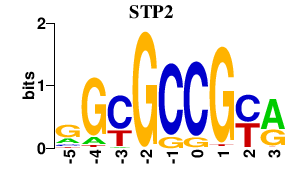
Results for STP2
Z-value: 0.75
Transcription factors associated with STP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STP2
|
S000001048 | Transcription factor |
Activity-expression correlation:
Activity profile of STP2 motif
Sorted Z-values of STP2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YOR348C | 6.39 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YCL025C | 5.79 |
AGP1
|
Low-affinity amino acid permease with broad substrate range, involved in uptake of asparagine, glutamine, and other amino acids; expression is regulated by the SPS plasma membrane amino acid sensor system (Ssy1p-Ptr3p-Ssy5p) |
|
| YKL217W | 4.68 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YDR343C | 4.25 |
HXT6
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt7p, expressed at high basal levels relative to other HXTs, repression of expression by high glucose requires SNF3 |
|
| YJL116C | 4.08 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YLL053C | 3.77 |
Putative protein; in the Sigma 1278B strain background YLL053C is contiguous with AQY2 which encodes an aquaporin |
||
| YDR277C | 3.64 |
MTH1
|
Negative regulator of the glucose-sensing signal transduction pathway, required for repression of transcription by Rgt1p; interacts with Rgt1p and the Snf3p and Rgt2p glucose sensors; phosphorylated by Yck1p, triggering Mth1p degradation |
|
| YHR033W | 3.47 |
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm |
||
| YBR296C | 3.33 |
PHO89
|
Na+/Pi cotransporter, active in early growth phase; similar to phosphate transporters of Neurospora crassa; transcription regulated by inorganic phosphate concentrations and Pho4p |
|
| YLL052C | 3.32 |
AQY2
|
Water channel that mediates the transport of water across cell membranes, only expressed in proliferating cells, controlled by osmotic signals, may be involved in freeze tolerance; disrupted by a stop codon in many S. cerevisiae strains |
|
| YFR053C | 3.32 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YDR536W | 3.31 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YLR327C | 3.19 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YMR251W | 2.85 |
GTO3
|
Omega class glutathione transferase; putative cytosolic localization |
|
| YJR115W | 2.79 |
Putative protein of unknown function |
||
| YOR072W | 2.71 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOR072W-A; diploid deletion strains are methotrexate, paraquat and wortmannin sensitive |
||
| YKR097W | 2.68 |
PCK1
|
Phosphoenolpyruvate carboxykinase, key enzyme in gluconeogenesis, catalyzes early reaction in carbohydrate biosynthesis, glucose represses transcription and accelerates mRNA degradation, regulated by Mcm1p and Cat8p, located in the cytosol |
|
| YAL039C | 2.59 |
CYC3
|
Cytochrome c heme lyase (holocytochrome c synthase), attaches heme to apo-cytochrome c (Cyc1p or Cyc7p) in the mitochondrial intermembrane space; human ortholog may have a role in microphthalmia with linear skin defects (MLS) |
|
| YCR021C | 2.56 |
HSP30
|
Hydrophobic plasma membrane localized, stress-responsive protein that negatively regulates the H(+)-ATPase Pma1p; induced by heat shock, ethanol treatment, weak organic acid, glucose limitation, and entry into stationary phase |
|
| YMR306C-A | 2.38 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR071C | 2.35 |
NRT1
|
High-affinity nicotinamide riboside transporter; also transports thiamine with low affinity; shares sequence similarity with Thi7p and Thi72p; proposed to be involved in 5-fluorocytosine sensitivity |
|
| YER065C | 2.29 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YML089C | 2.21 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression induced by calcium shortage |
||
| YKL148C | 2.20 |
SDH1
|
Flavoprotein subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
|
| YOR072W-A | 2.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YOR072W; originally identified by fungal homology and RT-PCR |
||
| YEL065W | 2.17 |
SIT1
|
Ferrioxamine B transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; transcription is induced during iron deprivation and diauxic shift; potentially phosphorylated by Cdc28p |
|
| YMR081C | 2.17 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YIL055C | 2.13 |
Putative protein of unknown function |
||
| YER185W | 2.12 |
PUG1
|
Plasma membrane protein with roles in the uptake of protoprophyrin IX and the efflux of heme; expression is induced under both low-heme and low-oxygen conditions |
|
| YBL075C | 2.10 |
SSA3
|
ATPase involved in protein folding and the response to stress; plays a role in SRP-dependent cotranslational protein-membrane targeting and translocation; member of the heat shock protein 70 (HSP70) family; localized to the cytoplasm |
|
| YLR267W | 2.08 |
BOP2
|
Protein of unknown function, overproduction suppresses a pam1 slv3 double null mutation |
|
| YDR216W | 2.07 |
ADR1
|
Carbon source-responsive zinc-finger transcription factor, required for transcription of the glucose-repressed gene ADH2, of peroxisomal protein genes, and of genes required for ethanol, glycerol, and fatty acid utilization |
|
| YNL125C | 2.07 |
ESBP6
|
Protein with similarity to monocarboxylate permeases, appears not to be involved in transport of monocarboxylates such as lactate, pyruvate or acetate across the plasma membrane |
|
| YOL081W | 2.02 |
IRA2
|
GTPase-activating protein that negatively regulates RAS by converting it from the GTP- to the GDP-bound inactive form, required for reducing cAMP levels under nutrient limiting conditions, has similarity to Ira1p and human neurofibromin |
|
| YLR122C | 2.01 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR123C |
||
| YNL305C | 2.01 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YNL305C is not an essential gene |
||
| YOR100C | 2.01 |
CRC1
|
Mitochondrial inner membrane carnitine transporter, required for carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria during fatty acid beta-oxidation |
|
| YKL093W | 2.00 |
MBR1
|
Protein involved in mitochondrial functions and stress response; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants |
|
| YPL024W | 1.96 |
RMI1
|
Involved in response to DNA damage; null mutants have increased rates of recombination and delayed S phase; interacts physically and genetically with Sgs1p (RecQ family member) and Top3p (topoisomerase III) |
|
| YJL045W | 1.95 |
Minor succinate dehydrogenase isozyme; homologous to Sdh1p, the major isozyme reponsible for the oxidation of succinate and transfer of electrons to ubiquinone; induced during the diauxic shift in a Cat8p-dependent manner |
||
| YMR135C | 1.94 |
GID8
|
Protein of unknown function, involved in proteasome-dependent catabolite inactivation of fructose-1,6-bisphosphatase; contains LisH and CTLH domains, like Vid30p; dosage-dependent regulator of START |
|
| YPL223C | 1.92 |
GRE1
|
Hydrophilin of unknown function; stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YKL109W | 1.91 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YMR194C-B | 1.87 |
Putative protein of unknown function |
||
| YIL160C | 1.85 |
POT1
|
3-ketoacyl-CoA thiolase with broad chain length specificity, cleaves 3-ketoacyl-CoA into acyl-CoA and acetyl-CoA during beta-oxidation of fatty acids |
|
| YGR067C | 1.81 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YLR296W | 1.81 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPL134C | 1.81 |
ODC1
|
Mitochondrial inner membrane transporter, exports 2-oxoadipate and 2-oxoglutarate from the mitochondrial matrix to the cytosol for lysine and glutamate biosynthesis and lysine catabolism; suppresses, in multicopy, an fmc1 null mutation |
|
| YBR132C | 1.77 |
AGP2
|
High affinity polyamine permease, preferentially uses spermidine over putrescine; expression is down-regulated by osmotic stress; plasma membrane carnitine transporter, also functions as a low-affinity amino acid permease |
|
| YEL044W | 1.77 |
IES6
|
Protein that associates with the INO80 chromatin remodeling complex under low-salt conditions |
|
| YDR096W | 1.76 |
GIS1
|
JmjC domain-containing histone demethylase; transcription factor involved in the expression of genes during nutrient limitation; also involved in the negative regulation of DPP1 and PHR1 |
|
| YCR010C | 1.75 |
ADY2
|
Acetate transporter required for normal sporulation; phosphorylated in mitochondria |
|
| YJR128W | 1.72 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF RSF2 |
||
| YCL042W | 1.71 |
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm |
||
| YDR406W | 1.71 |
PDR15
|
Plasma membrane ATP binding cassette (ABC) transporter, multidrug transporter and general stress response factor implicated in cellular detoxification; regulated by Pdr1p, Pdr3p and Pdr8p; promoter contains a PDR responsive element |
|
| YOR343C | 1.71 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YDR342C | 1.70 |
HXT7
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels |
|
| YLR123C | 1.70 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR122C; contains characteristic aminoacyl-tRNA motif |
||
| YNR064C | 1.69 |
Epoxide hydrolase, member of the alpha/beta hydrolase fold family; may have a role in detoxification of epoxides |
||
| YPL026C | 1.68 |
SKS1
|
Putative serine/threonine protein kinase; involved in the adaptation to low concentrations of glucose independent of the SNF3 regulated pathway |
|
| YCL040W | 1.61 |
GLK1
|
Glucokinase, catalyzes the phosphorylation of glucose at C6 in the first irreversible step of glucose metabolism; one of three glucose phosphorylating enzymes; expression regulated by non-fermentable carbon sources |
|
| YPR184W | 1.60 |
GDB1
|
Glycogen debranching enzyme containing glucanotranferase and alpha-1,6-amyloglucosidase activities, required for glycogen degradation; phosphorylated in mitochondria |
|
| YCR005C | 1.57 |
CIT2
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate, peroxisomal isozyme involved in glyoxylate cycle; expression is controlled by Rtg1p and Rtg2p transcription factors |
|
| YER184C | 1.57 |
Putative zinc cluster protein; deletion confers sensitivity to Calcufluor white, and prevents growth on glycerol or lactate as sole carbon source |
||
| YER004W | 1.55 |
FMP52
|
Protein of unknown function, localized to the mitochondrial outer membrane; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YMR181C | 1.54 |
Protein of unknown function; mRNA transcribed as part of a bicistronic transcript with a predicted transcriptional repressor RGM1/YMR182C; mRNA is destroyed by nonsense-mediated decay (NMD); YMR181C is not an essential gene |
||
| YDL169C | 1.53 |
UGX2
|
Protein of unknown function, transcript accumulates in response to any combination of stress conditions |
|
| YLR295C | 1.53 |
ATP14
|
Subunit h of the F0 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YEL011W | 1.53 |
GLC3
|
Glycogen branching enzyme, involved in glycogen accumulation; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YCR091W | 1.52 |
KIN82
|
Putative serine/threonine protein kinase, most similar to cyclic nucleotide-dependent protein kinase subfamily and the protein kinase C subfamily |
|
| YDL194W | 1.48 |
SNF3
|
Plasma membrane glucose sensor that regulates glucose transport; has 12 predicted transmembrane segments; long cytoplasmic C-terminal tail is required for low glucose induction of hexose transporter genes HXT2 and HXT4 |
|
| YDR542W | 1.48 |
PAU10
|
Hypothetical protein |
|
| YPR027C | 1.48 |
Putative protein of unknown function |
||
| YPL119C | 1.48 |
DBP1
|
Putative ATP-dependent RNA helicase of the DEAD-box protein family; mutants show reduced stability of the 40S ribosomal subunit scanning through 5' untranslated regions of mRNAs |
|
| YLR332W | 1.47 |
MID2
|
O-glycosylated plasma membrane protein that acts as a sensor for cell wall integrity signaling and activates the pathway; interacts with Rom2p, a guanine nucleotide exchange factor for Rho1p, and with cell integrity pathway protein Zeo1p |
|
| YGL192W | 1.45 |
IME4
|
Probable mRNA N6-adenosine methyltransferase that is required for IME1 transcript accumulation and for sporulation; expression is induced in starved MATa/MAT alpha diploid cells |
|
| YGR032W | 1.43 |
GSC2
|
Catalytic subunit of 1,3-beta-glucan synthase, involved in formation of the inner layer of the spore wall; activity positively regulated by Rho1p and negatively by Smk1p; has similarity to an alternate catalytic subunit, Fks1p (Gsc1p) |
|
| YGL193C | 1.43 |
Haploid-specific gene repressed by a1-alpha2, turned off in sir3 null strains, absence enhances the sensitivity of rad52-327 cells to campothecin almost 100-fold |
||
| YPL058C | 1.42 |
PDR12
|
Plasma membrane ATP-binding cassette (ABC) transporter, weak-acid-inducible multidrug transporter required for weak organic acid resistance; induced by sorbate and benzoate and regulated by War1p; mutants exhibit sorbate hypersensitivity |
|
| YFL059W | 1.41 |
SNZ3
|
Member of a stationary phase-induced gene family; transcription of SNZ2 is induced prior to diauxic shift, and also in the absence of thiamin in a Thi2p-dependent manner; forms a coregulated gene pair with SNO3 |
|
| YLR331C | 1.40 |
JIP3
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 98% of ORF overlaps the verified gene MID2 |
|
| YLR438W | 1.38 |
CAR2
|
L-ornithine transaminase (OTAse), catalyzes the second step of arginine degradation, expression is dually-regulated by allophanate induction and a specific arginine induction process; not nitrogen catabolite repression sensitive |
|
| YFL030W | 1.37 |
AGX1
|
Alanine:glyoxylate aminotransferase (AGT), catalyzes the synthesis of glycine from glyoxylate, which is one of three pathways for glycine biosynthesis in yeast; has similarity to mammalian and plant alanine:glyoxylate aminotransferases |
|
| YOR383C | 1.36 |
FIT3
|
Mannoprotein that is incorporated into the cell wall via a glycosylphosphatidylinositol (GPI) anchor, involved in the retention of siderophore-iron in the cell wall |
|
| YPR196W | 1.32 |
Putative maltose activator |
||
| YJR078W | 1.31 |
BNA2
|
Putative tryptophan 2,3-dioxygenase or indoleamine 2,3-dioxygenase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p and Aft2p |
|
| YDL223C | 1.31 |
HBT1
|
Substrate of the Hub1p ubiquitin-like protein that localizes to the shmoo tip (mating projection); mutants are defective for mating projection formation, thereby implicating Hbt1p in polarized cell morphogenesis |
|
| YIL162W | 1.29 |
SUC2
|
Invertase, sucrose hydrolyzing enzyme; a secreted, glycosylated form is regulated by glucose repression, and an intracellular, nonglycosylated enzyme is produced constitutively |
|
| YNL333W | 1.29 |
SNZ2
|
Member of a stationary phase-induced gene family; transcription of SNZ2 is induced prior to diauxic shift, and also in the absence of thiamin in a Thi2p-dependent manner; forms a coregulated gene pair with SNO2; interacts with Thi11p |
|
| YFL064C | 1.28 |
Putative protein of unknown function |
||
| YEL028W | 1.28 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YML090W | 1.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YML089C; exhibits growth defect on a non-fermentable (respiratory) carbon source |
||
| YMR306W | 1.25 |
FKS3
|
Protein involved in spore wall assembly, has similarity to 1,3-beta-D-glucan synthase catalytic subunits Fks1p and Gsc2p; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YNL242W | 1.25 |
ATG2
|
Peripheral membrane protein required for vesicle formation during autophagy, pexophagy, and the cytoplasm-to-vaucole targeting (Cvt) pathway; involved in Atg9p cycling between the pre-autophagosomal structure and mitochondria |
|
| YKL085W | 1.24 |
MDH1
|
Mitochondrial malate dehydrogenase, catalyzes interconversion of malate and oxaloacetate; involved in the tricarboxylic acid (TCA) cycle; phosphorylated |
|
| YPR065W | 1.23 |
ROX1
|
Heme-dependent repressor of hypoxic genes; contains an HMG domain that is responsible for DNA bending activity |
|
| YDL215C | 1.23 |
GDH2
|
NAD(+)-dependent glutamate dehydrogenase, degrades glutamate to ammonia and alpha-ketoglutarate; expression sensitive to nitrogen catabolite repression and intracellular ammonia levels |
|
| YBR077C | 1.22 |
SLM4
|
Component of the EGO complex, which is involved in the regulation of microautophagy, and of the GSE complex, which is required for proper sorting of amino acid permease Gap1p; gene exhibits synthetic genetic interaction with MSS4 |
|
| YCL054W | 1.22 |
SPB1
|
AdoMet-dependent methyltransferase involved in rRNA processing and 60S ribosomal subunit maturation; methylates G2922 in the tRNA docking site of the large subunit rRNA and in the absence of snR52, U2921; suppressor of PAB1 mutants |
|
| YFL024C | 1.22 |
EPL1
|
Component of NuA4, which is an essential histone H4/H2A acetyltransferase complex; homologous to Drosophila Enhancer of Polycomb |
|
| YOR178C | 1.21 |
GAC1
|
Regulatory subunit for Glc7p type-1 protein phosphatase (PP1), tethers Glc7p to Gsy2p glycogen synthase, binds Hsf1p heat shock transcription factor, required for induction of some HSF-regulated genes under heat shock |
|
| YOL084W | 1.21 |
PHM7
|
Protein of unknown function, expression is regulated by phosphate levels; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery and vacuole |
|
| YPL135W | 1.21 |
ISU1
|
Conserved protein of the mitochondrial matrix, performs a scaffolding function during assembly of iron-sulfur clusters, interacts physically and functionally with yeast frataxin (Yfh1p); isu1 isu2 double mutant is inviable |
|
| YLR402W | 1.21 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDL011C | 1.20 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the uncharacterized ORF YDL010W |
||
| YPR013C | 1.20 |
Putative zinc finger protein; YPR013C is not an essential gene |
||
| YNL179C | 1.17 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YJL199C | 1.16 |
MBB1
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; protein detected in large-scale protein-protein interaction studies |
|
| YDL010W | 1.15 |
GRX6
|
Cis-golgi localized monothiol glutaredoxin that binds an iron-sulfur cluster; more similar in activity to dithiol than other monothiol glutaredoxins; involved in the oxidative stress response; functional overlap with GRX6 |
|
| YIL113W | 1.15 |
SDP1
|
Stress-inducible dual-specificity MAP kinase phosphatase, negatively regulates Slt2p MAP kinase by direct dephosphorylation, diffuse localization under normal conditions shifts to punctate localization after heat shock |
|
| YMR206W | 1.13 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YLR437C-A | 1.12 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF CAR2/YLR438W |
||
| YDL210W | 1.12 |
UGA4
|
Permease that serves as a gamma-aminobutyrate (GABA) transport protein involved in the utilization of GABA as a nitrogen source; catalyzes the transport of putrescine and delta-aminolevulinic acid (ALA); localized to the vacuolar membrane |
|
| YNL180C | 1.12 |
RHO5
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely involved in protein kinase C (Pkc1p)-dependent signal transduction pathway that controls cell integrity |
|
| YBL064C | 1.11 |
PRX1
|
Mitochondrial peroxiredoxin (1-Cys Prx) with thioredoxin peroxidase activity, has a role in reduction of hydroperoxides; induced during respiratory growth and under conditions of oxidative stress; phosphorylated |
|
| YAR069C | 1.11 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR036W-A | 1.11 |
Protein of unknown function; transcription is regulated by Pdr1p |
||
| YLR403W | 1.10 |
SFP1
|
Transcription factor that controls expression of many ribosome biogenesis genes in response to nutrients and stress, regulates G2/M transitions during mitotic cell cycle and DNA-damage response, involved in cell size modulation |
|
| YDR092W | 1.10 |
UBC13
|
Ubiquitin-conjugating enzyme involved in the error-free DNA postreplication repair pathway; interacts with Mms2p to assemble ubiquitin chains at the Ub Lys-63 residue; DNA damage triggers redistribution from the cytoplasm to the nucleus |
|
| YLR304C | 1.10 |
ACO1
|
Aconitase, required for the tricarboxylic acid (TCA) cycle and also independently required for mitochondrial genome maintenance; phosphorylated; component of the mitochondrial nucleoid; mutation leads to glutamate auxotrophy |
|
| YNL014W | 1.10 |
HEF3
|
Translational elongation factor EF-3; paralog of YEF3 and member of the ABC superfamily; stimulates EF-1 alpha-dependent binding of aminoacyl-tRNA by the ribosome; normally expressed in zinc deficient cells |
|
| YNL019C | 1.10 |
Putative protein of unknown function |
||
| YIL057C | 1.10 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YMR195W | 1.10 |
ICY1
|
Protein of unknown function, required for viability in rich media of cells lacking mitochondrial DNA; mutants have an invasive growth defect with elongated morphology; induced by amino acid starvation |
|
| YFL063W | 1.09 |
Dubious open reading frame, based on available experimental and comparative sequence data |
||
| YKL147C | 1.09 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; partially overlaps the verified gene AVT3 |
||
| YAR070C | 1.09 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPL092W | 1.08 |
SSU1
|
Plasma membrane sulfite pump involved in sulfite metabolism and required for efficient sulfite efflux; major facilitator superfamily protein |
|
| YNL036W | 1.07 |
NCE103
|
Carbonic anhydrase; poorly transcribed under aerobic conditions and at an undetectable level under anaerobic conditions; involved in non-classical protein export pathway |
|
| YPR150W | 1.07 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene SUE1/YPR151C |
||
| YMR250W | 1.07 |
GAD1
|
Glutamate decarboxylase, converts glutamate into gamma-aminobutyric acid (GABA) during glutamate catabolism; involved in response to oxidative stress |
|
| YGL255W | 1.07 |
ZRT1
|
High-affinity zinc transporter of the plasma membrane, responsible for the majority of zinc uptake; transcription is induced under low-zinc conditions by the Zap1p transcription factor |
|
| YAR053W | 1.06 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YMR256C | 1.06 |
COX7
|
Subunit VII of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain |
|
| YKR009C | 1.06 |
FOX2
|
Multifunctional enzyme of the peroxisomal fatty acid beta-oxidation pathway; has 3-hydroxyacyl-CoA dehydrogenase and enoyl-CoA hydratase activities |
|
| YNR002C | 1.05 |
ATO2
|
Putative transmembrane protein involved in export of ammonia, a starvation signal that promotes cell death in aging colonies; phosphorylated in mitochondria; member of the TC 9.B.33 YaaH family; homolog of Ady2p and Y. lipolytica Gpr1p |
|
| YHR211W | 1.04 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YKL146W | 1.04 |
AVT3
|
Vacuolar transporter, exports large neutral amino acids from the vacuole; member of a family of seven S. cerevisiae genes (AVT1-7) related to vesicular GABA-glycine transporters |
|
| YJL093C | 1.03 |
TOK1
|
Outward-rectifier potassium channel of the plasma membrane with two pore domains in tandem, each of which forms a functional channel permeable to potassium; carboxy tail functions to prevent inner gate closures; target of K1 toxin |
|
| YIL045W | 1.03 |
PIG2
|
Putative type-1 protein phosphatase targeting subunit that tethers Glc7p type-1 protein phosphatase to Gsy2p glycogen synthase |
|
| YMR272C | 1.03 |
SCS7
|
Sphingolipid alpha-hydroxylase, functions in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids, has both cytochrome b5-like and hydroxylase/desaturase domains, not essential for growth |
|
| YLR125W | 1.03 |
Putative protein of unknown function; mutant has decreased Ty3 transposition; YLR125W is not an essential gene |
||
| YER158C | 1.02 |
Protein of unknown function, has similarity to Afr1p; potentially phosphorylated by Cdc28p |
||
| YBR047W | 1.02 |
FMP23
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YPR030W | 1.02 |
CSR2
|
Nuclear protein with a potential regulatory role in utilization of galactose and nonfermentable carbon sources; overproduction suppresses the lethality at high temperature of a chs5 spa2 double null mutation; potential Cdc28p substrate |
|
| YBR147W | 1.02 |
RTC2
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays fluconazole resistance and suppresses cdc13-1 temperature sensitivity |
|
| YMR316C-B | 1.01 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR541C | 1.01 |
Putative dihydrokaempferol 4-reductase |
||
| YFL060C | 1.01 |
SNO3
|
Protein of unknown function, nearly identical to Sno2p; expression is induced before the diauxic shift and also in the absence of thiamin |
|
| YMR279C | 1.00 |
Putative protein of unknown function; identified as a heat-induced gene in a high-throughout screen; YMR279C is not an essential gene |
||
| YER014C-A | 0.99 |
BUD25
|
Protein involved in bud-site selection; diploid mutants display a random budding pattern instead of the wild-type bipolar pattern |
|
| YDL214C | 0.99 |
PRR2
|
Serine/threonine protein kinase that inhibits pheromone induced signalling downstream of MAPK, possibly at the level of the Ste12p transcription factor |
|
| YKL163W | 0.99 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YPL111W | 0.99 |
CAR1
|
Arginase, responsible for arginine degradation, expression responds to both induction by arginine and nitrogen catabolite repression; disruption enhances freeze tolerance |
|
| YPL017C | 0.98 |
IRC15
|
Putative S-adenosylmethionine-dependent methyltransferase of the seven beta-strand family, required for accurate meiotic chromosome segregation; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YOR346W | 0.97 |
REV1
|
Deoxycytidyl transferase, forms a complex with the subunits of DNA polymerase zeta, Rev3p and Rev7p; involved in repair of abasic sites in damaged DNA |
|
| YLR236C | 0.97 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNL277W | 0.96 |
MET2
|
L-homoserine-O-acetyltransferase, catalyzes the conversion of homoserine to O-acetyl homoserine which is the first step of the methionine biosynthetic pathway |
|
| YOR345C | 0.96 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene REV1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YCL038C | 0.96 |
ATG22
|
Vacuolar integral membrane protein required for efflux of amino acids during autophagic body breakdown in the vacuole; null mutation causes a gradual loss of viability during starvation |
|
| YBR051W | 0.95 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partiallly overlaps the REG2/YBR050C regulatory subunit of the Glc7p type-1 protein phosphatase |
||
| YOR152C | 0.94 |
Putative protein of unknown function; has no similarity to any known protein; YOR152C is not an essential gene |
||
| YBR105C | 0.94 |
VID24
|
Peripheral membrane protein located at Vid (vacuole import and degradation) vesicles; regulates fructose-1,6-bisphosphatase (FBPase) targeting to the vacuole; involved in proteasome-dependent catabolite degradation of FBPase |
|
| YNL334C | 0.94 |
SNO2
|
Protein of unknown function, nearly identical to Sno3p; expression is induced before the diauxic shift and also in the absence of thiamin |
|
| YLR094C | 0.92 |
GIS3
|
Protein of unknown function |
|
| YDL012C | 0.92 |
Plasma membrane protein of unknown function; YDL012C is not an essential gene |
||
| YOL051W | 0.92 |
GAL11
|
Subunit of the RNA polymerase II mediator complex; associates with core polymerase subunits to form the RNA polymerase II holoenzyme; affects transcription by acting as target of activators and repressors |
|
| YBR046C | 0.92 |
ZTA1
|
Zeta-crystallin homolog, found in the cytoplasm and nucleus; has similarity to E. coli quinone oxidoreductase and to human zeta-crystallin, which has quinone oxidoreductase activity |
|
| YLR124W | 0.91 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YBR050C | 0.91 |
REG2
|
Regulatory subunit of the Glc7p type-1 protein phosphatase; involved with Reg1p, Glc7p, and Snf1p in regulation of glucose-repressible genes, also involved in glucose-induced proteolysis of maltose permease |
|
| YFL062W | 0.90 |
COS4
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YFR026C | 0.90 |
Putative protein of unknown function |
||
| YLR235C | 0.89 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene TOP3 |
||
| YJL133C-A | 0.89 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YDR178W | 0.87 |
SDH4
|
Membrane anchor subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
|
| YGL184C | 0.87 |
STR3
|
Cystathionine beta-lyase, converts cystathionine into homocysteine |
|
| YPL222W | 0.87 |
FMP40
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YML091C | 0.87 |
RPM2
|
Protein subunit of mitochondrial RNase P, has roles in nuclear transcription, cytoplasmic and mitochondrial RNA processing, and mitochondrial translation; distributed to mitochondria, cytoplasmic processing bodies, and the nucleus |
|
| YLR152C | 0.87 |
Putative protein of unknown function; YLR152C is not an essential gene |
||
| YMR096W | 0.87 |
SNZ1
|
Protein involved in vitamin B6 biosynthesis; member of a stationary phase-induced gene family; coregulated with SNO1; interacts with Sno1p and with Yhr198p, perhaps as a multiprotein complex containing other Snz and Sno proteins |
|
| YEL024W | 0.86 |
RIP1
|
Ubiquinol-cytochrome-c reductase, a Rieske iron-sulfur protein of the mitochondrial cytochrome bc1 complex; transfers electrons from ubiquinol to cytochrome c1 during respiration |
|
| YMR040W | 0.86 |
YET2
|
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; homolog of human BAP31 protein |
|
| YMR317W | 0.85 |
Putative protein of unknown function with some similarity to sialidase from Trypanosoma; YMR317W is not an essential gene |
||
| YER035W | 0.85 |
EDC2
|
RNA-binding protein, activates mRNA decapping directly by binding to the mRNA substrate and enhancing the activity of the decapping proteins Dcp1p and Dcp2p |
|
| YER096W | 0.84 |
SHC1
|
Sporulation-specific activator of Chs3p (chitin synthase III), required for the synthesis of the chitosan layer of ascospores; has similarity to Skt5p, which activates Chs3p during vegetative growth; transcriptionally induced at alkaline pH |
|
| YGR065C | 0.84 |
VHT1
|
High-affinity plasma membrane H+-biotin (vitamin H) symporter; mutation results in fatty acid auxotrophy; 12 transmembrane domain containing major facilitator subfamily member; mRNA levels negatively regulated by iron deprivation and biotin |
|
| YDR102C | 0.84 |
Dubious open reading frame; homozygous diploid deletion strain exhibits high budding index |
||
| YLR237W | 0.84 |
THI7
|
Plasma membrane transporter responsible for the uptake of thiamine, member of the major facilitator superfamily of transporters; mutation of human ortholog causes thiamine-responsive megaloblastic anemia |
|
| YMR118C | 0.83 |
Protein of unknown function with similarity to succinate dehydrogenase cytochrome b subunit; YMR118C is not an essential gene |
||
| YNR034W | 0.83 |
SOL1
|
Protein with a possible role in tRNA export; shows similarity to 6-phosphogluconolactonase non-catalytic domains but does not exhibit this enzymatic activity; homologous to Sol2p, Sol3p, and Sol4p |
|
| YDR298C | 0.83 |
ATP5
|
Subunit 5 of the stator stalk of mitochondrial F1F0 ATP synthase, which is an evolutionarily conserved enzyme complex required for ATP synthesis; homologous to bovine subunit OSCP (oligomycin sensitivity-conferring protein); phosphorylated |
|
| YGR146C | 0.83 |
Putative protein of unknown function; induced by iron homeostasis transcription factor Aft2p; multicopy suppressor of a temperature sensitive hsf1 mutant; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YJL161W | 0.82 |
FMP33
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YJL127C-B | 0.82 |
Putative protein of unknown function; identified based on homology to the filamentous fungus, Ashbya gossypii |
||
| YGR256W | 0.80 |
GND2
|
6-phosphogluconate dehydrogenase (decarboxylating), catalyzes an NADPH regenerating reaction in the pentose phosphate pathway; required for growth on D-glucono-delta-lactone |
|
| YOL085W-A | 0.80 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YOL085C |
||
| YLR377C | 0.79 |
FBP1
|
Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway, required for glucose metabolism |
|
| YPL089C | 0.79 |
RLM1
|
MADS-box transcription factor, component of the protein kinase C-mediated MAP kinase pathway involved in the maintenance of cell integrity; phosphorylated and activated by the MAP-kinase Slt2p |
|
| YGR239C | 0.79 |
PEX21
|
Peroxin required for targeting of peroxisomal matrix proteins containing PTS2; interacts with Pex7p; partially redundant with Pex18p |
|
| YAR050W | 0.79 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
Network of associatons between targets according to the STRING database.
First level regulatory network of STP2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:0006848 | pyruvate transport(GO:0006848) |
| 1.4 | 5.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 1.4 | 4.1 | GO:0015755 | fructose transport(GO:0015755) |
| 1.3 | 3.9 | GO:0015888 | thiamine transport(GO:0015888) |
| 1.0 | 2.0 | GO:0045980 | negative regulation of nucleotide metabolic process(GO:0045980) |
| 0.9 | 2.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.9 | 2.7 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.8 | 6.7 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.8 | 2.4 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.8 | 1.6 | GO:0046323 | glucose import(GO:0046323) |
| 0.8 | 2.3 | GO:0006068 | ethanol catabolic process(GO:0006068) primary alcohol catabolic process(GO:0034310) |
| 0.8 | 3.8 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.7 | 2.8 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.7 | 2.0 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.6 | 3.0 | GO:0032974 | amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 0.6 | 2.9 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.6 | 2.9 | GO:0051099 | positive regulation of binding(GO:0051099) |
| 0.6 | 2.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.6 | 3.3 | GO:0015758 | glucose transport(GO:0015758) |
| 0.5 | 2.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.5 | 3.9 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.5 | 1.8 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.4 | 4.0 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.4 | 1.7 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 0.4 | 1.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.4 | 2.1 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.4 | 0.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.4 | 2.0 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.4 | 1.2 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.4 | 4.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.4 | 5.2 | GO:0008645 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.4 | 1.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.4 | 1.1 | GO:0035786 | protein complex oligomerization(GO:0035786) |
| 0.4 | 0.4 | GO:0072350 | tricarboxylic acid metabolic process(GO:0072350) |
| 0.4 | 1.4 | GO:1903339 | inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.4 | 3.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 1.0 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.3 | 1.0 | GO:0010994 | free ubiquitin chain polymerization(GO:0010994) |
| 0.3 | 0.3 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.3 | 1.2 | GO:0036498 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) IRE1-mediated unfolded protein response(GO:0036498) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) |
| 0.3 | 0.9 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.3 | 4.9 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.3 | 0.9 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.3 | 0.9 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.3 | 0.8 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 1.9 | GO:0034762 | regulation of transmembrane transport(GO:0034762) |
| 0.3 | 2.5 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.3 | 1.1 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.3 | 0.8 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 0.3 | 0.8 | GO:0036257 | multivesicular body organization(GO:0036257) |
| 0.3 | 6.1 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.3 | 0.8 | GO:0031135 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.2 | 0.7 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.2 | 0.6 | GO:0055078 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.2 | 0.6 | GO:0032874 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.2 | 1.8 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 1.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.2 | 2.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 0.4 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.2 | 3.0 | GO:0019740 | nitrogen utilization(GO:0019740) |
| 0.2 | 1.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.2 | 0.9 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.2 | 1.1 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.2 | 1.6 | GO:0046352 | disaccharide catabolic process(GO:0046352) |
| 0.2 | 1.1 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.2 | 0.4 | GO:0015691 | cadmium ion transport(GO:0015691) |
| 0.2 | 1.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 2.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.2 | 2.4 | GO:0000753 | cell morphogenesis involved in conjugation with cellular fusion(GO:0000753) |
| 0.2 | 0.5 | GO:0019543 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 0.2 | 1.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 1.6 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.2 | 0.8 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.2 | 0.5 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.2 | 0.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 2.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 2.6 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.1 | 0.4 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) negative regulation of developmental process(GO:0051093) |
| 0.1 | 1.4 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 1.1 | GO:0015893 | drug transport(GO:0015893) |
| 0.1 | 1.1 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.4 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.1 | 5.7 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 0.4 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.1 | 0.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 1.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.7 | GO:0015851 | nucleobase transport(GO:0015851) |
| 0.1 | 0.2 | GO:0043200 | response to amino acid(GO:0043200) |
| 0.1 | 0.5 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 0.6 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.8 | GO:0000436 | carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.1 | 1.0 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 3.5 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.1 | 0.6 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.1 | 0.5 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.1 | 0.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 2.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.2 | GO:2000284 | positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 2.2 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 0.1 | GO:0034755 | high-affinity iron ion transmembrane transport(GO:0006827) iron ion transmembrane transport(GO:0034755) |
| 0.1 | 2.2 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 0.5 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.1 | 0.5 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.7 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 0.3 | GO:0071050 | snoRNA polyadenylation(GO:0071050) |
| 0.1 | 0.5 | GO:0006077 | (1->6)-beta-D-glucan metabolic process(GO:0006077) |
| 0.1 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.5 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.4 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 0.5 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.3 | GO:0007234 | osmosensory signaling via phosphorelay pathway(GO:0007234) |
| 0.1 | 0.2 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 1.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.1 | 0.2 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.1 | 0.2 | GO:0000411 | regulation of transcription by galactose(GO:0000409) positive regulation of transcription by galactose(GO:0000411) |
| 0.1 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.3 | GO:0046464 | triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.1 | 0.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.3 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.6 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 0.3 | GO:0000196 | MAPK cascade involved in cell wall organization or biogenesis(GO:0000196) |
| 0.1 | 1.4 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.3 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.1 | GO:0032069 | regulation of nuclease activity(GO:0032069) regulation of deoxyribonuclease activity(GO:0032070) regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.1 | 1.1 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 0.2 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 0.2 | GO:0036294 | response to anoxia(GO:0034059) cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to anoxia(GO:0071454) |
| 0.1 | 0.3 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of vesicle targeting, to, from or within Golgi(GO:0048209) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 0.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 0.2 | GO:0071495 | cellular response to biotic stimulus(GO:0071216) cellular response to endogenous stimulus(GO:0071495) response to cell cycle checkpoint signaling(GO:0072396) response to mitotic cell cycle checkpoint signaling(GO:0072414) response to spindle checkpoint signaling(GO:0072417) response to mitotic spindle checkpoint signaling(GO:0072476) response to mitotic cell cycle spindle assembly checkpoint signaling(GO:0072479) response to spindle assembly checkpoint signaling(GO:0072485) negative regulation of protein import into nucleus during spindle assembly checkpoint(GO:1901925) |
| 0.1 | 0.3 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.1 | 0.2 | GO:0034395 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.1 | 0.3 | GO:0009065 | glutamine family amino acid catabolic process(GO:0009065) |
| 0.1 | 0.1 | GO:0045923 | positive regulation of fatty acid metabolic process(GO:0045923) |
| 0.1 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.6 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.1 | 0.4 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.2 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.2 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.5 | GO:0046834 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 1.0 | GO:0010324 | membrane invagination(GO:0010324) lysosomal microautophagy(GO:0016237) single-organism membrane invagination(GO:1902534) |
| 0.0 | 1.1 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 0.1 | GO:1900544 | positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0071041 | antisense RNA transcript catabolic process(GO:0071041) |
| 0.0 | 0.2 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.0 | 0.2 | GO:0071322 | cellular response to carbohydrate stimulus(GO:0071322) |
| 0.0 | 0.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.0 | 0.2 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0040031 | snRNA pseudouridine synthesis(GO:0031120) snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.0 | 0.1 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) regulation of membrane lipid metabolic process(GO:1905038) |
| 0.0 | 0.2 | GO:0032048 | cardiolipin metabolic process(GO:0032048) |
| 0.0 | 0.1 | GO:0033993 | response to lipid(GO:0033993) response to oleic acid(GO:0034201) response to fatty acid(GO:0070542) cellular response to lipid(GO:0071396) cellular response to fatty acid(GO:0071398) cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) regulation of vacuole organization(GO:0044088) |
| 0.0 | 0.3 | GO:0000321 | re-entry into mitotic cell cycle after pheromone arrest(GO:0000321) |
| 0.0 | 0.0 | GO:0070482 | response to decreased oxygen levels(GO:0036293) response to oxygen levels(GO:0070482) |
| 0.0 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0042743 | hydrogen peroxide metabolic process(GO:0042743) hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.2 | GO:0009231 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.0 | 0.1 | GO:0016241 | regulation of macroautophagy(GO:0016241) |
| 0.0 | 0.0 | GO:0015688 | iron chelate transport(GO:0015688) |
| 0.0 | 0.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 2.2 | GO:0048468 | ascospore formation(GO:0030437) cell development(GO:0048468) |
| 0.0 | 0.1 | GO:0019563 | alditol catabolic process(GO:0019405) glycerol catabolic process(GO:0019563) |
| 0.0 | 0.1 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.2 | GO:0010525 | regulation of transposition, RNA-mediated(GO:0010525) |
| 0.0 | 0.8 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.6 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.2 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.3 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.2 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 1.1 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.1 | GO:0016562 | receptor recycling(GO:0001881) protein import into peroxisome matrix, receptor recycling(GO:0016562) receptor metabolic process(GO:0043112) |
| 0.0 | 0.1 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.0 | 0.2 | GO:0090054 | regulation of chromatin silencing at silent mating-type cassette(GO:0090054) |
| 0.0 | 0.1 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.0 | GO:1903513 | retrograde protein transport, ER to cytosol(GO:0030970) endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:0031134 | sister chromatid biorientation(GO:0031134) |
| 0.0 | 0.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.5 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.1 | GO:0046020 | negative regulation of transcription by pheromones(GO:0045996) negative regulation of transcription from RNA polymerase II promoter by pheromones(GO:0046020) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.0 | GO:0043687 | C-terminal protein amino acid modification(GO:0018410) post-translational protein modification(GO:0043687) |
| 0.0 | 0.0 | GO:0070623 | regulation of vitamin metabolic process(GO:0030656) positive regulation of vitamin metabolic process(GO:0046136) positive regulation of sulfur metabolic process(GO:0051176) regulation of thiamine biosynthetic process(GO:0070623) positive regulation of thiamine biosynthetic process(GO:0090180) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 | 0.3 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.7 | 2.8 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.7 | 2.0 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.7 | 2.0 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.6 | 1.8 | GO:0000274 | mitochondrial proton-transporting ATP synthase, stator stalk(GO:0000274) proton-transporting ATP synthase, stator stalk(GO:0045265) |
| 0.5 | 2.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.5 | 1.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.5 | 3.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.4 | 1.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.3 | 2.0 | GO:0045263 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.3 | 2.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 0.8 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 2.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 2.0 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 1.0 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 1.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 1.7 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.2 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 0.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.4 | GO:0000111 | nucleotide-excision repair factor 2 complex(GO:0000111) |
| 0.1 | 0.4 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 1.0 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.1 | 0.6 | GO:0072379 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 0.1 | 1.9 | GO:0031307 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.6 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.1 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 4.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.5 | GO:0034448 | EGO complex(GO:0034448) |
| 0.1 | 1.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.5 | GO:0070772 | PAS complex(GO:0070772) |
| 0.1 | 0.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 1.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.2 | GO:0032176 | split septin rings(GO:0032176) cellular bud neck split septin rings(GO:0032177) |
| 0.1 | 28.2 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.1 | 1.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 1.1 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.6 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.1 | 4.7 | GO:0000329 | fungal-type vacuole membrane(GO:0000329) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:0016587 | Isw1 complex(GO:0016587) |
| 0.0 | 0.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.0 | 5.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.7 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.1 | GO:0005825 | half bridge of spindle pole body(GO:0005825) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 1.0 | GO:0000322 | storage vacuole(GO:0000322) lytic vacuole(GO:0000323) fungal-type vacuole(GO:0000324) |
| 0.0 | 0.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 1.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 12.8 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.8 | GO:0061645 | actin cortical patch(GO:0030479) endocytic patch(GO:0061645) |
| 0.0 | 0.1 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.0 | GO:0000818 | MIS12/MIND type complex(GO:0000444) nuclear MIS12/MIND complex(GO:0000818) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 9.2 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 1.5 | 6.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 1.1 | 4.4 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.9 | 2.8 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.8 | 3.8 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.7 | 3.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.7 | 2.7 | GO:0015603 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.7 | 2.0 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.6 | 2.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.6 | 1.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.5 | 3.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.5 | 2.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.5 | 7.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.5 | 3.0 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.5 | 1.5 | GO:0005536 | glucose binding(GO:0005536) |
| 0.5 | 4.6 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.4 | 1.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.4 | 0.8 | GO:0015489 | putrescine transmembrane transporter activity(GO:0015489) |
| 0.4 | 1.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.4 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.3 | 1.0 | GO:0015186 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.3 | 1.7 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.3 | 1.3 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.3 | 1.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 1.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.3 | 2.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.3 | 3.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 3.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.3 | 0.8 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.3 | 0.5 | GO:0035591 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.3 | 0.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 0.8 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.3 | 3.1 | GO:0022838 | substrate-specific channel activity(GO:0022838) |
| 0.3 | 0.8 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 0.2 | 0.7 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.2 | 0.9 | GO:0016405 | CoA-ligase activity(GO:0016405) acid-thiol ligase activity(GO:0016878) |
| 0.2 | 3.2 | GO:0015578 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.2 | 0.9 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.2 | 1.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.2 | 1.7 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.2 | 2.3 | GO:0004888 | transmembrane signaling receptor activity(GO:0004888) signaling receptor activity(GO:0038023) transmembrane receptor activity(GO:0099600) |
| 0.2 | 0.8 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 0.6 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) |
| 0.2 | 0.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 0.7 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.2 | 3.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 1.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.2 | 1.6 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.2 | 1.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 0.8 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.2 | 0.8 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.2 | 2.1 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.2 | 0.5 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.2 | 1.2 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.2 | 0.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.2 | 1.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 2.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 2.6 | GO:0016769 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.4 | GO:0015085 | calcium ion transmembrane transporter activity(GO:0015085) |
| 0.1 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.9 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.4 | GO:0016406 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.1 | 0.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.4 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.4 | GO:0033592 | RNA strand annealing activity(GO:0033592) annealing activity(GO:0097617) |
| 0.1 | 1.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.9 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.1 | 0.2 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 0.2 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.1 | 0.2 | GO:0042947 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.1 | 0.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.3 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.1 | 1.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 1.0 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.7 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.1 | 0.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.3 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.1 | 0.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 3.1 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 0.6 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 0.1 | GO:0019903 | phosphatase binding(GO:0019902) protein phosphatase binding(GO:0019903) |
| 0.1 | 0.2 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 0.3 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.1 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.3 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.1 | 0.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.2 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.1 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.8 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.6 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.2 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) sulfurtransferase activity(GO:0016783) |
| 0.0 | 1.1 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.2 | GO:0016885 | ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.5 | GO:0001099 | RNA polymerase II core binding(GO:0000993) basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.2 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) drug transporter activity(GO:0090484) |
| 0.0 | 0.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0015216 | purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.9 | GO:0061733 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0016872 | intramolecular lyase activity(GO:0016872) |
| 0.0 | 0.4 | GO:0020037 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.0 | 0.1 | GO:0043492 | ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.1 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0015405 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.3 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.0 | 0.1 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.3 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.1 | GO:0005102 | receptor binding(GO:0005102) |
| 0.0 | 0.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0016903 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) oxidoreductase activity, acting on the aldehyde or oxo group of donors(GO:0016903) |
| 0.0 | 0.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.0 | GO:0015079 | potassium ion transmembrane transporter activity(GO:0015079) |
| 0.0 | 0.2 | GO:0016298 | lipase activity(GO:0016298) |
| 0.0 | 0.1 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 1.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.4 | 2.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 2.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 0.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.2 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 0.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 0.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 40.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.5 | 1.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.3 | 2.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.2 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 0.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 38.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |