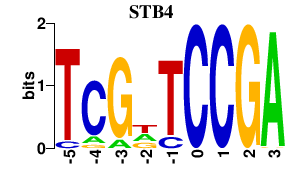
Results for STB4
Z-value: 1.42
Transcription factors associated with STB4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STB4
|
S000004621 | Putative transcription factor |
Activity-expression correlation:
Activity profile of STB4 motif
Sorted Z-values of STB4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YKR097W | 15.58 |
PCK1
|
Phosphoenolpyruvate carboxykinase, key enzyme in gluconeogenesis, catalyzes early reaction in carbohydrate biosynthesis, glucose represses transcription and accelerates mRNA degradation, regulated by Mcm1p and Cat8p, located in the cytosol |
|
| YNL117W | 10.63 |
MLS1
|
Malate synthase, enzyme of the glyoxylate cycle, involved in utilization of non-fermentable carbon sources; expression is subject to carbon catabolite repression; localizes in peroxisomes during growth in oleic acid medium |
|
| YPL262W | 8.48 |
FUM1
|
Fumarase, converts fumaric acid to L-malic acid in the TCA cycle; cytosolic and mitochondrial localization determined by the N-terminal mitochondrial targeting sequence and protein conformation; phosphorylated in mitochondria |
|
| YMR081C | 7.26 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YLR307C-A | 7.21 |
Putative protein of unknown function |
||
| YER024W | 7.00 |
YAT2
|
Carnitine acetyltransferase; has similarity to Yat1p, which is a carnitine acetyltransferase associated with the mitochondrial outer membrane |
|
| YJR078W | 6.34 |
BNA2
|
Putative tryptophan 2,3-dioxygenase or indoleamine 2,3-dioxygenase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p and Aft2p |
|
| YMR034C | 5.38 |
Putative transporter, member of the SLC10 carrier family; identified in a transposon mutagenesis screen as a gene involved in azole resistance; YMR034C is not an essential gene |
||
| YBR072W | 5.19 |
HSP26
|
Small heat shock protein (sHSP) with chaperone activity; forms hollow, sphere-shaped oligomers that suppress unfolded proteins aggregation; oligomer activation requires a heat-induced conformational change; not expressed in unstressed cells |
|
| YGR067C | 4.97 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YJR146W | 4.89 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene HMS2 |
||
| YLR122C | 4.78 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR123C |
||
| YIL057C | 4.61 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YBR222C | 4.48 |
PCS60
|
Peroxisomal AMP-binding protein, localizes to both the peroxisomal peripheral membrane and matrix, expression is highly inducible by oleic acid, similar to E. coli long chain acyl-CoA synthetase |
|
| YDR406W | 4.44 |
PDR15
|
Plasma membrane ATP binding cassette (ABC) transporter, multidrug transporter and general stress response factor implicated in cellular detoxification; regulated by Pdr1p, Pdr3p and Pdr8p; promoter contains a PDR responsive element |
|
| YNL144C | 4.37 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YNL144C is not an essential gene |
||
| YJR155W | 4.35 |
AAD10
|
Putative aryl-alcohol dehydrogenase with similarity to P. chrysosporium aryl-alcohol dehydrogenase; mutational analysis has not yet revealed a physiological role |
|
| YER065C | 4.35 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YBR051W | 4.33 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partiallly overlaps the REG2/YBR050C regulatory subunit of the Glc7p type-1 protein phosphatase |
||
| YBR050C | 4.30 |
REG2
|
Regulatory subunit of the Glc7p type-1 protein phosphatase; involved with Reg1p, Glc7p, and Snf1p in regulation of glucose-repressible genes, also involved in glucose-induced proteolysis of maltose permease |
|
| YLL060C | 4.30 |
GTT2
|
Glutathione S-transferase capable of homodimerization; functional overlap with Gtt2p, Grx1p, and Grx2p |
|
| YAL062W | 4.28 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YGL255W | 4.22 |
ZRT1
|
High-affinity zinc transporter of the plasma membrane, responsible for the majority of zinc uptake; transcription is induced under low-zinc conditions by the Zap1p transcription factor |
|
| YMR271C | 4.16 |
URA10
|
Minor orotate phosphoribosyltransferase (OPRTase) isozyme that catalyzes the fifth enzymatic step in the de novo biosynthesis of pyrimidines, converting orotate into orotidine-5'-phosphate; major OPRTase encoded by URA5 |
|
| YLL053C | 4.15 |
Putative protein; in the Sigma 1278B strain background YLL053C is contiguous with AQY2 which encodes an aquaporin |
||
| YLR123C | 3.92 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR122C; contains characteristic aminoacyl-tRNA motif |
||
| YDL169C | 3.89 |
UGX2
|
Protein of unknown function, transcript accumulates in response to any combination of stress conditions |
|
| YPL054W | 3.86 |
LEE1
|
Zinc-finger protein of unknown function |
|
| YPR002W | 3.83 |
PDH1
|
Mitochondrial protein that participates in respiration, induced by diauxic shift; homologous to E. coli PrpD, may take part in the conversion of 2-methylcitrate to 2-methylisocitrate |
|
| YJR077C | 3.79 |
MIR1
|
Mitochondrial phosphate carrier, imports inorganic phosphate into mitochondria; functionally redundant with Pic2p but more abundant than Pic2p under normal conditions; phosphorylated |
|
| YLL052C | 3.67 |
AQY2
|
Water channel that mediates the transport of water across cell membranes, only expressed in proliferating cells, controlled by osmotic signals, may be involved in freeze tolerance; disrupted by a stop codon in many S. cerevisiae strains |
|
| YGL184C | 3.67 |
STR3
|
Cystathionine beta-lyase, converts cystathionine into homocysteine |
|
| YAR023C | 3.62 |
Putative integral membrane protein, member of DUP240 gene family |
||
| YIL166C | 3.57 |
Putative protein with similarity to the allantoate permease (Dal5p) subfamily of the major facilitator superfamily; mRNA expression is elevated by sulfur limitation; YIL166C is a non-essential gene |
||
| YLR312C | 3.34 |
QNQ1
|
Protein of unknown function; null mutant quiescent and non-quiescent cells exhibit reduced reproductive capacity |
|
| YEL028W | 3.34 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR296W | 3.32 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR006C | 3.32 |
ICL2
|
2-methylisocitrate lyase of the mitochondrial matrix, functions in the methylcitrate cycle to catalyze the conversion of 2-methylisocitrate to succinate and pyruvate; ICL2 transcription is repressed by glucose and induced by ethanol |
|
| YOR208W | 3.31 |
PTP2
|
Phosphotyrosine-specific protein phosphatase involved in the inactivation of mitogen-activated protein kinase (MAPK) during osmolarity sensing; dephosporylates Hog1p MAPK and regulates its localization; localized to the nucleus |
|
| YGL208W | 3.28 |
SIP2
|
One of three beta subunits of the Snf1 serine/threonine protein kinase complex involved in the response to glucose starvation; null mutants exhibit accelerated aging; N-myristoylprotein localized to the cytoplasm and the plasma membrane |
|
| YKL065W-A | 3.17 |
Putative protein of unknown function |
||
| YFL055W | 3.17 |
AGP3
|
Low-affinity amino acid permease, may act to supply the cell with amino acids as nitrogen source in nitrogen-poor conditions; transcription is induced under conditions of sulfur limitation; plays a role in regulating Ty1 transposition |
|
| YMR056C | 3.11 |
AAC1
|
Mitochondrial inner membrane ADP/ATP translocator, exchanges cytosolic ADP for mitochondrially synthesized ATP; phosphorylated; Aac1p is a minor isoform while Pet9p is the major ADP/ATP translocator |
|
| YNR073C | 3.08 |
Putative mannitol dehydrogenase |
||
| YBR047W | 3.06 |
FMP23
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YLR295C | 3.05 |
ATP14
|
Subunit h of the F0 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YHR048W | 3.04 |
YHK8
|
Presumed antiporter of the DHA1 family of multidrug resistance transporters; contains 12 predicted transmembrane spans; expression of gene is up-regulated in cells exhibiting reduced susceptibility to azoles |
|
| YJR120W | 3.03 |
Protein of unknown function; essential for growth under anaerobic conditions; mutation causes decreased expression of ATP2, impaired respiration, defective sterol uptake, and altered levels/localization of ABC transporters Aus1p and Pdr11p |
||
| YOR147W | 3.03 |
MDM32
|
Mitochondrial inner membrane protein with similarity to Mdm31p, required for normal mitochondrial morphology and inheritance; interacts genetically with MMM1, MDM10, MDM12, and MDM34 |
|
| YFR022W | 3.03 |
ROG3
|
Protein that binds to Rsp5p, which is a hect-type ubiquitin ligase, via its 2 PY motifs; has similarity to Rod1p; mutation suppresses the temperature sensitivity of an mck1 rim11 double mutant |
|
| YNL142W | 3.01 |
MEP2
|
Ammonium permease involved in regulation of pseudohyphal growth; belongs to a ubiquitous family of cytoplasmic membrane proteins that transport only ammonium (NH4+); expression is under the nitrogen catabolite repression regulation |
|
| YOR343C | 3.00 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YDR343C | 2.97 |
HXT6
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt7p, expressed at high basal levels relative to other HXTs, repression of expression by high glucose requires SNF3 |
|
| YBR045C | 2.90 |
GIP1
|
Meiosis-specific regulatory subunit of the Glc7p protein phosphatase, regulates spore wall formation and septin organization, required for expression of some late meiotic genes and for normal localization of Glc7p |
|
| YFL024C | 2.79 |
EPL1
|
Component of NuA4, which is an essential histone H4/H2A acetyltransferase complex; homologous to Drosophila Enhancer of Polycomb |
|
| YOL154W | 2.73 |
ZPS1
|
Putative GPI-anchored protein; transcription is induced under low-zinc conditions, as mediated by the Zap1p transcription factor, and at alkaline pH |
|
| YDL170W | 2.72 |
UGA3
|
Transcriptional activator necessary for gamma-aminobutyrate (GABA)-dependent induction of GABA genes (such as UGA1, UGA2, UGA4); zinc-finger transcription factor of the Zn(2)-Cys(6) binuclear cluster domain type; localized to the nucleus |
|
| YKR067W | 2.70 |
GPT2
|
Glycerol-3-phosphate/dihydroxyacetone phosphate dual substrate-specific sn-1 acyltransferase located in lipid particles and the ER; involved in the stepwise acylation of glycerol-3-phosphate and dihydroxyacetone in lipid biosynthesis |
|
| YMR035W | 2.66 |
IMP2
|
Catalytic subunit of the mitochondrial inner membrane peptidase complex, required for maturation of mitochondrial proteins of the intermembrane space; complex contains Imp1p and Imp2p (both catalytic subunits), and Som1p |
|
| YPL223C | 2.65 |
GRE1
|
Hydrophilin of unknown function; stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YBR046C | 2.64 |
ZTA1
|
Zeta-crystallin homolog, found in the cytoplasm and nucleus; has similarity to E. coli quinone oxidoreductase and to human zeta-crystallin, which has quinone oxidoreductase activity |
|
| YML089C | 2.61 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression induced by calcium shortage |
||
| YLR125W | 2.55 |
Putative protein of unknown function; mutant has decreased Ty3 transposition; YLR125W is not an essential gene |
||
| YKR066C | 2.55 |
CCP1
|
Mitochondrial cytochrome-c peroxidase; degrades reactive oxygen species in mitochondria, involved in the response to oxidative stress |
|
| YCL040W | 2.55 |
GLK1
|
Glucokinase, catalyzes the phosphorylation of glucose at C6 in the first irreversible step of glucose metabolism; one of three glucose phosphorylating enzymes; expression regulated by non-fermentable carbon sources |
|
| YAR050W | 2.51 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YLR124W | 2.47 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YIL023C | 2.44 |
YKE4
|
Zinc transporter; localizes to the ER; null mutant is sensitive to calcofluor white, leads to zinc accumulation in cytosol; ortholog of the mouse KE4 and member of the ZIP (ZRT, IRT-like Protein) family |
|
| YBL015W | 2.43 |
ACH1
|
Acetyl-coA hydrolase, primarily localized to mitochondria; phosphorylated; required for acetate utilization and for diploid pseudohyphal growth |
|
| YDR102C | 2.43 |
Dubious open reading frame; homozygous diploid deletion strain exhibits high budding index |
||
| YGR065C | 2.40 |
VHT1
|
High-affinity plasma membrane H+-biotin (vitamin H) symporter; mutation results in fatty acid auxotrophy; 12 transmembrane domain containing major facilitator subfamily member; mRNA levels negatively regulated by iron deprivation and biotin |
|
| YPL021W | 2.38 |
ECM23
|
Non-essential protein of unconfirmed function; affects pre-rRNA processing, may act as a negative regulator of the transcription of genes involved in pseudohyphal growth; homologous to Srd1p |
|
| YCL042W | 2.38 |
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm |
||
| YAR053W | 2.37 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YBL102W | 2.34 |
SFT2
|
Non-essential tetra-spanning membrane protein found mostly in the late Golgi, can suppress some sed5 alleles; may be part of the transport machinery, but precise function is unknown; similar to mammalian syntaxin 5 |
|
| YGR023W | 2.33 |
MTL1
|
Protein with both structural and functional similarity to Mid2p, which is a plasma membrane sensor required for cell integrity signaling during pheromone-induced morphogenesis; suppresses rgd1 null mutations |
|
| YLR252W | 2.30 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the verified gene SYM1, a mitochondrial protein involved in ethanol metabolism |
||
| YDR421W | 2.29 |
ARO80
|
Zinc finger transcriptional activator of the Zn2Cys6 family; activates transcription of aromatic amino acid catabolic genes in the presence of aromatic amino acids |
|
| YPR010C-A | 2.28 |
Putative protein of unknown function; conserved among Saccharomyces sensu stricto species |
||
| YPR184W | 2.27 |
GDB1
|
Glycogen debranching enzyme containing glucanotranferase and alpha-1,6-amyloglucosidase activities, required for glycogen degradation; phosphorylated in mitochondria |
|
| YFL052W | 2.25 |
Putative zinc cluster protein that contains a DNA binding domain; null mutant sensitive to calcofluor white, low osmolarity and heat, suggesting a role for YFL052Wp in cell wall integrity |
||
| YDR533C | 2.23 |
HSP31
|
Possible chaperone and cysteine protease with similarity to E. coli Hsp31; member of the DJ-1/ThiJ/PfpI superfamily, which includes human DJ-1 involved in Parkinson's disease; exists as a dimer and contains a putative metal-binding site |
|
| YJR150C | 2.23 |
DAN1
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YGR022C | 2.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF MTL1/YGR023W |
||
| YBL043W | 2.18 |
ECM13
|
Non-essential protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YJL116C | 2.15 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YPL024W | 2.14 |
RMI1
|
Involved in response to DNA damage; null mutants have increased rates of recombination and delayed S phase; interacts physically and genetically with Sgs1p (RecQ family member) and Top3p (topoisomerase III) |
|
| YGL083W | 2.12 |
SCY1
|
Putative kinase, suppressor of GTPase mutant, similar to bovine rhodopsin kinase |
|
| YHR092C | 2.10 |
HXT4
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YJR147W | 2.10 |
HMS2
|
Protein with similarity to heat shock transcription factors; overexpression suppresses the pseudohyphal filamentation defect of a diploid mep1 mep2 homozygous null mutant |
|
| YHL046W-A | 2.08 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YAR047C | 2.08 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR124W | 2.08 |
Putative protein of unknown function; non-essential gene; expression is strongly induced by alpha factor |
||
| YKL162C | 2.06 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the mitochondrion |
||
| YDR505C | 2.06 |
PSP1
|
Asn and gln rich protein of unknown function; high-copy suppressor of POL1 (DNA polymerase alpha) and partial suppressor of CDC2 (polymerase delta) and CDC6 (pre-RC loading factor) mutations; overexpression results in growth inhibition |
|
| YBL030C | 2.06 |
PET9
|
Major ADP/ATP carrier of the mitochondrial inner membrane, exchanges cytosolic ADP for mitochondrially synthesized ATP; phosphorylated; required for viability in many common lab strains carrying a mutation in the polymorphic SAL1 gene |
|
| YGL165C | 2.05 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF CUP2/YGL166W |
||
| YEL020C | 2.04 |
Hypothetical protein with low sequence identity to Pdc1p |
||
| YDR148C | 2.03 |
KGD2
|
Dihydrolipoyl transsuccinylase, component of the mitochondrial alpha-ketoglutarate dehydrogenase complex, which catalyzes the oxidative decarboxylation of alpha-ketoglutarate to succinyl-CoA in the TCA cycle; phosphorylated |
|
| YNL274C | 2.03 |
GOR1
|
Glyoxylate reductase; null mutation results in increased biomass after diauxic shift; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YKR015C | 2.02 |
Putative protein of unknown function |
||
| YLR326W | 1.99 |
Hypothetical protein |
||
| YMR206W | 1.99 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YPL222C-A | 1.99 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YPR140W | 1.99 |
TAZ1
|
Lyso-phosphatidylcholine acyltransferase, required for normal phospholipid content of mitochondrial membranes; may remodel acyl groups of cardiolipin in the inner membrane; similar to human tafazzin, which is implicated in Barth syndrome |
|
| YBR067C | 1.98 |
TIP1
|
Major cell wall mannoprotein with possible lipase activity; transcription is induced by heat- and cold-shock; member of the Srp1p/Tip1p family of serine-alanine-rich proteins |
|
| YBR296C | 1.94 |
PHO89
|
Na+/Pi cotransporter, active in early growth phase; similar to phosphate transporters of Neurospora crassa; transcription regulated by inorganic phosphate concentrations and Pho4p |
|
| YDR103W | 1.90 |
STE5
|
Pheromone-response scaffold protein; binds kinases Ste11p, Ste7p, and Fus3p to form a MAPK cascade complex that interacts with the plasma membrane, via a PH (pleckstrin homology) and PM/NLS domain, and with Ste4p-Ste18p, during signaling |
|
| YAR060C | 1.89 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR393W | 1.88 |
ATP10
|
Mitochondrial inner membrane protein required for assembly of the F0 sector of mitochondrial F1F0 ATP synthase, interacts genetically with ATP6 |
|
| YEL049W | 1.87 |
PAU2
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YNL134C | 1.85 |
Putative protein of unknown function with similarity to dehydrogenases from other model organisms; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and nucleus and is induced by the DNA-damaging agent MMS |
||
| YHL047C | 1.85 |
ARN2
|
Transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; responsible for uptake of iron bound to the siderophore triacetylfusarinine C |
|
| YNL036W | 1.85 |
NCE103
|
Carbonic anhydrase; poorly transcribed under aerobic conditions and at an undetectable level under anaerobic conditions; involved in non-classical protein export pathway |
|
| YMR110C | 1.84 |
HFD1
|
Putative fatty aldehyde dehydrogenase, located in the mitochondrial outer membrane and also in lipid particles; has similarity to human fatty aldehyde dehydrogenase (FALDH) which is implicated in Sjogren-Larsson syndrome |
|
| YLR402W | 1.83 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YCR005C | 1.80 |
CIT2
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate, peroxisomal isozyme involved in glyoxylate cycle; expression is controlled by Rtg1p and Rtg2p transcription factors |
|
| YJR020W | 1.80 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR110W | 1.78 |
Putative protein of unknown function; transcription is increased in response to genotoxic stress; plays a role in restricting Ty1 transposition |
||
| YLR403W | 1.78 |
SFP1
|
Transcription factor that controls expression of many ribosome biogenesis genes in response to nutrients and stress, regulates G2/M transitions during mitotic cell cycle and DNA-damage response, involved in cell size modulation |
|
| YNL143C | 1.78 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR277C | 1.78 |
MTH1
|
Negative regulator of the glucose-sensing signal transduction pathway, required for repression of transcription by Rgt1p; interacts with Rgt1p and the Snf3p and Rgt2p glucose sensors; phosphorylated by Yck1p, triggering Mth1p degradation |
|
| YJR046W | 1.77 |
TAH11
|
DNA replication licensing factor, required for pre-replication complex assembly |
|
| YGL131C | 1.77 |
SNT2
|
DNA binding protein with similarity to the S. pombe Snt2 protein |
|
| YDR536W | 1.76 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YJR019C | 1.75 |
TES1
|
Peroxisomal acyl-CoA thioesterase likely to be involved in fatty acid oxidation rather than fatty acid synthesis; conserved protein also found in human peroxisomes; TES1 mRNA levels increase during growth on fatty acids |
|
| YKR102W | 1.73 |
FLO10
|
Lectin-like protein with similarity to Flo1p, thought to be involved in flocculation |
|
| YER033C | 1.73 |
ZRG8
|
Protein of unknown function; authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; GFP-fusion protein is localized to the cytoplasm; transcription induced under conditions of zinc deficiency |
|
| YGL084C | 1.72 |
GUP1
|
Plasma membrane protein involved in remodeling GPI anchors; member of the MBOAT family of putative membrane-bound O-acyltransferases; proposed to be involved in glycerol transport |
|
| YJR045C | 1.70 |
SSC1
|
Mitochondrial matrix ATPase, subunit of the presequence translocase-associated protein import motor (PAM) and of SceI endonuclease; involved in protein folding and translocation into the matrix; phosphorylated; member of HSP70 family |
|
| YAR035W | 1.67 |
YAT1
|
Outer mitochondrial carnitine acetyltransferase, minor ethanol-inducible enzyme involved in transport of activated acyl groups from the cytoplasm into the mitochondrial matrix; phosphorylated |
|
| YMR103C | 1.67 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR178C | 1.67 |
GAC1
|
Regulatory subunit for Glc7p type-1 protein phosphatase (PP1), tethers Glc7p to Gsy2p glycogen synthase, binds Hsf1p heat shock transcription factor, required for induction of some HSF-regulated genes under heat shock |
|
| YDR298C | 1.66 |
ATP5
|
Subunit 5 of the stator stalk of mitochondrial F1F0 ATP synthase, which is an evolutionarily conserved enzyme complex required for ATP synthesis; homologous to bovine subunit OSCP (oligomycin sensitivity-conferring protein); phosphorylated |
|
| YLR342W | 1.66 |
FKS1
|
Catalytic subunit of 1,3-beta-D-glucan synthase, functionally redundant with alternate catalytic subunit Gsc2p; binds to regulatory subunit Rho1p; involved in cell wall synthesis and maintenance; localizes to sites of cell wall remodeling |
|
| YBL017C | 1.66 |
PEP1
|
Type I transmembrane sorting receptor for multiple vacuolar hydrolases; cycles between the late-Golgi and prevacuolar endosome-like compartments |
|
| YDR043C | 1.66 |
NRG1
|
Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters; mediates glucose repression and negatively regulates a variety of processes including filamentous growth and alkaline pH response |
|
| YNL115C | 1.65 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YNL115C is not an essential gene |
||
| YOR023C | 1.64 |
AHC1
|
Subunit of the Ada histone acetyltransferase complex, required for structural integrity of the complex |
|
| YOR231W | 1.62 |
MKK1
|
Mitogen-activated kinase kinase involved in protein kinase C signaling pathway that controls cell integrity; upon activation by Bck1p phosphorylates downstream target, Slt2p; functionally redundant with Mkk2p |
|
| YIL125W | 1.61 |
KGD1
|
Component of the mitochondrial alpha-ketoglutarate dehydrogenase complex, which catalyzes a key step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation of alpha-ketoglutarate to form succinyl-CoA |
|
| YOR378W | 1.60 |
Putative protein of unknown function; belongs to the DHA2 family of drug:H+ antiporters; YOR378W is not an essential gene |
||
| YDR178W | 1.59 |
SDH4
|
Membrane anchor subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
|
| YNR034W-A | 1.58 |
Putative protein of unknown function; expression is regulated by Msn2p/Msn4p |
||
| YDR542W | 1.56 |
PAU10
|
Hypothetical protein |
|
| YAR027W | 1.55 |
UIP3
|
Putative integral membrane protein of unknown function; interacts with Ulp1p at the nuclear periphery; member of DUP240 gene family |
|
| YBR077C | 1.55 |
SLM4
|
Component of the EGO complex, which is involved in the regulation of microautophagy, and of the GSE complex, which is required for proper sorting of amino acid permease Gap1p; gene exhibits synthetic genetic interaction with MSS4 |
|
| YIL042C | 1.54 |
PKP1
|
Mitochondrial protein kinase involved in negative regulation of pyruvate dehydrogenase complex activity by phosphorylating the ser-133 residue of the Pda1p subunit; acts in concert with kinase Pkp2p and phosphatases Ptc5p and Ptc6p |
|
| YHR159W | 1.52 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; potential Cdc28p substrate |
||
| YML090W | 1.52 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YML089C; exhibits growth defect on a non-fermentable (respiratory) carbon source |
||
| YER066W | 1.51 |
Putative protein of unknown function; YER066W is not an essential gene |
||
| YLR304C | 1.46 |
ACO1
|
Aconitase, required for the tricarboxylic acid (TCA) cycle and also independently required for mitochondrial genome maintenance; phosphorylated; component of the mitochondrial nucleoid; mutation leads to glutamate auxotrophy |
|
| YFR023W | 1.46 |
PES4
|
Poly(A) binding protein, suppressor of DNA polymerase epsilon mutation, similar to Mip6p |
|
| YKR016W | 1.46 |
AIM28
|
Protein of unknown function; non-tagged protein is detected in purified mitochondria in high-throughput studies; null mutant displays decreased frequency of mitochondrial genome loss and severe growth defect in minimal glycerol media |
|
| YFR053C | 1.45 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YHL021C | 1.44 |
AIM17
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) |
|
| YPL026C | 1.44 |
SKS1
|
Putative serine/threonine protein kinase; involved in the adaptation to low concentrations of glucose independent of the SNF3 regulated pathway |
|
| YLR297W | 1.43 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YLR297W is not an essential gene; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YPR196W | 1.43 |
Putative maltose activator |
||
| YCR068W | 1.42 |
ATG15
|
Lipase, required for intravacuolar lysis of autophagosomes; located in the endoplasmic reticulum membrane and targeted to intravacuolar vesicles during autophagy via the multivesicular body (MVB) pathway |
|
| YOL128C | 1.42 |
YGK3
|
Protein kinase related to mammalian glycogen synthase kinases of the GSK-3 family; GSK-3 homologs (Mck1p, Rim11p, Mrk1p, Ygk3p) are involved in control of Msn2p-dependent transcription of stress responsive genes and in protein degradation |
|
| YER150W | 1.40 |
SPI1
|
GPI-anchored cell wall protein involved in weak acid resistance; basal expression requires Msn2p/Msn4p; expression is induced under conditions of stress and during the diauxic shift; similar to Sed1p |
|
| YFR033C | 1.40 |
QCR6
|
Subunit 6 of the ubiquinol cytochrome-c reductase complex, which is a component of the mitochondrial inner membrane electron transport chain; highly acidic protein; required for maturation of cytochrome c1 |
|
| YJL100W | 1.40 |
LSB6
|
Type II phosphatidylinositol 4-kinase that binds Las17p, which is a homolog of human Wiskott-Aldrich Syndrome protein involved in actin patch assembly and actin polymerization |
|
| YAL067C | 1.39 |
SEO1
|
Putative permease, member of the allantoate transporter subfamily of the major facilitator superfamily; mutation confers resistance to ethionine sulfoxide |
|
| YLR294C | 1.37 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene ATP14 |
||
| YMR316C-B | 1.34 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBR293W | 1.34 |
VBA2
|
Permease of basic amino acids in the vacuolar membrane |
|
| YDR014W-A | 1.34 |
HED1
|
Meiosis-specific protein that down-regulates Rad51p-mediated mitotic recombination when the meiotic recombination machinery is impaired; early meiotic gene, transcribed specifically during meiotic prophase |
|
| YPL183W-A | 1.32 |
RTC6
|
Homolog of the prokaryotic ribosomal protein L36, likely to be a mitochondrial ribosomal protein coded in the nuclear genome; null mutation suppresses cdc13-1 temperature sensitivity |
|
| YPL111W | 1.32 |
CAR1
|
Arginase, responsible for arginine degradation, expression responds to both induction by arginine and nitrogen catabolite repression; disruption enhances freeze tolerance |
|
| YML054C | 1.31 |
CYB2
|
Cytochrome b2 (L-lactate cytochrome-c oxidoreductase), component of the mitochondrial intermembrane space, required for lactate utilization; expression is repressed by glucose and anaerobic conditions |
|
| YPR191W | 1.30 |
QCR2
|
Subunit 2 of the ubiquinol cytochrome-c reductase complex, which is a component of the mitochondrial inner membrane electron transport chain; phosphorylated; transcription is regulated by Hap1p, Hap2p/Hap3p, and heme |
|
| YOL082W | 1.30 |
ATG19
|
Receptor protein for the cytoplasm-to-vacuole targeting (Cvt) pathway; recognizes cargo proteins aminopeptidase I (Lap4p) and alpha-mannosidase (Ams1p) and delivers them to the preautophagosomal structure for packaging into Cvt vesicles |
|
| YDR387C | 1.29 |
Putative transporter, member of the sugar porter family; YDR387C is not an essential gene |
||
| YOR387C | 1.29 |
Putative protein of unknown function; regulated by the metal-responsive Aft1p transcription factor; highly inducible in zinc-depleted conditions; localizes to the soluble fraction |
||
| YKL163W | 1.28 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YOR142W | 1.27 |
LSC1
|
Alpha subunit of succinyl-CoA ligase, which is a mitochondrial enzyme of the TCA cycle that catalyzes the nucleotide-dependent conversion of succinyl-CoA to succinate; phosphorylated |
|
| YBR284W | 1.26 |
Putative protein of unknown function; YBR284W is not an essential gene; null mutant exhibits decreased resistance to rapamycin and wortmannin |
||
| YGL134W | 1.25 |
PCL10
|
Pho85p cyclin; recruits, activates, and targets Pho85p cyclin-dependent protein kinase to its substrate |
|
| YIL162W | 1.25 |
SUC2
|
Invertase, sucrose hydrolyzing enzyme; a secreted, glycosylated form is regulated by glucose repression, and an intracellular, nonglycosylated enzyme is produced constitutively |
|
| YBR292C | 1.25 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR292C is not an essential gene |
||
| YOL051W | 1.23 |
GAL11
|
Subunit of the RNA polymerase II mediator complex; associates with core polymerase subunits to form the RNA polymerase II holoenzyme; affects transcription by acting as target of activators and repressors |
|
| YNR055C | 1.23 |
HOL1
|
Putative transporter in the major facilitator superfamily (DHA1 family) of multidrug resistance transporters; mutations in membrane-spanning domains permit cation and histidinol uptake |
|
| YLL055W | 1.22 |
YCT1
|
High-affinity cysteine-specific transporter with similarity to the Dal5p family of transporters; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; YCT1 is not an essential gene |
|
| YMR317W | 1.22 |
Putative protein of unknown function with some similarity to sialidase from Trypanosoma; YMR317W is not an essential gene |
||
| YOR394W | 1.20 |
PAU21
|
Hypothetical protein |
|
| YLR047C | 1.19 |
FRE8
|
Protein with sequence similarity to iron/copper reductases, involved in iron homeostasis; deletion mutant has iron deficiency/accumulation growth defects; expression increased in the absence of copper-responsive transcription factor Mac1p |
|
| YMR194C-B | 1.19 |
Putative protein of unknown function |
||
| YPL222W | 1.18 |
FMP40
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YDL174C | 1.18 |
DLD1
|
D-lactate dehydrogenase, oxidizes D-lactate to pyruvate, transcription is heme-dependent, repressed by glucose, and derepressed in ethanol or lactate; located in the mitochondrial inner membrane |
|
| YDR541C | 1.18 |
Putative dihydrokaempferol 4-reductase |
||
| YPL274W | 1.18 |
SAM3
|
High-affinity S-adenosylmethionine permease, required for utilization of S-adenosylmethionine as a sulfur source; has similarity to S-methylmethionine permease Mmp1p |
|
| YKL141W | 1.17 |
SDH3
|
Cytochrome b subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
|
| YOR024W | 1.16 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR212C | 1.15 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR281C | 1.14 |
PHM6
|
Protein of unknown function, expression is regulated by phosphate levels |
|
| YMR171C | 1.13 |
EAR1
|
Specificity factor required for Rsp5p-dependent ubiquitination and sorting of specific cargo proteins at the multivesicular body; mRNA is targeted to the bud via the mRNA transport system involving She2p: YMR171C is not an essential gene |
|
| YPL171C | 1.12 |
OYE3
|
Widely conserved NADPH oxidoreductase containing flavin mononucleotide (FMN), homologous to Oye2p with slight differences in ligand binding and catalytic properties; may be involved in sterol metabolism |
|
| YLR035C | 1.12 |
MLH2
|
Protein involved in the mismatch repair of certain frameshift intermediates and involved in meiotic recombination; forms a complex with Mlh1p |
Network of associatons between targets according to the STRING database.
First level regulatory network of STB4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 2.4 | 7.2 | GO:0019543 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 1.9 | 18.8 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 1.4 | 7.1 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 1.3 | 5.2 | GO:0015886 | heme transport(GO:0015886) |
| 1.1 | 19.2 | GO:0006101 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 1.0 | 3.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 1.0 | 3.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.0 | 5.9 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.9 | 3.7 | GO:0019346 | transsulfuration(GO:0019346) |
| 0.9 | 0.9 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.9 | 2.7 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.8 | 2.5 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.8 | 3.4 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.8 | 3.3 | GO:1903137 | inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.8 | 3.2 | GO:0046323 | glucose import(GO:0046323) |
| 0.8 | 2.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.8 | 6.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.7 | 2.7 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.7 | 0.7 | GO:0035786 | protein complex oligomerization(GO:0035786) |
| 0.7 | 19.1 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.6 | 2.6 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.6 | 3.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.6 | 2.5 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.6 | 1.7 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.6 | 1.7 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.6 | 1.7 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.6 | 2.8 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.5 | 2.7 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.5 | 2.1 | GO:0051101 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.5 | 2.7 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.5 | 4.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.5 | 1.4 | GO:0036257 | multivesicular body organization(GO:0036257) |
| 0.5 | 2.4 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.5 | 1.8 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.5 | 2.3 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.4 | 1.3 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.4 | 1.3 | GO:0019627 | urea metabolic process(GO:0019627) |
| 0.4 | 1.3 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.4 | 1.2 | GO:0031135 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.4 | 1.2 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.4 | 2.0 | GO:0032048 | cardiolipin metabolic process(GO:0032048) |
| 0.4 | 4.4 | GO:0015893 | drug transport(GO:0015893) |
| 0.4 | 0.8 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.4 | 1.1 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 0.4 | 2.3 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.4 | 2.6 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.4 | 2.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.4 | 11.6 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.4 | 3.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.4 | 1.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of metal ion(GO:0051238) sequestering of iron ion(GO:0097577) |
| 0.4 | 4.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.3 | 2.0 | GO:0044205 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.3 | 0.3 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.3 | 2.3 | GO:0015758 | glucose transport(GO:0015758) |
| 0.3 | 1.9 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.3 | 0.9 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.3 | 1.9 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.3 | 0.9 | GO:0034090 | maintenance of meiotic sister chromatid cohesion(GO:0034090) |
| 0.3 | 0.6 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.3 | 12.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.3 | 2.2 | GO:0070272 | proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.3 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.3 | 0.8 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) regulation of membrane lipid metabolic process(GO:1905038) |
| 0.3 | 2.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 0.8 | GO:0006343 | establishment of chromatin silencing(GO:0006343) |
| 0.3 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 1.0 | GO:0000431 | regulation of transcription from RNA polymerase II promoter by galactose(GO:0000431) positive regulation of transcription from RNA polymerase II promoter by galactose(GO:0000435) |
| 0.2 | 1.5 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.2 | 1.0 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.2 | 1.0 | GO:0090337 | regulation of formin-nucleated actin cable assembly(GO:0090337) positive regulation of formin-nucleated actin cable assembly(GO:0090338) |
| 0.2 | 1.2 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.2 | 0.5 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.2 | 1.4 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.2 | 0.7 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.2 | 0.7 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.2 | 0.6 | GO:0010942 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.2 | 0.8 | GO:0032102 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.2 | 0.6 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.2 | 0.6 | GO:0071454 | response to anoxia(GO:0034059) cellular response to anoxia(GO:0071454) |
| 0.2 | 0.6 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.2 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 2.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.2 | 1.5 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.2 | 0.7 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.2 | 5.2 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.2 | 0.7 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.2 | 1.4 | GO:0005987 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.2 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.2 | 1.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 0.5 | GO:0010994 | free ubiquitin chain polymerization(GO:0010994) |
| 0.2 | 2.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 0.6 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 0.2 | 0.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.2 | 0.5 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.2 | 1.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.4 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.7 | GO:0051274 | beta-glucan biosynthetic process(GO:0051274) |
| 0.1 | 0.3 | GO:0051098 | regulation of binding(GO:0051098) |
| 0.1 | 1.7 | GO:0046834 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.0 | GO:0033993 | response to lipid(GO:0033993) response to oleic acid(GO:0034201) response to fatty acid(GO:0070542) cellular response to lipid(GO:0071396) cellular response to fatty acid(GO:0071398) cellular response to oleic acid(GO:0071400) |
| 0.1 | 0.3 | GO:0000304 | response to singlet oxygen(GO:0000304) |
| 0.1 | 0.4 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.1 | 0.4 | GO:0032071 | regulation of deoxyribonuclease activity(GO:0032070) regulation of endodeoxyribonuclease activity(GO:0032071) positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.1 | 0.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.4 | GO:0000736 | double-strand break repair via single-strand annealing, removal of nonhomologous ends(GO:0000736) |
| 0.1 | 0.4 | GO:0010969 | regulation of pheromone-dependent signal transduction involved in conjugation with cellular fusion(GO:0010969) regulation of signal transduction involved in conjugation with cellular fusion(GO:0060238) |
| 0.1 | 0.5 | GO:0001080 | nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) nitrogen catabolite activation of transcription(GO:0090294) |
| 0.1 | 0.2 | GO:0006798 | polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.4 | GO:0090282 | positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.1 | 1.8 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 1.4 | GO:0001101 | response to acid chemical(GO:0001101) |
| 0.1 | 3.2 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.1 | 0.8 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 | 0.6 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.5 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.1 | 2.0 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.1 | 0.3 | GO:0042843 | D-xylose catabolic process(GO:0042843) |
| 0.1 | 0.3 | GO:2000284 | positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 1.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.3 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) negative regulation of cell cycle checkpoint(GO:1901977) regulation of DNA damage checkpoint(GO:2000001) negative regulation of DNA damage checkpoint(GO:2000002) negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.1 | 0.8 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.3 | GO:0071825 | lipid tube assembly(GO:0060988) protein-lipid complex assembly(GO:0065005) protein-lipid complex subunit organization(GO:0071825) |
| 0.1 | 0.3 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 1.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.3 | GO:0060188 | regulation of protein desumoylation(GO:0060188) |
| 0.1 | 0.3 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) |
| 0.1 | 0.8 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.2 | GO:0007119 | budding cell isotropic bud growth(GO:0007119) |
| 0.1 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.4 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.1 | 1.3 | GO:0012501 | apoptotic process(GO:0006915) cell death(GO:0008219) programmed cell death(GO:0012501) |
| 0.1 | 0.1 | GO:0010674 | negative regulation of transcription from RNA polymerase II promoter involved in meiotic cell cycle(GO:0010674) |
| 0.1 | 1.3 | GO:0015918 | sterol transport(GO:0015918) |
| 0.1 | 0.3 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of vesicle targeting, to, from or within Golgi(GO:0048209) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 1.3 | GO:0031503 | protein complex localization(GO:0031503) |
| 0.1 | 0.6 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.5 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.6 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 1.1 | GO:0016237 | membrane invagination(GO:0010324) lysosomal microautophagy(GO:0016237) single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.2 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.1 | 0.5 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 1.0 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.0 | GO:0001324 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline involved in chronological cell aging(GO:0001323) age-dependent response to oxidative stress involved in chronological cell aging(GO:0001324) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.5 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.0 | 0.3 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.2 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.0 | 0.2 | GO:0045033 | peroxisome inheritance(GO:0045033) |
| 0.0 | 0.3 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.1 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 0.1 | GO:0009107 | lipoate metabolic process(GO:0009106) lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0046174 | alditol catabolic process(GO:0019405) glycerol catabolic process(GO:0019563) polyol catabolic process(GO:0046174) |
| 0.0 | 0.2 | GO:0000436 | carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 | 0.3 | GO:0051653 | establishment of spindle localization(GO:0051293) spindle localization(GO:0051653) |
| 0.0 | 0.1 | GO:0090295 | nitrogen catabolite repression of transcription(GO:0090295) |
| 0.0 | 1.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.1 | GO:0043270 | positive regulation of ion transport(GO:0043270) |
| 0.0 | 0.5 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.4 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) UDP-glucose transport(GO:0015786) |
| 0.0 | 0.6 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.8 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.9 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.2 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.7 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 0.3 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.1 | GO:0000388 | spliceosome conformational change to release U4 (or U4atac) and U1 (or U11)(GO:0000388) |
| 0.0 | 0.2 | GO:0009187 | cyclic nucleotide metabolic process(GO:0009187) |
| 0.0 | 0.1 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 1.1 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0001881 | receptor recycling(GO:0001881) protein import into peroxisome matrix, receptor recycling(GO:0016562) receptor metabolic process(GO:0043112) |
| 0.0 | 0.1 | GO:0043007 | maintenance of rDNA(GO:0043007) |
| 0.0 | 0.2 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.8 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.2 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.0 | 0.3 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.0 | GO:2000045 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) regulation of cell cycle G1/S phase transition(GO:1902806) negative regulation of cell cycle G1/S phase transition(GO:1902807) positive regulation of cell cycle G1/S phase transition(GO:1902808) regulation of G1/S transition of mitotic cell cycle(GO:2000045) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.7 | GO:0072666 | protein targeting to vacuole(GO:0006623) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 0.2 | GO:0034401 | chromatin organization involved in regulation of transcription(GO:0034401) |
| 0.0 | 0.1 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.1 | GO:0019400 | glycerol metabolic process(GO:0006071) alditol metabolic process(GO:0019400) |
| 0.0 | 0.2 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 0.2 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 0.1 | GO:0060237 | regulation of fungal-type cell wall organization(GO:0060237) |
| 0.0 | 0.9 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.0 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0010525 | regulation of transposition, RNA-mediated(GO:0010525) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 13.5 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 1.3 | 15.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 1.1 | 8.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.9 | 2.7 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.8 | 2.3 | GO:0045265 | mitochondrial proton-transporting ATP synthase, stator stalk(GO:0000274) proton-transporting ATP synthase, stator stalk(GO:0045265) |
| 0.7 | 2.1 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.7 | 3.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.6 | 1.9 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.6 | 3.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.6 | 2.5 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.6 | 1.8 | GO:0070211 | Snt2C complex(GO:0070211) |
| 0.6 | 2.8 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.5 | 1.5 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.5 | 2.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.4 | 1.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.4 | 2.9 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.4 | 1.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.4 | 1.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 1.7 | GO:0001400 | mating projection base(GO:0001400) |
| 0.3 | 1.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 1.5 | GO:0034448 | EGO complex(GO:0034448) |
| 0.3 | 4.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 0.9 | GO:0030287 | cell wall-bounded periplasmic space(GO:0030287) |
| 0.2 | 1.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 5.1 | GO:0098573 | integral component of mitochondrial membrane(GO:0032592) intrinsic component of mitochondrial membrane(GO:0098573) |
| 0.2 | 1.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 4.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.2 | 1.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 3.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 6.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.8 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.1 | 0.9 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.3 | GO:0071561 | nucleus-vacuole junction(GO:0071561) |
| 0.1 | 1.8 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 1.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 3.1 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 2.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.6 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.9 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 0.3 | GO:0032126 | eisosome(GO:0032126) |
| 0.1 | 0.2 | GO:0032176 | split septin rings(GO:0032176) cellular bud neck split septin rings(GO:0032177) |
| 0.1 | 9.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 0.8 | GO:0097346 | Swr1 complex(GO:0000812) INO80-type complex(GO:0097346) |
| 0.1 | 0.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 4.7 | GO:0019867 | outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.1 | 0.6 | GO:1902555 | ribonuclease P complex(GO:0030677) endoribonuclease complex(GO:1902555) |
| 0.1 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 2.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 15.2 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 0.3 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.0 | 0.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.9 | GO:0005934 | cellular bud tip(GO:0005934) |
| 0.0 | 0.2 | GO:0035361 | Cul8-RING ubiquitin ligase complex(GO:0035361) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.3 | GO:0033698 | Rpd3L complex(GO:0033698) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 9.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.0 | GO:0031310 | intrinsic component of vacuolar membrane(GO:0031310) vacuolar transporter chaperone complex(GO:0033254) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0016406 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 1.7 | 5.2 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 1.7 | 5.2 | GO:0016208 | AMP binding(GO:0016208) |
| 1.5 | 5.9 | GO:0005537 | mannose binding(GO:0005537) |
| 1.3 | 5.2 | GO:0005471 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 1.2 | 13.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.1 | 5.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 1.1 | 7.7 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.9 | 2.8 | GO:0048038 | quinone binding(GO:0048038) |
| 0.9 | 7.0 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.9 | 4.3 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 0.9 | 6.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.8 | 15.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.8 | 3.3 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.8 | 2.5 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.8 | 1.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.8 | 3.1 | GO:0016898 | lactate dehydrogenase activity(GO:0004457) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.8 | 3.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.7 | 2.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.7 | 2.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.7 | 3.4 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.7 | 3.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.6 | 1.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.6 | 4.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.6 | 5.4 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.6 | 2.4 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.6 | 1.2 | GO:0015489 | putrescine transmembrane transporter activity(GO:0015489) |
| 0.6 | 3.5 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.5 | 13.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.5 | 1.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.5 | 1.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.5 | 5.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.5 | 7.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.5 | 1.8 | GO:0015603 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.4 | 4.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.4 | 1.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 1.1 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.4 | 2.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.3 | 1.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.3 | 4.2 | GO:0022838 | substrate-specific channel activity(GO:0022838) |
| 0.3 | 0.3 | GO:0015151 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.3 | 0.3 | GO:0052742 | phosphatidylinositol kinase activity(GO:0052742) |
| 0.3 | 2.7 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 1.2 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 0.3 | 1.4 | GO:0050253 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.3 | 1.4 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.3 | 2.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.3 | 3.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.3 | 0.3 | GO:0016878 | CoA-ligase activity(GO:0016405) acid-thiol ligase activity(GO:0016878) |
| 0.2 | 2.0 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.2 | 1.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 1.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 0.7 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 1.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 2.0 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.2 | 3.1 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.2 | 0.8 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 0.6 | GO:0015186 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.2 | 0.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 0.8 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.2 | 0.6 | GO:0031406 | amino acid binding(GO:0016597) carboxylic acid binding(GO:0031406) organic acid binding(GO:0043177) |
| 0.2 | 0.9 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.2 | 4.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.2 | 2.4 | GO:0016684 | peroxidase activity(GO:0004601) oxidoreductase activity, acting on peroxide as acceptor(GO:0016684) |
| 0.2 | 1.5 | GO:0016723 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.2 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 1.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 0.6 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.2 | 2.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 0.6 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 2.5 | GO:0016298 | lipase activity(GO:0016298) |
| 0.1 | 0.6 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 2.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 7.2 | GO:0043492 | ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.1 | 0.4 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 2.2 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 10.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.1 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.5 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.1 | 0.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.5 | GO:1901618 | alcohol transmembrane transporter activity(GO:0015665) organic hydroxy compound transmembrane transporter activity(GO:1901618) |
| 0.1 | 0.5 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.1 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.1 | 1.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.4 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.8 | GO:0016769 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.5 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.1 | 0.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 2.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.3 | GO:0001025 | transcription factor activity, core RNA polymerase III binding(GO:0000995) RNA polymerase III transcription factor binding(GO:0001025) TFIIIB-type transcription factor activity(GO:0001026) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 0.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 1.0 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.1 | 0.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 0.4 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 0.2 | GO:0051864 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) histone demethylase activity(GO:0032452) histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.5 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 0.2 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.1 | 0.2 | GO:0004691 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.4 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.5 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 0.9 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.1 | 0.3 | GO:0016830 | carbon-carbon lyase activity(GO:0016830) |
| 0.1 | 0.7 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 0.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 2.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 1.1 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.1 | 0.7 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0015405 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) |
| 0.0 | 0.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.6 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.2 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.7 | 2.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.6 | 1.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.4 | 1.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 0.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 1.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 0.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 87.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 17.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.8 | 2.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.4 | 1.5 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.3 | 1.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.2 | 2.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 0.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.7 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 0.8 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.1 | 87.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME INNATE IMMUNE SYSTEM | Genes involved in Innate Immune System |