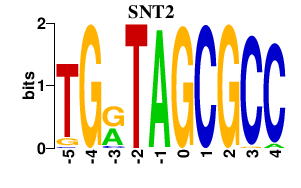
Results for SNT2
Z-value: 0.73
Transcription factors associated with SNT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SNT2
|
S000003099 | DNA binding protein with similarity to the S. pombe Snt2 protein |
Activity-expression correlation:
Activity profile of SNT2 motif
Sorted Z-values of SNT2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YMR107W | 9.05 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YJR095W | 5.93 |
SFC1
|
Mitochondrial succinate-fumarate transporter, transports succinate into and fumarate out of the mitochondrion; required for ethanol and acetate utilization |
|
| YLR327C | 4.74 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YIL160C | 4.25 |
POT1
|
3-ketoacyl-CoA thiolase with broad chain length specificity, cleaves 3-ketoacyl-CoA into acyl-CoA and acetyl-CoA during beta-oxidation of fatty acids |
|
| YBL075C | 3.63 |
SSA3
|
ATPase involved in protein folding and the response to stress; plays a role in SRP-dependent cotranslational protein-membrane targeting and translocation; member of the heat shock protein 70 (HSP70) family; localized to the cytoplasm |
|
| YJR078W | 3.44 |
BNA2
|
Putative tryptophan 2,3-dioxygenase or indoleamine 2,3-dioxygenase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p and Aft2p |
|
| YAL039C | 3.20 |
CYC3
|
Cytochrome c heme lyase (holocytochrome c synthase), attaches heme to apo-cytochrome c (Cyc1p or Cyc7p) in the mitochondrial intermembrane space; human ortholog may have a role in microphthalmia with linear skin defects (MLS) |
|
| YOR343C | 3.05 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YAR050W | 2.84 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YHR138C | 2.60 |
Putative protein of unknown function; has similarity to Pbi2p; double null mutant lacking Pbi2p and Yhr138p exhibits highly fragmented vacuoles |
||
| YOR348C | 2.47 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YMR096W | 2.39 |
SNZ1
|
Protein involved in vitamin B6 biosynthesis; member of a stationary phase-induced gene family; coregulated with SNO1; interacts with Sno1p and with Yhr198p, perhaps as a multiprotein complex containing other Snz and Sno proteins |
|
| YDR536W | 2.37 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YLR149C | 2.35 |
Putative protein of unknown function; YLR149C is not an essential gene |
||
| YBR145W | 2.35 |
ADH5
|
Alcohol dehydrogenase isoenzyme V; involved in ethanol production |
|
| YAR047C | 2.26 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YCR091W | 2.13 |
KIN82
|
Putative serine/threonine protein kinase, most similar to cyclic nucleotide-dependent protein kinase subfamily and the protein kinase C subfamily |
|
| YPR184W | 2.11 |
GDB1
|
Glycogen debranching enzyme containing glucanotranferase and alpha-1,6-amyloglucosidase activities, required for glycogen degradation; phosphorylated in mitochondria |
|
| YGL121C | 2.09 |
GPG1
|
Proposed gamma subunit of the heterotrimeric G protein that interacts with the receptor Grp1p; involved in regulation of pseudohyphal growth; requires Gpb1p or Gpb2p to interact with Gpa2p |
|
| YPR027C | 2.06 |
Putative protein of unknown function |
||
| YIL055C | 2.06 |
Putative protein of unknown function |
||
| YPR010C-A | 2.06 |
Putative protein of unknown function; conserved among Saccharomyces sensu stricto species |
||
| YLR205C | 1.99 |
HMX1
|
ER localized, heme-binding peroxidase involved in the degradation of heme; does not exhibit heme oxygenase activity despite similarity to heme oxygenases; expression regulated by AFT1 |
|
| YDL169C | 1.98 |
UGX2
|
Protein of unknown function, transcript accumulates in response to any combination of stress conditions |
|
| YDL214C | 1.83 |
PRR2
|
Serine/threonine protein kinase that inhibits pheromone induced signalling downstream of MAPK, possibly at the level of the Ste12p transcription factor |
|
| YPR120C | 1.78 |
CLB5
|
B-type cyclin involved in DNA replication during S phase; activates Cdc28p to promote initiation of DNA synthesis; functions in formation of mitotic spindles along with Clb3p and Clb4p; most abundant during late G1 phase |
|
| YMR141C | 1.73 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR438W | 1.66 |
CAR2
|
L-ornithine transaminase (OTAse), catalyzes the second step of arginine degradation, expression is dually-regulated by allophanate induction and a specific arginine induction process; not nitrogen catabolite repression sensitive |
|
| YBR144C | 1.61 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR144C is not an essential gene |
||
| YHR211W | 1.60 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YMR090W | 1.60 |
Putative protein of unknown function with similarity to DTDP-glucose 4,6-dehydratases; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YMR090W is not an essential gene |
||
| YJR128W | 1.59 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF RSF2 |
||
| YCL038C | 1.55 |
ATG22
|
Vacuolar integral membrane protein required for efflux of amino acids during autophagic body breakdown in the vacuole; null mutation causes a gradual loss of viability during starvation |
|
| YPL017C | 1.51 |
IRC15
|
Putative S-adenosylmethionine-dependent methyltransferase of the seven beta-strand family, required for accurate meiotic chromosome segregation; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YNL133C | 1.48 |
FYV6
|
Protein of unknown function, required for survival upon exposure to K1 killer toxin; proposed to regulate double-strand break repair via non-homologous end-joining |
|
| YNL274C | 1.47 |
GOR1
|
Glyoxylate reductase; null mutation results in increased biomass after diauxic shift; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YDR542W | 1.47 |
PAU10
|
Hypothetical protein |
|
| YGL073W | 1.47 |
HSF1
|
Trimeric heat shock transcription factor, activates multiple genes in response to stresses that include hyperthermia; recognizes variable heat shock elements (HSEs) consisting of inverted NGAAN repeats; posttranslationally regulated |
|
| YMR095C | 1.43 |
SNO1
|
Protein of unconfirmed function, involved in pyridoxine metabolism; expression is induced during stationary phase; forms a putative glutamine amidotransferase complex with Snz1p, with Sno1p serving as the glutaminase |
|
| YJL037W | 1.43 |
IRC18
|
Putative protein of unknown function; expression induced in respiratory-deficient cells and in carbon-limited chemostat cultures; similar to adjacent ORF, YJL038C; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YOL025W | 1.41 |
LAG2
|
Protein involved in determination of longevity; LAG2 gene is preferentially expressed in young cells; overexpression extends the mean and maximum life span of cells |
|
| YDL138W | 1.39 |
RGT2
|
Plasma membrane glucose receptor, highly similar to Snf3p; both Rgt2p and Snf3p serve as transmembrane glucose sensors generating an intracellular signal that induces expression of glucose transporter (HXT) genes |
|
| YGR043C | 1.39 |
NQM1
|
Protein of unknown function; transcription is repressed by Mot1p and induced by alpha-factor and during diauxic shift; null mutant non-quiescent cells exhibit reduced reproductive capacity |
|
| YJR077C | 1.38 |
MIR1
|
Mitochondrial phosphate carrier, imports inorganic phosphate into mitochondria; functionally redundant with Pic2p but more abundant than Pic2p under normal conditions; phosphorylated |
|
| YLR437C-A | 1.37 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF CAR2/YLR438W |
||
| YER084W | 1.36 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YKL020C | 1.33 |
SPT23
|
ER membrane protein involved in regulation of OLE1 transcription, acts with homolog Mga2p; inactive ER form dimerizes and one subunit is then activated by ubiquitin/proteasome-dependent processing followed by nuclear targeting |
|
| YBR117C | 1.33 |
TKL2
|
Transketolase, similar to Tkl1p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YFL007W | 1.28 |
BLM10
|
Proteosome activator subunit; found in association with core particles, with and without the 19S regulatory particle; required for resistance to bleomycin, may be involved in protecting against oxidative damage; similar to mammalian PA200 |
|
| YDR277C | 1.27 |
MTH1
|
Negative regulator of the glucose-sensing signal transduction pathway, required for repression of transcription by Rgt1p; interacts with Rgt1p and the Snf3p and Rgt2p glucose sensors; phosphorylated by Yck1p, triggering Mth1p degradation |
|
| YHR160C | 1.27 |
PEX18
|
Peroxin required for targeting of peroxisomal matrix proteins containing PTS2; interacts with Pex7p; partially redundant with Pex21p |
|
| YMR118C | 1.27 |
Protein of unknown function with similarity to succinate dehydrogenase cytochrome b subunit; YMR118C is not an essential gene |
||
| YPL164C | 1.24 |
MLH3
|
Protein involved in DNA mismatch repair and crossing-over during meiotic recombination; forms a complex with Mlh1p; mammalian homolog is implicated mammalian microsatellite instability |
|
| YEL009C | 1.24 |
GCN4
|
Transcriptional activator of amino acid biosynthetic genes in response to amino acid starvation; expression is tightly regulated at both the transcriptional and translational levels |
|
| YDL114W | 1.24 |
Putative protein of unknown function with similarity to acyl-carrier-protein reductases; YDL114W is not an essential gene |
||
| YML120C | 1.22 |
NDI1
|
NADH:ubiquinone oxidoreductase, transfers electrons from NADH to ubiquinone in the respiratory chain but does not pump protons, in contrast to the higher eukaryotic multisubunit respiratory complex I; phosphorylated; homolog of human AMID |
|
| YEL008W | 1.20 |
Hypothetical protein predicted to be involved in metabolism |
||
| YOR011W-A | 1.20 |
Putative protein of unknown function |
||
| YEL028W | 1.18 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YAR019W-A | 1.16 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YMR316C-B | 1.15 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL093W | 1.15 |
SPC105
|
Protein required for accurate chromosome segregation, localizes to the nuclear side of the spindle pole body; forms a complex with Ydr532cp |
|
| YGL074C | 1.15 |
Dubious open reading frame unlikely to encode a functional protein; overlaps 5' end of essential HSF1 gene encoding heat shock transcription factor |
||
| YLR111W | 1.14 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR307W | 1.13 |
CDA1
|
Chitin deacetylase, together with Cda2p involved in the biosynthesis ascospore wall component, chitosan; required for proper rigidity of the ascospore wall |
|
| YBR180W | 1.13 |
DTR1
|
Putative dityrosine transporter, required for spore wall synthesis; expressed during sporulation; member of the major facilitator superfamily (DHA1 family) of multidrug resistance transporters |
|
| YER121W | 1.13 |
Putative protein of unknown function; may be involved in phosphatase regulation and/or generation of precursor metabolites and energy |
||
| YMR017W | 1.12 |
SPO20
|
Meiosis-specific subunit of the t-SNARE complex, required for prospore membrane formation during sporulation; similar to but not functionally redundant with Sec9p; SNAP-25 homolog |
|
| YBR179C | 1.12 |
FZO1
|
Mitochondrial integral membrane protein involved in mitochondrial fusion and maintenance of the mitochondrial genome; contains N-terminal GTPase domain |
|
| YHR008C | 1.12 |
SOD2
|
Mitochondrial superoxide dismutase, protects cells against oxygen toxicity; phosphorylated |
|
| YFR053C | 1.11 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YBR208C | 1.10 |
DUR1,2
|
Urea amidolyase, contains both urea carboxylase and allophanate hydrolase activities, degrades urea to CO2 and NH3; expression sensitive to nitrogen catabolite repression and induced by allophanate, an intermediate in allantoin degradation |
|
| YOR152C | 1.10 |
Putative protein of unknown function; has no similarity to any known protein; YOR152C is not an essential gene |
||
| YOR148C | 1.08 |
SPP2
|
Essential protein that promotes the first step of splicing and is required for the final stages of spliceosome maturation; interacts with Prp2p, which may release Spp2p from the spliceosome following the first cleavage reaction |
|
| YJR151C | 1.06 |
DAN4
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YPL258C | 1.03 |
THI21
|
Hydroxymethylpyrimidine phosphate kinase, involved in the last steps in thiamine biosynthesis; member of a gene family with THI20 and THI22; functionally redundant with Thi20p |
|
| YAR019C | 1.03 |
CDC15
|
Protein kinase of the Mitotic Exit Network that is localized to the spindle pole bodies at late anaphase; promotes mitotic exit by directly switching on the kinase activity of Dbf2p |
|
| YMR317W | 1.02 |
Putative protein of unknown function with some similarity to sialidase from Trypanosoma; YMR317W is not an essential gene |
||
| YOR186W | 1.02 |
Putative protein of unknown function; proper regulation of expression during heat stress is sphingolipid-dependent |
||
| YBR071W | 1.02 |
Putative protein of unknown function; (GFP)-fusion and epitope-tagged proteins localize to the cytoplasm; mRNA expression may be regulated by the cell cycle and/or cell wall stress |
||
| YFL055W | 1.02 |
AGP3
|
Low-affinity amino acid permease, may act to supply the cell with amino acids as nitrogen source in nitrogen-poor conditions; transcription is induced under conditions of sulfur limitation; plays a role in regulating Ty1 transposition |
|
| YLR142W | 1.01 |
PUT1
|
Proline oxidase, nuclear-encoded mitochondrial protein involved in utilization of proline as sole nitrogen source; PUT1 transcription is induced by Put3p in the presence of proline and the absence of a preferred nitrogen source |
|
| YML054C | 1.01 |
CYB2
|
Cytochrome b2 (L-lactate cytochrome-c oxidoreductase), component of the mitochondrial intermembrane space, required for lactate utilization; expression is repressed by glucose and anaerobic conditions |
|
| YLR377C | 1.00 |
FBP1
|
Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway, required for glucose metabolism |
|
| YJR106W | 0.98 |
ECM27
|
Non-essential protein of unknown function |
|
| YDR343C | 0.96 |
HXT6
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt7p, expressed at high basal levels relative to other HXTs, repression of expression by high glucose requires SNF3 |
|
| YER187W | 0.96 |
Putative protein of unknown function; induced in respiratory-deficient cells |
||
| YOR065W | 0.95 |
CYT1
|
Cytochrome c1, component of the mitochondrial respiratory chain; expression is regulated by the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex |
|
| YDR446W | 0.92 |
ECM11
|
Non-essential protein apparently involved in meiosis, GFP fusion protein is present in discrete clusters in the nucleus throughout mitosis; may be involved in maintaining chromatin structure |
|
| YGL170C | 0.91 |
SPO74
|
Component of the meiotic outer plaque of the spindle pole body, involved in modifying the meiotic outer plaque that is required prior to prospore membrane formation |
|
| YDR070C | 0.90 |
FMP16
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YOL067C | 0.89 |
RTG1
|
Transcription factor (bHLH) involved in interorganelle communication between mitochondria, peroxisomes, and nucleus |
|
| YIL107C | 0.89 |
PFK26
|
6-phosphofructo-2-kinase, inhibited by phosphoenolpyruvate and sn-glycerol 3-phosphate, has negligible fructose-2,6-bisphosphatase activity, transcriptional regulation involves protein kinase A |
|
| YAR035W | 0.89 |
YAT1
|
Outer mitochondrial carnitine acetyltransferase, minor ethanol-inducible enzyme involved in transport of activated acyl groups from the cytoplasm into the mitochondrial matrix; phosphorylated |
|
| YEL009C-A | 0.89 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNR002C | 0.89 |
ATO2
|
Putative transmembrane protein involved in export of ammonia, a starvation signal that promotes cell death in aging colonies; phosphorylated in mitochondria; member of the TC 9.B.33 YaaH family; homolog of Ady2p and Y. lipolytica Gpr1p |
|
| YGL033W | 0.87 |
HOP2
|
Meiosis-specific protein that localizes to chromosomes, preventing synapsis between nonhomologous chromosomes and ensuring synapsis between homologs; complexes with Mnd1p to promote homolog pairing and meiotic double-strand break repair |
|
| YOL100W | 0.87 |
PKH2
|
Serine/threonine protein kinase involved in sphingolipid-mediated signaling pathway that controls endocytosis; activates Ypk1p and Ykr2p, components of signaling cascade required for maintenance of cell wall integrity; redundant with Pkh1p |
|
| YHR082C | 0.87 |
KSP1
|
Nonessential putative serine/threonine protein kinase of unknown cellular role; overproduction causes allele-specific suppression of the prp20-10 mutation |
|
| YLR228C | 0.84 |
ECM22
|
Sterol regulatory element binding protein, regulates transcription of the sterol biosynthetic genes ERG2 and ERG3; member of the fungus-specific Zn[2]-Cys[6] binuclear cluster family of transcription factors; homologous to Upc2p |
|
| YEL007W | 0.84 |
Putative protein with sequence similarity to S. pombe gti1+ (gluconate transport inducer 1) |
||
| YLR176C | 0.83 |
RFX1
|
Major transcriptional repressor of DNA-damage-regulated genes, recruits repressors Tup1p and Cyc8p to their promoters; involved in DNA damage and replication checkpoint pathway; similar to a family of mammalian DNA binding RFX1-4 proteins |
|
| YJL038C | 0.83 |
LOH1
|
Putative protein of unknown function; expression induced during sporulation and repressed during vegetative growth by Sum1p and Hst1p; similar to adjacent open reading frame, YJL037W |
|
| YBR077C | 0.83 |
SLM4
|
Component of the EGO complex, which is involved in the regulation of microautophagy, and of the GSE complex, which is required for proper sorting of amino acid permease Gap1p; gene exhibits synthetic genetic interaction with MSS4 |
|
| YDR216W | 0.82 |
ADR1
|
Carbon source-responsive zinc-finger transcription factor, required for transcription of the glucose-repressed gene ADH2, of peroxisomal protein genes, and of genes required for ethanol, glycerol, and fatty acid utilization |
|
| YOL060C | 0.82 |
MAM3
|
Protein required for normal mitochondrial morphology, has similarity to hemolysins |
|
| YNL240C | 0.81 |
NAR1
|
Nuclear architecture related protein; component of the cytosolic iron-sulfur (FeS) protein assembly machinery, required for maturation of cytosolic and nuclear FeS proteins; homologous to human Narf |
|
| YBR212W | 0.81 |
NGR1
|
RNA binding protein that negatively regulates growth rate; interacts with the 3' UTR of the mitochondrial porin (POR1) mRNA and enhances its degradation; overexpression impairs mitochondrial function; expressed in stationary phase |
|
| YER167W | 0.81 |
BCK2
|
Protein rich in serine and threonine residues involved in protein kinase C signaling pathway, which controls cell integrity; overproduction suppresses pkc1 mutations |
|
| YLR402W | 0.81 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDL139C | 0.79 |
SCM3
|
Nonhistone component of centromeric chromatin that binds stoichiometrically to CenH3-H4 histones, required for kinetochore assembly; contains nuclear export signal (NES); required for G2/M progression and localization of Cse4p |
|
| YOR386W | 0.78 |
PHR1
|
DNA photolyase involved in photoreactivation, repairs pyrimidine dimers in the presence of visible light; induced by DNA damage; regulated by transcriptional repressor Rph1p |
|
| YLR403W | 0.78 |
SFP1
|
Transcription factor that controls expression of many ribosome biogenesis genes in response to nutrients and stress, regulates G2/M transitions during mitotic cell cycle and DNA-damage response, involved in cell size modulation |
|
| YLL056C | 0.78 |
Putative protein of unknown function, transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in pleiotropic drug resistance (PDR) phenomenon; YLL056C is not an essential gene |
||
| YMR279C | 0.77 |
Putative protein of unknown function; identified as a heat-induced gene in a high-throughout screen; YMR279C is not an essential gene |
||
| YJL093C | 0.77 |
TOK1
|
Outward-rectifier potassium channel of the plasma membrane with two pore domains in tandem, each of which forms a functional channel permeable to potassium; carboxy tail functions to prevent inner gate closures; target of K1 toxin |
|
| YHR002W | 0.76 |
LEU5
|
Mitochondrial carrier protein involved in the accumulation of CoA in the mitochondrial matrix; homolog of human Graves disease protein; does not encode an isozyme of Leu4p, as first hypothesized |
|
| YBR139W | 0.76 |
Putative serine type carboxypeptidase with a role in phytochelatin synthesis; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; expression induced by nitrogen limitation in a GLN3, GAT1-independent manner |
||
| YOR235W | 0.74 |
IRC13
|
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YDR218C | 0.72 |
SPR28
|
Sporulation-specific homolog of the yeast CDC3/10/11/12 family of bud neck microfilament genes; meiotic septin expressed at high levels during meiotic divisions and ascospore formation |
|
| YHR125W | 0.72 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR256W | 0.70 |
GND2
|
6-phosphogluconate dehydrogenase (decarboxylating), catalyzes an NADPH regenerating reaction in the pentose phosphate pathway; required for growth on D-glucono-delta-lactone |
|
| YDL130W-A | 0.70 |
STF1
|
Protein involved in regulation of the mitochondrial F1F0-ATP synthase; Stf1p and Stf2p may act as stabilizing factors that enhance inhibitory action of the Inh1p protein |
|
| YLR035C | 0.70 |
MLH2
|
Protein involved in the mismatch repair of certain frameshift intermediates and involved in meiotic recombination; forms a complex with Mlh1p |
|
| YCL069W | 0.67 |
VBA3
|
Permease of basic amino acids in the vacuolar membrane |
|
| YGR053C | 0.67 |
Putative protein of unknown function |
||
| YHR210C | 0.67 |
Putative protein of unknown function; non-essential gene; highly expressed under anaeorbic conditions; sequence similarity to aldose 1-epimerases such as GAL10 |
||
| YNL223W | 0.66 |
ATG4
|
Conserved cysteine protease required for autophagy; cleaves Atg8p to a form required for autophagosome and Cvt vesicle generation; mediates attachment of autophagosomes to microtubules through interactions with Tub1p and Tub2p |
|
| YKL023W | 0.65 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YOL026C | 0.65 |
MIM1
|
Mitochondrial outer membrane protein, required for assembly of the translocase of the outer membrane (TOM) complex and thereby for mitochondrial protein import; N terminus is exposed to the cytosol: transmembrane segment is highly conserved |
|
| YGR110W | 0.65 |
Putative protein of unknown function; transcription is increased in response to genotoxic stress; plays a role in restricting Ty1 transposition |
||
| YJR115W | 0.63 |
Putative protein of unknown function |
||
| YEL010W | 0.60 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YKR021W | 0.60 |
ALY1
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
|
| YPR023C | 0.59 |
EAF3
|
Esa1p-associated factor, nonessential component of the NuA4 acetyltransferase complex, homologous to Drosophila dosage compensation protein MSL3; plays a role in regulating Ty1 transposition |
|
| YCL025C | 0.59 |
AGP1
|
Low-affinity amino acid permease with broad substrate range, involved in uptake of asparagine, glutamine, and other amino acids; expression is regulated by the SPS plasma membrane amino acid sensor system (Ssy1p-Ptr3p-Ssy5p) |
|
| YAL034C | 0.59 |
FUN19
|
Non-essential protein of unknown function |
|
| YGL094C | 0.58 |
PAN2
|
Essential subunit of the Pan2p-Pan3p poly(A)-ribonuclease complex, which acts to control poly(A) tail length and regulate the stoichiometry and activity of postreplication repair complexes |
|
| YGL096W | 0.58 |
TOS8
|
Homeodomain-containing transcription factor; SBF regulated target gene that in turn regulates expression of genes involved in G1/S phase events such as bud site selection, bud emergence and cell cycle progression; similarity to Cup9p |
|
| YOR062C | 0.58 |
Protein of unknown function; similar to YKR075Cp and Reg1p; expression regulated by glucose and Rgt1p; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YDR247W | 0.58 |
VHS1
|
Cytoplasmic serine/threonine protein kinase; identified as a high-copy suppressor of the synthetic lethality of a sis2 sit4 double mutant, suggesting a role in G1/S phase progression; homolog of Sks1p |
|
| YBL100C | 0.58 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; almost completely overlaps the 5' end of ATP1 |
||
| YOR072W | 0.57 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOR072W-A; diploid deletion strains are methotrexate, paraquat and wortmannin sensitive |
||
| YJR085C | 0.57 |
Putative protein of unknown function; GFP-fusion protein is induced in response to the DNA-damaging agent MMS; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YBL099W | 0.57 |
ATP1
|
Alpha subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; phosphorylated |
|
| YDR287W | 0.56 |
INM2
|
Inositol monophosphatase, involved in biosynthesis of inositol; enzymatic activity requires magnesium ions and is inhibited by lithium and sodium ions; inm1 inm2 double mutant lacks inositol auxotrophy |
|
| YPR054W | 0.56 |
SMK1
|
Middle sporulation-specific mitogen-activated protein kinase (MAPK) required for production of the outer spore wall layers; negatively regulates activity of the glucan synthase subunit Gsc2p |
|
| YEL059W | 0.56 |
Dubious open reading frame unlikely to encode a functional protein |
||
| YBR039W | 0.55 |
ATP3
|
Gamma subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YJL116C | 0.55 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YIR014W | 0.55 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; expression directly regulated by the metabolic and meiotic transcriptional regulator Ume6p; YIR014W is a non-essential gene |
||
| YKL217W | 0.55 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YDR090C | 0.54 |
Putative protein of unknown function |
||
| YGL197W | 0.54 |
MDS3
|
Protein with an N-terminal kelch-like domain, putative negative regulator of early meiotic gene expression; required, with Pmd1p, for growth under alkaline conditions |
|
| YMR053C | 0.54 |
STB2
|
Protein that interacts with Sin3p in a two-hybrid assay and is part of a large protein complex with Sin3p and Stb1p |
|
| YPL214C | 0.53 |
THI6
|
Bifunctional enzyme with thiamine-phosphate pyrophosphorylase and 4-methyl-5-beta-hydroxyethylthiazole kinase activities, required for thiamine biosynthesis; GFP-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YLR122C | 0.53 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR123C |
||
| YHR075C | 0.52 |
PPE1
|
Protein with carboxyl methyl esterase activity that may have a role in demethylation of the phosphoprotein phosphatase catalytic subunit; also identified as a small subunit mitochondrial ribosomal protein |
|
| YDR541C | 0.52 |
Putative dihydrokaempferol 4-reductase |
||
| YAR053W | 0.50 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR041W | 0.50 |
BUD9
|
Protein involved in bud-site selection; diploid mutants display a unipolar budding pattern instead of the wild-type bipolar pattern, and bud at the distal pole |
|
| YDL054C | 0.50 |
MCH1
|
Protein with similarity to mammalian monocarboxylate permeases, which are involved in transport of monocarboxylic acids across the plasma membrane; mutant is not deficient in monocarboxylate transport |
|
| YMR170C | 0.49 |
ALD2
|
Cytoplasmic aldehyde dehydrogenase, involved in ethanol oxidation and beta-alanine biosynthesis; uses NAD+ as the preferred coenzyme; expression is stress induced and glucose repressed; very similar to Ald3p |
|
| YIL057C | 0.48 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YAL001C | 0.48 |
TFC3
|
Largest of six subunits of the RNA polymerase III transcription initiation factor complex (TFIIIC); part of the TauB domain of TFIIIC that binds DNA at the BoxB promoter sites of tRNA and similar genes; cooperates with Tfc6p in DNA binding |
|
| YDR014W-A | 0.48 |
HED1
|
Meiosis-specific protein that down-regulates Rad51p-mediated mitotic recombination when the meiotic recombination machinery is impaired; early meiotic gene, transcribed specifically during meiotic prophase |
|
| YDR282C | 0.48 |
Putative protein of unknown function |
||
| YLL055W | 0.48 |
YCT1
|
High-affinity cysteine-specific transporter with similarity to the Dal5p family of transporters; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; YCT1 is not an essential gene |
|
| YKL028W | 0.47 |
TFA1
|
TFIIE large subunit, involved in recruitment of RNA polymerase II to the promoter, activation of TFIIH, and promoter opening |
|
| YKR040C | 0.47 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YKR041W |
||
| YBR209W | 0.47 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR209W is not an essential gene |
||
| YKR049C | 0.47 |
FMP46
|
Putative redox protein containing a thioredoxin fold; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YNL025C | 0.47 |
SSN8
|
Cyclin-like component of the RNA polymerase II holoenzyme, involved in phosphorylation of the RNA polymerase II C-terminal domain; involved in glucose repression and telomere maintenance |
|
| YKL171W | 0.46 |
Putative protein of unknown function; predicted protein kinase; implicated in proteasome function; epitope-tagged protein localizes to the cytoplasm |
||
| YER098W | 0.46 |
UBP9
|
Ubiquitin carboxyl-terminal hydrolase, ubiquitin-specific protease that cleaves ubiquitin-protein fusions |
|
| YLR346C | 0.46 |
Putative protein of unknown function found in mitochondria; expression is regulated by transcription factors involved in pleiotropic drug resistance, Pdr1p and Yrr1p; YLR346C is not an essential gene |
||
| YLR123C | 0.46 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR122C; contains characteristic aminoacyl-tRNA motif |
||
| YLR425W | 0.46 |
TUS1
|
Guanine nucleotide exchange factor (GEF) that functions to modulate Rho1p activity as part of the cell integrity signaling pathway; multicopy suppressor of tor2 mutation and ypk1 ypk2 double mutation; potential Cdc28p substrate |
|
| YOR382W | 0.46 |
FIT2
|
Mannoprotein that is incorporated into the cell wall via a glycosylphosphatidylinositol (GPI) anchor, involved in the retention of siderophore-iron in the cell wall |
|
| YFR007W | 0.46 |
YFH7
|
Putative kinase with similarity to the phosphoribulokinase/uridine kinase/bacterial pantothenate kinase (PRK/URK/PANK) subfamily of P-loop kinases; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) |
|
| YPR169W-A | 0.45 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps two other dubious ORFs: YPR170C and YPR170W-B |
||
| YKR105C | 0.45 |
VBA5
|
Putative transporter of the Major Facilitator Superfamily (MFS); proposed role as a basic amino acid permease based on phylogeny |
|
| YGL236C | 0.45 |
MTO1
|
Mitochondrial protein, forms a heterodimer complex with Mss1p that performs the 5-carboxymethylaminomethyl modification of the wobble uridine base in mitochondrial tRNAs; required for respiration in paromomycin-resistant 15S rRNA mutants |
|
| YMR041C | 0.45 |
ARA2
|
NAD-dependent arabinose dehydrogenase, involved in biosynthesis of erythroascorbic acid; similar to plant L-galactose dehydrogenase |
|
| YOR071C | 0.45 |
NRT1
|
High-affinity nicotinamide riboside transporter; also transports thiamine with low affinity; shares sequence similarity with Thi7p and Thi72p; proposed to be involved in 5-fluorocytosine sensitivity |
|
| YGL163C | 0.45 |
RAD54
|
DNA-dependent ATPase, stimulates strand exchange by modifying the topology of double-stranded DNA; involved in the recombinational repair of double-strand breaks in DNA during vegetative growth and meiosis; member of the SWI/SNF family |
|
| YDR445C | 0.45 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJL092W | 0.44 |
SRS2
|
DNA helicase and DNA-dependent ATPase involved in DNA repair, needed for proper timing of commitment to meiotic recombination and transition from Meiosis I to II; affects genome stability by suppressing unscheduled homologous recombination |
|
| YCR073W-A | 0.44 |
SOL2
|
Protein with a possible role in tRNA export; shows similarity to 6-phosphogluconolactonase non-catalytic domains but does not exhibit this enzymatic activity; homologous to Sol1p, Sol3p, and Sol4p |
|
| YGR048W | 0.44 |
UFD1
|
Protein that interacts with Cdc48p and Npl4p, involved in recognition of polyubiquitinated proteins and their presentation to the 26S proteasome for degradation; involved in transporting proteins from the ER to the cytosol |
|
| YPL006W | 0.44 |
NCR1
|
Vacuolar membrane protein that transits through the biosynthetic vacuolar protein sorting pathway, involved in sphingolipid metabolism; glycoprotein and functional orthologue of human Niemann Pick C1 (NPC1) protein |
|
| YNR019W | 0.43 |
ARE2
|
Acyl-CoA:sterol acyltransferase, isozyme of Are1p; endoplasmic reticulum enzyme that contributes the major sterol esterification activity in the presence of oxygen |
|
| YCL040W | 0.42 |
GLK1
|
Glucokinase, catalyzes the phosphorylation of glucose at C6 in the first irreversible step of glucose metabolism; one of three glucose phosphorylating enzymes; expression regulated by non-fermentable carbon sources |
|
| YLR070C | 0.42 |
XYL2
|
Xylitol dehydrogenase, converts xylitol to D-xylulose in the endogenous xylose utilization pathway |
|
| YLR454W | 0.42 |
FMP27
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YPR124W | 0.42 |
CTR1
|
High-affinity copper transporter of the plasma membrane, mediates nearly all copper uptake under low copper conditions; transcriptionally induced at low copper levels and degraded at high copper levels |
|
| YPR123C | 0.42 |
Hypothetical protein |
||
| YBR083W | 0.42 |
TEC1
|
Transcription factor required for full Ty1 epxression, Ty1-mediated gene activation, and haploid invasive and diploid pseudohyphal growth; TEA/ATTS DNA-binding domain family member |
|
| YDR085C | 0.42 |
AFR1
|
Alpha-factor pheromone receptor regulator, negatively regulates pheromone receptor signaling; required for normal mating projection (shmoo) formation; required for Spa2p to recruit Mpk1p to shmoo tip during mating; interacts with Cdc12p |
|
| YER097W | 0.41 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
Network of associatons between targets according to the STRING database.
First level regulatory network of SNT2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 1.1 | 3.2 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.7 | 4.4 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.7 | 1.5 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.7 | 3.4 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.6 | 2.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.6 | 2.2 | GO:0031138 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.5 | 2.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.5 | 1.9 | GO:0046323 | glucose import(GO:0046323) |
| 0.5 | 1.4 | GO:0009593 | detection of chemical stimulus(GO:0009593) detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) detection of stimulus(GO:0051606) |
| 0.5 | 2.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.4 | 1.3 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) unsaturated fatty acid metabolic process(GO:0033559) positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.4 | 2.0 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.4 | 3.3 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.4 | 1.2 | GO:0043065 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.4 | 3.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.4 | 1.2 | GO:0006000 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 1.6 | GO:0042357 | thiamine diphosphate biosynthetic process(GO:0009229) thiamine diphosphate metabolic process(GO:0042357) |
| 0.4 | 5.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.4 | 1.1 | GO:0043419 | urea metabolic process(GO:0019627) urea catabolic process(GO:0043419) |
| 0.4 | 1.1 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) |
| 0.4 | 1.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.4 | 2.2 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.4 | 1.8 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.3 | 1.0 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 2.3 | GO:0006007 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.3 | 1.5 | GO:0034486 | amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 0.3 | 0.9 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.3 | 1.2 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.3 | 1.2 | GO:0001315 | age-dependent response to reactive oxygen species(GO:0001315) |
| 0.3 | 3.7 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.3 | 0.8 | GO:2000058 | regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.2 | 0.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 1.4 | GO:0000272 | polysaccharide catabolic process(GO:0000272) |
| 0.2 | 1.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 0.9 | GO:0071396 | response to lipid(GO:0033993) response to oleic acid(GO:0034201) response to fatty acid(GO:0070542) cellular response to lipid(GO:0071396) cellular response to fatty acid(GO:0071398) cellular response to oleic acid(GO:0071400) |
| 0.2 | 1.5 | GO:0034762 | regulation of transmembrane transport(GO:0034762) |
| 0.2 | 1.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 0.8 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.2 | 1.0 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.2 | 0.6 | GO:0051222 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 0.2 | 0.8 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.2 | 0.6 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.2 | 0.9 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) |
| 0.2 | 0.9 | GO:0032105 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.2 | 1.2 | GO:0051322 | anaphase(GO:0051322) |
| 0.2 | 0.7 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.2 | 0.6 | GO:0009300 | antisense RNA transcription(GO:0009300) regulation of antisense RNA transcription(GO:0060194) negative regulation of antisense RNA transcription(GO:0060195) |
| 0.2 | 0.5 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.4 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.9 | GO:0051098 | regulation of binding(GO:0051098) |
| 0.1 | 0.7 | GO:0015758 | glucose transport(GO:0015758) |
| 0.1 | 0.7 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 1.5 | GO:0045931 | positive regulation of mitotic cell cycle(GO:0045931) |
| 0.1 | 0.5 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.1 | 1.2 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.1 | 0.4 | GO:1903339 | inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.1 | 0.4 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) tyrosine catabolic process(GO:0006572) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.5 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.1 | 0.8 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.9 | GO:0000411 | regulation of transcription by galactose(GO:0000409) positive regulation of transcription by galactose(GO:0000411) |
| 0.1 | 1.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 0.4 | GO:0032079 | positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.1 | 0.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.4 | GO:0019568 | arabinose metabolic process(GO:0019566) arabinose catabolic process(GO:0019568) |
| 0.1 | 0.6 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.3 | GO:0071281 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.1 | 0.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.8 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.1 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.8 | GO:0043487 | regulation of RNA stability(GO:0043487) regulation of mRNA stability(GO:0043488) |
| 0.1 | 1.3 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 0.1 | 0.5 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.4 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 1.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.2 | GO:0010994 | free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) |
| 0.1 | 0.3 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.1 | 0.2 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) regulation of metaphase/anaphase transition of cell cycle(GO:1902099) |
| 0.1 | 0.7 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 0.8 | GO:0015749 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.1 | 0.4 | GO:0070096 | mitochondrial outer membrane translocase complex assembly(GO:0070096) |
| 0.1 | 1.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.3 | GO:0031134 | sister chromatid biorientation(GO:0031134) |
| 0.1 | 0.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 0.4 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.8 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.9 | GO:0019740 | nitrogen utilization(GO:0019740) |
| 0.0 | 0.4 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) copper ion homeostasis(GO:0055070) |
| 0.0 | 1.1 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.2 | GO:0015883 | FAD transport(GO:0015883) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 2.0 | GO:0001403 | invasive growth in response to glucose limitation(GO:0001403) |
| 0.0 | 0.3 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.0 | 0.4 | GO:0000753 | cell morphogenesis involved in conjugation with cellular fusion(GO:0000753) |
| 0.0 | 1.8 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.2 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) |
| 0.0 | 0.2 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.3 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.2 | GO:0051569 | positive regulation of histone methylation(GO:0031062) regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 1.0 | GO:1903050 | regulation of proteasomal protein catabolic process(GO:0061136) regulation of proteolysis involved in cellular protein catabolic process(GO:1903050) regulation of cellular protein catabolic process(GO:1903362) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.0 | GO:1900544 | positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.0 | 0.9 | GO:0071940 | ascospore wall assembly(GO:0030476) spore wall assembly(GO:0042244) spore wall biogenesis(GO:0070590) ascospore wall biogenesis(GO:0070591) fungal-type cell wall assembly(GO:0071940) |
| 0.0 | 0.1 | GO:0016559 | mitochondrial fission(GO:0000266) peroxisome fission(GO:0016559) |
| 0.0 | 0.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.0 | 0.2 | GO:0000321 | re-entry into mitotic cell cycle after pheromone arrest(GO:0000321) |
| 0.0 | 0.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0001041 | transcription from RNA polymerase III type 2 promoter(GO:0001009) transcription from a RNA polymerase III hybrid type promoter(GO:0001041) |
| 0.0 | 0.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.0 | GO:0009746 | response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.1 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.1 | GO:0000161 | MAPK cascade involved in osmosensory signaling pathway(GO:0000161) |
| 0.0 | 0.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0045991 | carbon catabolite activation of transcription(GO:0045991) |
| 0.0 | 0.3 | GO:0016237 | membrane invagination(GO:0010324) lysosomal microautophagy(GO:0016237) single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 0.0 | GO:0055078 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.3 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 | 0.0 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.0 | GO:0000076 | DNA replication checkpoint(GO:0000076) replication fork protection(GO:0048478) |
| 0.0 | 0.4 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.3 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.1 | GO:0006101 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.0 | 0.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.4 | 4.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 1.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 0.7 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.2 | 0.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 1.1 | GO:0044462 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 0.2 | 1.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 0.8 | GO:0034448 | EGO complex(GO:0034448) |
| 0.2 | 1.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.4 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.1 | 0.5 | GO:0001400 | mating projection base(GO:0001400) |
| 0.1 | 2.2 | GO:0031307 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.6 | GO:0032221 | Rpd3S complex(GO:0032221) |
| 0.1 | 2.7 | GO:0042763 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.1 | 0.6 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 1.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.6 | GO:0032126 | eisosome(GO:0032126) |
| 0.1 | 1.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 11.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 0.3 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.1 | 3.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.4 | GO:0000799 | condensin complex(GO:0000796) nuclear condensin complex(GO:0000799) |
| 0.1 | 0.3 | GO:0016587 | Isw1 complex(GO:0016587) |
| 0.1 | 0.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.3 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.1 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.0 | 1.3 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.8 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.2 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 7.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.1 | GO:0031499 | TRAMP complex(GO:0031499) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.8 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 1.1 | 4.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.7 | 4.2 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.7 | 2.7 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.6 | 2.9 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.5 | 1.4 | GO:0005536 | glucose binding(GO:0005536) |
| 0.4 | 2.7 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.4 | 1.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.4 | 2.5 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.4 | 1.2 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 1.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.4 | 3.1 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.4 | 2.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.4 | 1.5 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.4 | 1.1 | GO:0008902 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.3 | 0.9 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.3 | 1.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.3 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 1.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.2 | 0.7 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.2 | 0.6 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.2 | 0.8 | GO:0005261 | cation channel activity(GO:0005261) |
| 0.2 | 0.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 6.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.2 | 2.0 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 2.1 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.2 | 0.5 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.1 | 0.4 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.1 | 0.8 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.4 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 1.3 | GO:0020037 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.1 | 1.3 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 1.5 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 0.4 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.4 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 0.8 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 1.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.5 | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen(GO:0016705) |
| 0.1 | 0.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.3 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) trehalose-phosphatase activity(GO:0004805) |
| 0.1 | 0.3 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 0.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.6 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.1 | 0.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 2.8 | GO:0004871 | signal transducer activity(GO:0004871) |
| 0.1 | 1.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.1 | 0.4 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.1 | 0.1 | GO:0042947 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.1 | 0.3 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.3 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 0.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.2 | GO:0001025 | transcription factor activity, core RNA polymerase III binding(GO:0000995) RNA polymerase III transcription factor binding(GO:0001025) TFIIIB-type transcription factor activity(GO:0001026) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.8 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.1 | 2.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.5 | GO:0016723 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 0.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.4 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.7 | GO:0016769 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 3.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 3.5 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.9 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.8 | GO:0061733 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.1 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.5 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.0 | 0.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.3 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.0 | 1.0 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.1 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) transcription factor activity, RNA polymerase III promoter sequence-specific binding, TFIIIB recruiting(GO:0001004) transcription factor activity, RNA polymerase III type 1 promoter sequence-specific binding, TFIIIB recruiting(GO:0001005) transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) transcription factor activity, RNA polymerase III type 2 promoter sequence-specific binding, TFIIIB recruiting(GO:0001008) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) transcription factor activity, RNA polymerase III transcription factor recruiting(GO:0001153) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0004364 | glutathione transferase activity(GO:0004364) glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.5 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 0.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 25.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.3 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 1.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 0.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 21.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME TRANSCRIPTION | Genes involved in Transcription |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |