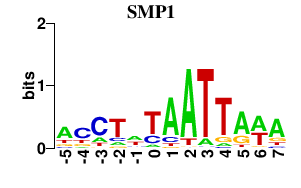
Results for SMP1
Z-value: 0.58
Transcription factors associated with SMP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMP1
|
S000000386 | Putative transcription factor of the MADS-box family |
Activity-expression correlation:
Activity profile of SMP1 motif
Sorted Z-values of SMP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YDR098C | 4.27 |
GRX3
|
Hydroperoxide and superoxide-radical responsive glutathione-dependent oxidoreductase; monothiol glutaredoxin subfamily member along with Grx4p and Grx5p; protects cells from oxidative damage |
|
| YLR154C | 2.69 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YJR094W-A | 2.65 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YLR388W | 2.51 |
RPS29A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps29Bp and has similarity to rat S29 and E. coli S14 ribosomal proteins |
|
| YLR154W-B | 2.22 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YHR181W | 2.21 |
SVP26
|
Integral membrane protein of the early Golgi apparatus and endoplasmic reticulum, involved in COP II vesicle transport; may also function to promote retention of proteins in the early Golgi compartment |
|
| YEL040W | 2.18 |
UTR2
|
Cell wall protein that functions in the transfer of chitin to beta(1-6)glucan; putative chitin transglycosidase; glycosylphosphatidylinositol (GPI)-anchored protein localized to the bud neck; has a role in cell wall maintenance |
|
| YLR154W-A | 2.16 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YPR157W | 2.10 |
Putative protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YMR116C | 2.10 |
ASC1
|
G-protein beta subunit and guanine nucleotide dissociation inhibitor for Gpa2p; ortholog of RACK1 that inhibits translation; core component of the small (40S) ribosomal subunit; represses Gcn4p in the absence of amino acid starvation |
|
| YDR417C | 2.09 |
Hypothetical protein |
||
| YIL096C | 1.98 |
Putative protein of unknown function; associates with precursors of the 60S ribosomal subunit |
||
| YHR180W-A | 1.85 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps dubious ORF YHR180C-B and long terminal repeat YHRCsigma3 |
||
| YNL162W | 1.78 |
RPL42A
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl42Bp and has similarity to rat L44 ribosomal protein |
|
| YKL164C | 1.73 |
PIR1
|
O-glycosylated protein required for cell wall stability; attached to the cell wall via beta-1,3-glucan; mediates mitochondrial translocation of Apn1p; expression regulated by the cell integrity pathway and by Swi5p during the cell cycle |
|
| YOR063W | 1.69 |
RPL3
|
Protein component of the large (60S) ribosomal subunit, has similarity to E. coli L3 and rat L3 ribosomal proteins; involved in the replication and maintenance of killer double stranded RNA virus |
|
| YLR154W-C | 1.68 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YPL273W | 1.67 |
SAM4
|
S-adenosylmethionine-homocysteine methyltransferase, functions along with Mht1p in the conversion of S-adenosylmethionine (AdoMet) to methionine to control the methionine/AdoMet ratio |
|
| YNR001W-A | 1.67 |
Dubious open reading frame unlikely to encode a functional protein; identified by homology |
||
| YMR142C | 1.54 |
RPL13B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl13Ap; not essential for viability; has similarity to rat L13 ribosomal protein |
|
| YDL055C | 1.49 |
PSA1
|
GDP-mannose pyrophosphorylase (mannose-1-phosphate guanyltransferase), synthesizes GDP-mannose from GTP and mannose-1-phosphate in cell wall biosynthesis; required for normal cell wall structure |
|
| YMR143W | 1.48 |
RPS16A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps16Bp and has similarity to E. coli S9 and rat S16 ribosomal proteins |
|
| YGL031C | 1.46 |
RPL24A
|
Ribosomal protein L30 of the large (60S) ribosomal subunit, nearly identical to Rpl24Bp and has similarity to rat L24 ribosomal protein; not essential for translation but may be required for normal translation rate |
|
| YDR033W | 1.36 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YGR085C | 1.25 |
RPL11B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl11Ap; involved in ribosomal assembly; depletion causes degradation of proteins and RNA of the 60S subunit; has similarity to E. coli L5 and rat L11 |
|
| YMR303C | 1.25 |
ADH2
|
Glucose-repressible alcohol dehydrogenase II, catalyzes the conversion of ethanol to acetaldehyde; involved in the production of certain carboxylate esters; regulated by ADR1 |
|
| YGL030W | 1.20 |
RPL30
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L30 ribosomal protein; involved in pre-rRNA processing in the nucleolus; autoregulates splicing of its transcript |
|
| YMR290C | 1.18 |
HAS1
|
ATP-dependent RNA helicase; localizes to both the nuclear periphery and nucleolus; highly enriched in nuclear pore complex fractions; constituent of 66S pre-ribosomal particles |
|
| YMR241W | 1.17 |
YHM2
|
Mitochondrial DNA-binding protein, component of the mitochondrial nucleoid structure, involved in mtDNA replication and segregation of mitochondrial genomes; member of the mitochondrial carrier protein family |
|
| YDL241W | 1.16 |
Putative protein of unknown function; YDL241W is not an essential gene |
||
| YGR271C-A | 1.15 |
EFG1
|
Protein of unknown function; null mutant is sensitive to hydroxyurea and is delayed in recovering from alpha-factor arrest; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus |
|
| YNL182C | 1.14 |
IPI3
|
Essential component of the Rix1 complex (Rix1p, Ipi1p, Ipi3p) that is required for processing of ITS2 sequences from 35S pre-rRNA; highly conserved and contains WD40 motifs; Rix1 complex associates with Mdn1p in pre-60S ribosomal particles |
|
| YKL218C | 1.13 |
SRY1
|
3-hydroxyaspartate dehydratase, deaminates L-threo-3-hydroxyaspartate to form oxaloacetate and ammonia; required for survival in the presence of hydroxyaspartate |
|
| YBR092C | 1.13 |
PHO3
|
Constitutively expressed acid phosphatase similar to Pho5p; brought to the cell surface by transport vesicles; hydrolyzes thiamin phosphates in the periplasmic space, increasing cellular thiamin uptake; expression is repressed by thiamin |
|
| YMR290W-A | 1.12 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; overlaps 5' end of essential HAS1 gene which encodes an ATP-dependent RNA helicase |
||
| YLR432W | 1.12 |
IMD3
|
Inosine monophosphate dehydrogenase, catalyzes the first step of GMP biosynthesis, member of a four-gene family in S. cerevisiae, constitutively expressed |
|
| YML073C | 1.11 |
RPL6A
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, has similarity to Rpl6Bp and to rat L6 ribosomal protein; binds to 5.8S rRNA |
|
| YDR509W | 1.11 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBR191W | 1.11 |
RPL21A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl21Bp and has similarity to rat L21 ribosomal protein |
|
| YIL052C | 1.10 |
RPL34B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl34Ap and has similarity to rat L34 ribosomal protein |
|
| YML063W | 1.08 |
RPS1B
|
Ribosomal protein 10 (rp10) of the small (40S) subunit; nearly identical to Rps1Ap and has similarity to rat S3a ribosomal protein |
|
| YDR508C | 1.07 |
GNP1
|
High-affinity glutamine permease, also transports Leu, Ser, Thr, Cys, Met and Asn; expression is fully dependent on Grr1p and modulated by the Ssy1p-Ptr3p-Ssy5p (SPS) sensor of extracellular amino acids |
|
| YML075C | 1.06 |
HMG1
|
One of two isozymes of HMG-CoA reductase that catalyzes the conversion of HMG-CoA to mevalonate, which is a rate-limiting step in sterol biosynthesis; localizes to the nuclear envelope; overproduction induces the formation of karmellae |
|
| YKL219W | 1.04 |
COS9
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YOL086C | 1.04 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YPR010C | 1.03 |
RPA135
|
RNA polymerase I subunit A135 |
|
| YGR148C | 1.03 |
RPL24B
|
Ribosomal protein L30 of the large (60S) ribosomal subunit, nearly identical to Rpl24Ap and has similarity to rat L24 ribosomal protein; not essential for translation but may be required for normal translation rate |
|
| YHR141C | 1.02 |
RPL42B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl42Ap and has similarity to rat L44; required for propagation of the killer toxin-encoding M1 double-stranded RNA satellite of the L-A double-stranded RNA virus |
|
| YGR035C | 0.96 |
Putative protein of unknown function, potential Cdc28p substrate; transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in multidrug resistance |
||
| YMR217W | 0.96 |
GUA1
|
GMP synthase, an enzyme that catalyzes the second step in the biosynthesis of GMP from inosine 5'-phosphate (IMP); transcription is not subject to regulation by guanine but is negatively regulated by nutrient starvation |
|
| YER007C-A | 0.95 |
TMA20
|
Protein of unknown function that associates with ribosomes and has a putative RNA binding domain; interacts with Tma22p; null mutant exhibits translation defects; has homology to human oncogene MCT-1 |
|
| YJL162C | 0.93 |
JJJ2
|
Protein of unknown function, contains a J-domain, which is a region with homology to the E. coli DnaJ protein |
|
| YMR246W | 0.92 |
FAA4
|
Long chain fatty acyl-CoA synthetase, regulates protein modification during growth in the presence of ethanol, functions to incorporate palmitic acid into phospholipids and neutral lipids |
|
| YFR055W | 0.91 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YDR041W | 0.90 |
RSM10
|
Mitochondrial ribosomal protein of the small subunit, has similarity to E. coli S10 ribosomal protein; essential for viability, unlike most other mitoribosomal proteins |
|
| YLL044W | 0.88 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; transcription of both YLL044W and the overlapping gene RPL8B is reduced in the gcr1 null mutant |
||
| YOR046C | 0.88 |
DBP5
|
Cytoplasmic ATP-dependent RNA helicase of the DEAD-box family involved in mRNA export from the nucleus; involved in translation termination |
|
| YFL022C | 0.86 |
FRS2
|
Alpha subunit of cytoplasmic phenylalanyl-tRNA synthetase, forms a tetramer with Frs1p to form active enzyme; evolutionarily distant from mitochondrial phenylalanyl-tRNA synthetase based on protein sequence, but substrate binding is similar |
|
| YLR048W | 0.84 |
RPS0B
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps0Ap; required for maturation of 18S rRNA along with Rps0Ap; deletion of either RPS0 gene reduces growth rate, deletion of both genes is lethal |
|
| YOR096W | 0.84 |
RPS7A
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps7Bp; interacts with Kti11p; deletion causes hypersensitivity to zymocin; has similarity to rat S7 and Xenopus S8 ribosomal proteins |
|
| YOR073W-A | 0.84 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially/completely overlaps the verified ORF CDC21/YOR074C; identified by RT-PCR |
||
| YGR090W | 0.83 |
UTP22
|
Possible U3 snoRNP protein involved in maturation of pre-18S rRNA, based on computational analysis of large-scale protein-protein interaction data |
|
| YER026C | 0.83 |
CHO1
|
Phosphatidylserine synthase, functions in phospholipid biosynthesis; catalyzes the reaction CDP-diaclyglycerol + L-serine = CMP + L-1-phosphatidylserine, transcriptionally repressed by myo-inositol and choline |
|
| YKR094C | 0.83 |
RPL40B
|
Fusion protein, identical to Rpl40Ap, that is cleaved to yield ubiquitin and a ribosomal protein of the large (60S) ribosomal subunit with similarity to rat L40; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes |
|
| YML071C | 0.83 |
COG8
|
Component of the conserved oligomeric Golgi complex (Cog1p through Cog8p), a cytosolic tethering complex that functions in protein trafficking to mediate fusion of transport vesicles to Golgi compartments |
|
| YER137C | 0.82 |
Putative protein of unknown function |
||
| YLR257W | 0.82 |
Putative protein of unknown function |
||
| YJL115W | 0.81 |
ASF1
|
Nucleosome assembly factor, involved in chromatin assembly and disassembly, anti-silencing protein that causes derepression of silent loci when overexpressed; plays a role in regulating Ty1 transposition |
|
| YKR013W | 0.81 |
PRY2
|
Protein of unknown function, has similarity to Pry1p and Pry3p and to the plant PR-1 class of pathogen related proteins |
|
| YML074C | 0.80 |
FPR3
|
Nucleolar peptidyl-prolyl cis-trans isomerase (PPIase); FK506 binding protein; phosphorylated by casein kinase II (Cka1p-Cka2p-Ckb1p-Ckb2p) and dephosphorylated by Ptp1p |
|
| YOR320C | 0.79 |
GNT1
|
N-acetylglucosaminyltransferase capable of modification of N-linked glycans in the Golgi apparatus |
|
| YMR106C | 0.78 |
YKU80
|
Subunit of the telomeric Ku complex (Yku70p-Yku80p), involved in telomere length maintenance, structure and telomere position effect; relocates to sites of double-strand cleavage to promote nonhomologous end joining during DSB repair |
|
| YFR056C | 0.77 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YLR196W | 0.77 |
PWP1
|
Protein with WD-40 repeats involved in rRNA processing; associates with trans-acting ribosome biogenesis factors; similar to beta-transducin superfamily |
|
| YGR040W | 0.77 |
KSS1
|
Mitogen-activated protein kinase (MAPK) involved in signal transduction pathways that control filamentous growth and pheromone response; the KSS1 gene is nonfunctional in S288C strains and functional in W303 strains |
|
| YLR453C | 0.77 |
RIF2
|
Protein that binds to the Rap1p C-terminus and acts synergistically with Rif1p to help control telomere length and establish telomeric silencing; deletion results in telomere elongation |
|
| YOR074C | 0.77 |
CDC21
|
Thymidylate synthase, required for de novo biosynthesis of pyrimidine deoxyribonucleotides; expression is induced at G1/S |
|
| YDR040C | 0.76 |
ENA1
|
P-type ATPase sodium pump, involved in Na+ and Li+ efflux to allow salt tolerance |
|
| YML043C | 0.76 |
RRN11
|
Protein required for rDNA transcription by RNA polymerase I, component of the core factor (CF) of rDNA transcription factor, which also contains Rrn6p and Rrn7p |
|
| YPL160W | 0.75 |
CDC60
|
Cytosolic leucyl tRNA synthetase, ligates leucine to the appropriate tRNA |
|
| YPL037C | 0.75 |
EGD1
|
Subunit beta1 of the nascent polypeptide-associated complex (NAC) involved in protein targeting, associated with cytoplasmic ribosomes; enhances DNA binding of the Gal4p activator; homolog of human BTF3b |
|
| YOR169C | 0.74 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; open reading frame overlaps the verified gene GLN4/YOR168W |
||
| YDL179W | 0.73 |
PCL9
|
Cyclin, forms a functional kinase complex with Pho85p cyclin-dependent kinase (Cdk), expressed in late M/early G1 phase, activated by Swi5p |
|
| YIL148W | 0.73 |
RPL40A
|
Fusion protein, identical to Rpl40Bp, that is cleaved to yield ubiquitin and a ribosomal protein of the large (60S) ribosomal subunit with similarity to rat L40; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes |
|
| YGR036C | 0.72 |
CAX4
|
Dolichyl pyrophosphate (Dol-P-P) phosphatase with a luminally oriented active site in the ER, cleaves the anhydride linkage in Dol-P-P, required for Dol-P-P-linked oligosaccharide intermediate synthesis and protein N-glycosylation |
|
| YBR104W | 0.72 |
YMC2
|
Mitochondrial protein, putative inner membrane transporter with a role in oleate metabolism and glutamate biosynthesis; member of the mitochondrial carrier (MCF) family; has similarity with Ymc1p |
|
| YOL039W | 0.72 |
RPP2A
|
Ribosomal protein P2 alpha, a component of the ribosomal stalk, which is involved in the interaction between translational elongation factors and the ribosome; regulates the accumulation of P1 (Rpp1Ap and Rpp1Bp) in the cytoplasm |
|
| YCR004C | 0.71 |
YCP4
|
Protein of unknown function, has sequence and structural similarity to flavodoxins; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YCR102W-A | 0.71 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YJR123W | 0.70 |
RPS5
|
Protein component of the small (40S) ribosomal subunit, the least basic of the non-acidic ribosomal proteins; phosphorylated in vivo; essential for viability; has similarity to E. coli S7 and rat S5 ribosomal proteins |
|
| YOL013W-A | 0.70 |
Putative protein of unknown function; identified by SAGE |
||
| YAL059W | 0.70 |
ECM1
|
Protein of unknown function, localized in the nucleoplasm and the nucleolus, genetically interacts with MTR2 in 60S ribosomal protein subunit export |
|
| YDL047W | 0.70 |
SIT4
|
Type 2A-related serine-threonine phosphatase that functions in the G1/S transition of the mitotic cycle; cytoplasmic and nuclear protein that modulates functions mediated by Pkc1p including cell wall and actin cytoskeleton organization |
|
| YER131W | 0.68 |
RPS26B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps26Ap and has similarity to rat S26 ribosomal protein |
|
| YNR054C | 0.67 |
ESF2
|
Essential nucleolar protein involved in pre-18S rRNA processing; binds to RNA and stimulates ATPase activity of Dbp8; involved in assembly of the small subunit (SSU) processome |
|
| YDR531W | 0.67 |
Pantothenate kinase (ATP:D-pantothenate 4'-phosphotransferase, EC 2.7.1.33); catalyzes the first committed step in the universal biosynthetic pathway leading to coenzyme A1 |
||
| YKR003W | 0.67 |
OSH6
|
Member of an oxysterol-binding protein family with overlapping, redundant functions in sterol metabolism and which collectively perform a function essential for viability; GFP-fusion protein localizes to the cell periphery |
|
| YBL108W | 0.67 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YBR158W | 0.67 |
AMN1
|
Protein required for daughter cell separation, multiple mitotic checkpoints, and chromosome stability; contains 12 degenerate leucine-rich repeat motifs; expression is induced by the Mitotic Exit Network (MEN) |
|
| YLR191W | 0.66 |
PEX13
|
Integral peroxisomal membrane required for the translocation of peroxisomal matrix proteins, interacts with the PTS1 signal recognition factor Pex5p and the PTS2 signal recognition factor Pex7p, forms a complex with Pex14p and Pex17p |
|
| YOL040C | 0.66 |
RPS15
|
Protein component of the small (40S) ribosomal subunit; has similarity to E. coli S19 and rat S15 ribosomal proteins |
|
| YNL178W | 0.65 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YKR012C | 0.65 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene PRY2 |
||
| YPL030W | 0.65 |
TRM44
|
tRNA(Ser) Um(44) 2'-O-methyltransferase; involved in maintaining levels of the tRNA-Ser species tS(CGA) and tS(UGA); conserved among metazoans and fungi but there does not appear to be a homolog in plants; TRM44 is a non-essential gene |
|
| YCL022C | 0.64 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps verified gene KCC4/YCL024W |
||
| YGR285C | 0.64 |
ZUO1
|
Cytosolic ribosome-associated chaperone that acts, together with Ssz1p and the Ssb proteins, as a chaperone for nascent polypeptide chains; contains a DnaJ domain and functions as a J-protein partner for Ssb1p and Ssb2p |
|
| YHL001W | 0.63 |
RPL14B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl14Ap and has similarity to rat L14 ribosomal protein |
|
| YPL108W | 0.63 |
Cytoplasmic protein of unknown function; non-essential gene that is induced in a GDH1 deleted strain with altered redox metabolism; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YNR061C | 0.63 |
Putative protein of unknown function |
||
| YIL118W | 0.63 |
RHO3
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins involved in the establishment of cell polarity; GTPase activity positively regulated by the GTPase activating protein (GAP) Rgd1p |
|
| YFR037C | 0.63 |
RSC8
|
Component of the RSC chromatin remodeling complex; essential for viability and mitotic growth; homolog of SWI/SNF subunit Swi3p, but unlike Swi3p, does not activate transcription of reporters |
|
| YNL058C | 0.62 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YNL058C is not an essential gene |
||
| YHR052W | 0.62 |
CIC1
|
Essential protein that interacts with proteasome components and has a potential role in proteasome substrate specificity; also copurifies with 66S pre-ribosomal particles |
|
| YJR063W | 0.62 |
RPA12
|
RNA polymerase I subunit A12.2; contains two zinc binding domains, and the N terminal domain is responsible for anchoring to the RNA pol I complex |
|
| YLR028C | 0.62 |
ADE16
|
Enzyme of 'de novo' purine biosynthesis containing both 5-aminoimidazole-4-carboxamide ribonucleotide transformylase and inosine monophosphate cyclohydrolase activities, isozyme of Ade17p; ade16 ade17 mutants require adenine and histidine |
|
| YLR060W | 0.61 |
FRS1
|
Beta subunit of cytoplasmic phenylalanyl-tRNA synthetase, forms a tetramer with Frs2p to generate active enzyme; sequence is evolutionarily distant from mitochondrial phenylalanyl-tRNA synthetase (Msf1p), but substrate binding is similar |
|
| YOR133W | 0.61 |
EFT1
|
Elongation factor 2 (EF-2), also encoded by EFT2; catalyzes ribosomal translocation during protein synthesis; contains diphthamide, the unique posttranslationally modified histidine residue specifically ADP-ribosylated by diphtheria toxin |
|
| YIR021W | 0.61 |
MRS1
|
Protein required for the splicing of two mitochondrial group I introns (BI3 in COB and AI5beta in COX1); forms a splicing complex, containing four subunits of Mrs1p and two subunits of the BI3-encoded maturase, that binds to the BI3 RNA |
|
| YML119W | 0.60 |
Putative protein of unknown funtion; YML119W is not an essential gene; potential Cdc28p substrate |
||
| YLR441C | 0.60 |
RPS1A
|
Ribosomal protein 10 (rp10) of the small (40S) subunit; nearly identical to Rps1Bp and has similarity to rat S3a ribosomal protein |
|
| YOR309C | 0.59 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene NOP58 |
||
| YDL240W | 0.59 |
LRG1
|
Putative GTPase-activating protein (GAP) involved in the Pkc1p-mediated signaling pathway that controls cell wall integrity; appears to specifically regulate 1,3-beta-glucan synthesis |
|
| YDL143W | 0.59 |
CCT4
|
Subunit of the cytosolic chaperonin Cct ring complex, related to Tcp1p, required for the assembly of actin and tubulins in vivo |
|
| YDR226W | 0.59 |
ADK1
|
Adenylate kinase, required for purine metabolism; localized to the cytoplasm and the mitochondria; lacks cleavable signal sequence |
|
| YLR076C | 0.58 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the essential gene RPL10 which encodes the ribosomal protein L10 |
||
| YLR025W | 0.58 |
SNF7
|
One of four subunits of the endosomal sorting complex required for transport III (ESCRT-III); involved in the sorting of transmembrane proteins into the multivesicular body (MVB) pathway; recruited from the cytoplasm to endosomal membranes |
|
| YOR340C | 0.58 |
RPA43
|
RNA polymerase I subunit A43 |
|
| YDL211C | 0.58 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YDL167C | 0.57 |
NRP1
|
Protein of unknown function, rich in asparagine residues |
|
| YIL018W | 0.57 |
RPL2B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl2Ap and has similarity to E. coli L2 and rat L8 ribosomal proteins; expression is upregulated at low temperatures |
|
| YAL029C | 0.57 |
MYO4
|
One of two type V myosin motors (along with MYO2) involved in actin-based transport of cargos; required for mRNA transport, including ASH1 mRNA, and facilitating the growth and movement of ER tubules into the growing bud along with She3p |
|
| YMR099C | 0.56 |
Glucose-6-phosphate 1-epimerase (hexose-6-phosphate mutarotase), likely involved in carbohydrate metabolism; GFP-fusion protein localizes to both the nucleus and cytoplasm and is induced in response to the DNA-damaging agent MMS |
||
| YNL175C | 0.55 |
NOP13
|
Protein of unknown function, localizes to the nucleolus and nucleoplasm; contains an RNA recognition motif (RRM) and has similarity to Nop12p, which is required for processing of pre-18S rRNA |
|
| YPR079W | 0.55 |
MRL1
|
Membrane protein with similarity to mammalian mannose-6-phosphate receptors, possibly functions as a sorting receptor in the delivery of vacuolar hydrolases |
|
| YNL057W | 0.55 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR004W | 0.54 |
PEX31
|
Peroxisomal integral membrane protein, involved in negative regulation of peroxisome size; partially functionally redundant with Pex30p and Pex32p; probably acts at a step downstream of steps mediated by Pex28p and Pex29p |
|
| YOL101C | 0.54 |
IZH4
|
Membrane protein involved in zinc metabolism, member of the four-protein IZH family, expression induced by fatty acids and altered zinc levels; deletion reduces sensitivity to excess zinc; possible role in sterol metabolism |
|
| YBR084C-A | 0.51 |
RPL19A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl19Bp and has similarity to rat L19 ribosomal protein; rpl19a and rpl19b single null mutations result in slow growth, while the double null mutation is lethal |
|
| YIL029C | 0.51 |
Putative protein of unknown function; deletion confers sensitivity to 4-(N-(S-glutathionylacetyl)amino) phenylarsenoxide (GSAO) |
||
| YBL087C | 0.51 |
RPL23A
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl23Bp and has similarity to E. coli L14 and rat L23 ribosomal proteins |
|
| YBR181C | 0.50 |
RPS6B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps6Ap and has similarity to rat S6 ribosomal protein |
|
| YGR251W | 0.50 |
Putative protein of unknown function; deletion mutant has defects in pre-rRNA processing; green fluorescent protein (GFP)-fusion protein localizes to both the nucleus and the nucleolus; YGR251W is an essential gene |
||
| YML065W | 0.50 |
ORC1
|
Largest subunit of the origin recognition complex, which directs DNA replication by binding to replication origins and is also involved in transcriptional silencing; exhibits ATPase activity |
|
| YHR094C | 0.49 |
HXT1
|
Low-affinity glucose transporter of the major facilitator superfamily, expression is induced by Hxk2p in the presence of glucose and repressed by Rgt1p when glucose is limiting |
|
| YMR141W-A | 0.49 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene RPL13B/YMR142C |
||
| YGR159C | 0.49 |
NSR1
|
Nucleolar protein that binds nuclear localization sequences, required for pre-rRNA processing and ribosome biogenesis |
|
| YML022W | 0.49 |
APT1
|
Adenine phosphoribosyltransferase, catalyzes the formation of AMP from adenine and 5-phosphoribosylpyrophosphate; involved in the salvage pathway of purine nucleotide biosynthesis |
|
| YDR023W | 0.49 |
SES1
|
Cytosolic seryl-tRNA synthetase, class II aminoacyl-tRNA synthetase that aminoacylates tRNA(Ser), displays tRNA-dependent amino acid recognition which enhances discrimination of the serine substrate, interacts with peroxin Pex21p |
|
| YDR450W | 0.48 |
RPS18A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps18Bp and has similarity to E. coli S13 and rat S18 ribosomal proteins |
|
| YNL189W | 0.48 |
SRP1
|
Karyopherin alpha homolog, forms a dimer with karyopherin beta Kap95p to mediate import of nuclear proteins, binds the nuclear localization signal of the substrate during import; may also play a role in regulation of protein degradation |
|
| YDL081C | 0.48 |
RPP1A
|
Ribosomal stalk protein P1 alpha, involved in the interaction between translational elongation factors and the ribosome; accumulation of P1 in the cytoplasm is regulated by phosphorylation and interaction with the P2 stalk component |
|
| YJL011C | 0.48 |
RPC17
|
RNA polymerase III subunit C17; physically interacts with C31, C11, and TFIIIB70; may be involved in the recruitment of pol III by the preinitiation complex |
|
| YDR050C | 0.48 |
TPI1
|
Triose phosphate isomerase, abundant glycolytic enzyme; mRNA half-life is regulated by iron availability; transcription is controlled by activators Reb1p, Gcr1p, and Rap1p through binding sites in the 5' non-coding region |
|
| YJL169W | 0.48 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YJL168C/SET2 |
||
| YOR293W | 0.48 |
RPS10A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps10Bp and has similarity to rat ribosomal protein S10 |
|
| YNL090W | 0.47 |
RHO2
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, involved in the establishment of cell polarity and in microtubule assembly |
|
| YGL158W | 0.47 |
RCK1
|
Protein kinase involved in the response to oxidative stress; identified as suppressor of S. pombe cell cycle checkpoint mutations |
|
| YBR048W | 0.47 |
RPS11B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps11Ap and has similarity to E. coli S17 and rat S11 ribosomal proteins |
|
| YEL068C | 0.47 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHR216W | 0.46 |
IMD2
|
Inosine monophosphate dehydrogenase, catalyzes the first step of GMP biosynthesis, expression is induced by mycophenolic acid resulting in resistance to the drug, expression is repressed by nutrient limitation |
|
| YCL018W | 0.46 |
LEU2
|
Beta-isopropylmalate dehydrogenase (IMDH), catalyzes the third step in the leucine biosynthesis pathway |
|
| YIL169C | 0.46 |
Putative protein of unknown function; serine/threonine rich and highly similar to YOL155C, a putative glucan alpha-1,4-glucosidase; transcript is induced in both high and low pH environments; YIL169C is a non-essential gene |
||
| YKL068W-A | 0.46 |
Putative protein of unknown function; identified by homology to Ashbya gossypii |
||
| YMR100W | 0.45 |
MUB1
|
Protein of unknown function, deletion causes multi-budding phenotype; has similarity to Aspergillus nidulans samB gene |
|
| YLL012W | 0.45 |
YEH1
|
Steryl ester hydrolase, one of three gene products (Yeh1p, Yeh2p, Tgl1p) responsible for steryl ester hydrolase activity and involved in sterol homeostasis; localized to lipid particle membranes |
|
| YIR042C | 0.45 |
Putative protein of unknown function; YIR042C is a non-essential gene |
||
| YHR021C | 0.45 |
RPS27B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps27Ap and has similarity to rat S27 ribosomal protein |
|
| YMR205C | 0.45 |
PFK2
|
Beta subunit of heterooctameric phosphofructokinase involved in glycolysis, indispensable for anaerobic growth, activated by fructose-2,6-bisphosphate and AMP, mutation inhibits glucose induction of cell cycle-related genes |
|
| YOR241W | 0.45 |
MET7
|
Folylpolyglutamate synthetase, catalyzes extension of the glutamate chains of the folate coenzymes, required for methionine synthesis and for maintenance of mitochondrial DNA |
|
| YOR357C | 0.44 |
SNX3
|
Sorting nexin required to maintain late-Golgi resident enzymes in their proper location by recycling molecules from the prevacuolar compartment; contains a PX domain and sequence similarity to human Snx3p |
|
| YDR326C | 0.44 |
YSP2
|
Protein involved in programmed cell death; mutant shows resistance to cell death induced by amiodarone or intracellular acidification |
|
| YOR376W-A | 0.44 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YNL043C | 0.44 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YIP3/YNL044W |
||
| YPR119W | 0.44 |
CLB2
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YNR003C | 0.44 |
RPC34
|
RNA polymerase III subunit C34; interacts with TFIIIB70 and is a key determinant in pol III recruitment by the preinitiation complex |
|
| YPL146C | 0.44 |
NOP53
|
Nucleolar protein; involved in biogenesis of the 60S subunit of the ribosome; interacts with rRNA processing factors Cbf5p and Nop2p; null mutant is viable but growth is severely impaired |
|
| YGR018C | 0.44 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YGR017W |
||
| YCR102C | 0.43 |
Putative protein of unknown function; involved in copper metabolism; similar to C.carbonum toxD gene; YCR102C is not an essential gene |
||
| YAR002W | 0.43 |
NUP60
|
Subunit of the nuclear pore complex (NPC), functions to anchor Nup2p to the NPC in a process controlled by the nucleoplasmic concentration of Gsp1p-GTP; potential Cdc28p substrate; involved in telomere maintenance |
|
| YFR032C | 0.43 |
Putative protein of unknown function; transcribed during sporulation; YFR032C is not an essential gene |
||
| YBR190W | 0.43 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the verified ribosomal protein gene RPL21A/YBR191W |
||
| YJL190C | 0.43 |
RPS22A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps22Bp and has similarity to E. coli S8 and rat S15a ribosomal proteins |
|
| YGL224C | 0.43 |
SDT1
|
Pyrimidine nucleotidase; overexpression suppresses the 6-AU sensitivity of transcription elongation factor S-II, as well as resistance to other pyrimidine derivatives |
|
| YBR267W | 0.43 |
REI1
|
Cytoplasmic pre-60S factor; required for the correct recycling of shuttling factors Alb1, Arx1 and Tif6 at the end of the ribosomal large subunit biogenesis; involved in bud growth in the mitotic signaling network |
|
| YHR043C | 0.43 |
DOG2
|
2-deoxyglucose-6-phosphate phosphatase, member of a family of low molecular weight phosphatases, similar to Dog1p, induced by oxidative and osmotic stress, confers 2-deoxyglucose resistance when overexpressed |
|
| YOR385W | 0.42 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YOR385W is not an essential gene |
||
| YNR072W | 0.42 |
HXT17
|
Hexose transporter, up-regulated in media containing raffinose and galactose at pH 7.7 versus pH 4.7, repressed by high levels of glucose |
|
| YGR035W-A | 0.42 |
Putative protein of unknown function |
||
| YAL007C | 0.42 |
ERP2
|
Protein that forms a heterotrimeric complex with Erp1p, Emp24p, and Erv25p; member, along with Emp24p and Erv25p, of the p24 family involved in ER to Golgi transport and localized to COPII-coated vesicles |
|
| YGR108W | 0.42 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YGR034W | 0.41 |
RPL26B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl26Ap and has similarity to E. coli L24 and rat L26 ribosomal proteins; binds to 5.8S rRNA |
|
| YMR123W | 0.41 |
PKR1
|
V-ATPase assembly factor, functions with other V-ATPase assembly factors in the ER to efficiently assemble the V-ATPase membrane sector (V0); overproduction confers resistance to Pichia farinosa killer toxin |
|
| YCR034W | 0.40 |
FEN1
|
Fatty acid elongase, involved in sphingolipid biosynthesis; acts on fatty acids of up to 24 carbons in length; mutations have regulatory effects on 1,3-beta-glucan synthase, vacuolar ATPase, and the secretory pathway |
|
| YMR304C-A | 0.40 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene SCW10 |
||
| YHR005C | 0.40 |
GPA1
|
GTP-binding alpha subunit of the heterotrimeric G protein that couples to pheromone receptors; negatively regulates the mating pathway by sequestering G(beta)gamma and by triggering an adaptive response; activates Vps34p at the endosome |
|
| YKL063C | 0.40 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the Golgi |
||
| YBR085W | 0.40 |
AAC3
|
Mitochondrial inner membrane ADP/ATP translocator, exchanges cytosolic ADP for mitochondrially synthesized ATP; expressed under anaerobic conditions; similar to Pet9p and Aac1p; has roles in maintenance of viability and in respiration |
|
| YPL250W-A | 0.40 |
Identified by fungal homology and RT-PCR |
||
| YDR099W | 0.40 |
BMH2
|
14-3-3 protein, minor isoform; controls proteome at post-transcriptional level, binds proteins and DNA, involved in regulation of many processes including exocytosis, vesicle transport, Ras/MAPK signaling, and rapamycin-sensitive signaling |
|
| YLR129W | 0.40 |
DIP2
|
Nucleolar protein, specifically associated with the U3 snoRNA, part of the large ribonucleoprotein complex known as the small subunit (SSU) processome, required for 18S rRNA biogenesis, part of the active pre-rRNA processing complex |
Network of associatons between targets according to the STRING database.
First level regulatory network of SMP1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0014070 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 0.5 | 1.6 | GO:0046039 | GTP biosynthetic process(GO:0006183) GTP metabolic process(GO:0046039) |
| 0.5 | 1.5 | GO:0019673 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.5 | 1.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.4 | 1.7 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.4 | 2.9 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.4 | 1.2 | GO:0048024 | regulation of RNA splicing(GO:0043484) regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.4 | 1.9 | GO:0009757 | cellular glucose homeostasis(GO:0001678) carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.4 | 1.1 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.3 | 1.0 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.3 | 1.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.3 | 5.5 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.3 | 0.9 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.3 | 0.3 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.3 | 0.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 1.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.3 | 0.8 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.2 | 1.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 1.7 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.2 | 1.0 | GO:0015909 | long-chain fatty acid transport(GO:0015909) fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.2 | 1.0 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 2.2 | GO:0000947 | amino acid catabolic process to alcohol via Ehrlich pathway(GO:0000947) |
| 0.2 | 33.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 2.5 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.2 | 1.5 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.2 | 0.6 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.2 | 3.3 | GO:0006037 | cell wall chitin metabolic process(GO:0006037) |
| 0.2 | 0.4 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.2 | 0.6 | GO:0071454 | response to anoxia(GO:0034059) cellular response to anoxia(GO:0071454) |
| 0.2 | 0.8 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.2 | 2.5 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.2 | 0.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.2 | 0.9 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.2 | 0.8 | GO:0017157 | regulation of exocytosis(GO:0017157) regulation of secretion(GO:0051046) regulation of secretion by cell(GO:1903530) |
| 0.2 | 0.5 | GO:0044209 | purine nucleobase salvage(GO:0043096) AMP salvage(GO:0044209) adenine metabolic process(GO:0046083) |
| 0.2 | 0.6 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.2 | 0.8 | GO:0007535 | donor selection(GO:0007535) |
| 0.2 | 1.4 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.2 | 1.1 | GO:0043102 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.1 | 0.4 | GO:0046901 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.1 | 0.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 1.2 | GO:0000080 | mitotic G1 phase(GO:0000080) G1 phase(GO:0051318) |
| 0.1 | 1.0 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 0.1 | 1.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.6 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 3.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.4 | GO:0071044 | histone mRNA metabolic process(GO:0008334) histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.3 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.8 | GO:0001402 | signal transduction involved in filamentous growth(GO:0001402) |
| 0.1 | 0.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.1 | GO:0040031 | snRNA pseudouridine synthesis(GO:0031120) snRNA modification(GO:0040031) |
| 0.1 | 0.1 | GO:0071482 | cellular response to radiation(GO:0071478) cellular response to light stimulus(GO:0071482) |
| 0.1 | 0.3 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin structure organization(GO:0031032) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 1.9 | GO:0042797 | tRNA transcription(GO:0009304) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 0.2 | GO:0050000 | telomere localization(GO:0034397) chromosome localization(GO:0050000) |
| 0.1 | 0.8 | GO:0046040 | IMP biosynthetic process(GO:0006188) 'de novo' IMP biosynthetic process(GO:0006189) IMP metabolic process(GO:0046040) |
| 0.1 | 0.4 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 0.4 | GO:0060188 | regulation of protein desumoylation(GO:0060188) |
| 0.1 | 0.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.6 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 0.2 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 0.3 | GO:0032239 | regulation of mRNA export from nucleus(GO:0010793) regulation of nucleobase-containing compound transport(GO:0032239) regulation of RNA export from nucleus(GO:0046831) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.1 | 0.4 | GO:0045141 | meiotic telomere clustering(GO:0045141) establishment of chromosome localization(GO:0051303) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.1 | 0.4 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.1 | 0.4 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 1.7 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.3 | GO:0046495 | nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) |
| 0.1 | 0.6 | GO:0000376 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.1 | 0.3 | GO:0000296 | spermine transport(GO:0000296) |
| 0.1 | 1.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.3 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 0.1 | 0.3 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.7 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) meiotic recombination checkpoint(GO:0051598) |
| 0.1 | 0.9 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.1 | 0.2 | GO:0042990 | regulation of transcription factor import into nucleus(GO:0042990) |
| 0.1 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.4 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.1 | 2.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.1 | GO:2001021 | regulation of cell cycle checkpoint(GO:1901976) negative regulation of cell cycle checkpoint(GO:1901977) regulation of DNA damage checkpoint(GO:2000001) negative regulation of DNA damage checkpoint(GO:2000002) negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.1 | 0.4 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.1 | 0.9 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.3 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.3 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.1 | 0.3 | GO:1902750 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.1 | 1.7 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.2 | GO:0019419 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.1 | 0.6 | GO:0016233 | telomere capping(GO:0016233) |
| 0.1 | 0.6 | GO:0048309 | endoplasmic reticulum inheritance(GO:0048309) |
| 0.1 | 1.1 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.9 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 1.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.7 | GO:0046939 | nucleotide phosphorylation(GO:0046939) |
| 0.1 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) protein-lipid complex assembly(GO:0065005) protein-lipid complex subunit organization(GO:0071825) |
| 0.1 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.6 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine metabolic process(GO:0006047) UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.3 | GO:0030030 | cell projection organization(GO:0030030) cell projection assembly(GO:0030031) |
| 0.1 | 0.2 | GO:0000751 | mitotic cell cycle arrest in response to pheromone(GO:0000751) mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.3 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.1 | 0.2 | GO:0007234 | osmosensory signaling via phosphorelay pathway(GO:0007234) |
| 0.1 | 0.2 | GO:0035950 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 0.1 | 0.2 | GO:0032211 | negative regulation of telomere maintenance(GO:0032205) negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of homeostatic process(GO:0032845) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 0.2 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.1 | 0.2 | GO:0000771 | agglutination involved in conjugation with cellular fusion(GO:0000752) agglutination involved in conjugation(GO:0000771) heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) cell-cell adhesion(GO:0098609) adhesion between unicellular organisms(GO:0098610) multi organism cell adhesion(GO:0098740) cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.1 | 0.3 | GO:0072668 | obsolete tubulin complex biogenesis(GO:0072668) |
| 0.1 | 0.1 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) |
| 0.1 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.7 | GO:0001100 | negative regulation of exit from mitosis(GO:0001100) |
| 0.1 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.6 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 2.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.1 | GO:0043902 | positive regulation of multi-organism process(GO:0043902) |
| 0.0 | 0.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.3 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.0 | GO:0051196 | regulation of nucleoside metabolic process(GO:0009118) regulation of nucleotide catabolic process(GO:0030811) regulation of cofactor metabolic process(GO:0051193) regulation of coenzyme metabolic process(GO:0051196) regulation of ATP metabolic process(GO:1903578) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.0 | 0.1 | GO:0000092 | mitotic anaphase B(GO:0000092) |
| 0.0 | 0.1 | GO:1901073 | aminoglycan biosynthetic process(GO:0006023) chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 4.2 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.3 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.1 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0097502 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) regulation of transcription-coupled nucleotide-excision repair(GO:0090262) regulation of nucleotide-excision repair(GO:2000819) regulation of histone H2B ubiquitination(GO:2001166) regulation of histone H2B conserved C-terminal lysine ubiquitination(GO:2001173) regulation of transcription elongation from RNA polymerase I promoter(GO:2001207) positive regulation of transcription elongation from RNA polymerase I promoter(GO:2001209) |
| 0.0 | 0.9 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0071035 | nuclear polyadenylation-dependent rRNA catabolic process(GO:0071035) |
| 0.0 | 0.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0009423 | chorismate biosynthetic process(GO:0009423) |
| 0.0 | 0.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.6 | GO:0006614 | cotranslational protein targeting to membrane(GO:0006613) SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.3 | GO:0042181 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) ketone biosynthetic process(GO:0042181) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0009262 | deoxyribonucleotide metabolic process(GO:0009262) deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0000162 | tryptophan biosynthetic process(GO:0000162) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.0 | 0.5 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 1.6 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 0.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0032042 | mitochondrial DNA metabolic process(GO:0032042) |
| 0.0 | 0.0 | GO:0043038 | amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.0 | 0.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.0 | GO:0007232 | osmosensory signaling pathway via Sho1 osmosensor(GO:0007232) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.5 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.0 | 0.3 | GO:0044088 | regulation of vacuole fusion, non-autophagic(GO:0032889) regulation of vacuole organization(GO:0044088) |
| 0.0 | 0.3 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.0 | GO:0043470 | regulation of carbohydrate catabolic process(GO:0043470) regulation of cellular carbohydrate catabolic process(GO:0043471) |
| 0.0 | 0.3 | GO:0043094 | cellular metabolic compound salvage(GO:0043094) |
| 0.0 | 0.1 | GO:0034517 | ribophagy(GO:0034517) |
| 0.0 | 0.0 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 | 0.2 | GO:0015918 | sterol transport(GO:0015918) |
| 0.0 | 0.1 | GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity(GO:0051090) |
| 0.0 | 0.5 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.0 | GO:0000200 | inactivation of MAPK activity(GO:0000188) inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) negative regulation of MAP kinase activity(GO:0043407) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.0 | 0.2 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.2 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.4 | GO:0051647 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) nucleus localization(GO:0051647) |
| 0.0 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.2 | GO:0044089 | positive regulation of cellular component biogenesis(GO:0044089) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0043007 | maintenance of rDNA(GO:0043007) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) regulation of cellular amine catabolic process(GO:0033241) |
| 0.0 | 0.6 | GO:0008202 | steroid metabolic process(GO:0008202) sterol metabolic process(GO:0016125) |
| 0.0 | 0.2 | GO:0000105 | histidine biosynthetic process(GO:0000105) histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.0 | 0.0 | GO:0070786 | positive regulation of cell growth(GO:0030307) positive regulation of growth of unicellular organism as a thread of attached cells(GO:0070786) positive regulation of pseudohyphal growth by positive regulation of transcription from RNA polymerase II promoter(GO:1900461) positive regulation of pseudohyphal growth(GO:2000222) |
| 0.0 | 0.1 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.1 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.0 | GO:0036170 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.0 | 0.2 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.0 | GO:0035459 | cargo loading into vesicle(GO:0035459) cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:0060277 | obsolete negative regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0060277) |
| 0.0 | 0.0 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.0 | GO:2000221 | negative regulation of cell growth(GO:0030308) negative regulation of pseudohyphal growth(GO:2000221) |
| 0.0 | 0.2 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.0 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:0048279 | vesicle fusion with endoplasmic reticulum(GO:0048279) |
| 0.0 | 0.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.0 | 0.1 | GO:0007009 | plasma membrane organization(GO:0007009) |
| 0.0 | 0.0 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0006797 | polyphosphate metabolic process(GO:0006797) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0060195 | antisense RNA transcription(GO:0009300) regulation of antisense RNA transcription(GO:0060194) negative regulation of antisense RNA transcription(GO:0060195) |
| 0.0 | 0.1 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 1.1 | GO:0097344 | Rix1 complex(GO:0097344) |
| 0.3 | 1.0 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 1.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 26.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 17.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 1.1 | GO:0030287 | cell wall-bounded periplasmic space(GO:0030287) |
| 0.2 | 2.2 | GO:0000144 | cellular bud neck septin ring(GO:0000144) |
| 0.2 | 3.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 0.8 | GO:0070860 | RNA polymerase I core factor complex(GO:0070860) |
| 0.2 | 0.8 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 0.8 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.2 | 0.9 | GO:0032545 | CURI complex(GO:0032545) |
| 0.2 | 0.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.0 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 2.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 0.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.5 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.4 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.2 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.1 | 0.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.4 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 3.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.6 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 0.4 | GO:0005940 | septin ring(GO:0005940) |
| 0.1 | 0.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 0.3 | GO:0033551 | monopolin complex(GO:0033551) |
| 0.1 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.4 | GO:0031499 | TRAMP complex(GO:0031499) |
| 0.1 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.9 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 0.3 | GO:0070772 | PAS complex(GO:0070772) |
| 0.1 | 0.1 | GO:0000133 | polarisome(GO:0000133) |
| 0.1 | 1.4 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.2 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.1 | 0.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.4 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.3 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.3 | GO:0031933 | nuclear heterochromatin(GO:0005720) nuclear telomeric heterochromatin(GO:0005724) telomeric heterochromatin(GO:0031933) |
| 0.0 | 7.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.0 | GO:0071561 | nucleus-vacuole junction(GO:0071561) |
| 0.0 | 0.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0000111 | nucleotide-excision repair factor 2 complex(GO:0000111) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) Elg1 RFC-like complex(GO:0031391) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 1.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.7 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0030892 | mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) |
| 0.0 | 0.1 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.4 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.8 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.0 | 0.1 | GO:0016587 | Isw1 complex(GO:0016587) |
| 0.0 | 0.2 | GO:0033180 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 4.1 | GO:0005933 | cellular bud(GO:0005933) |
| 0.0 | 0.1 | GO:0033254 | vacuolar transporter chaperone complex(GO:0033254) |
| 0.0 | 0.1 | GO:0034044 | exomer complex(GO:0034044) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.1 | GO:0071819 | DUBm complex(GO:0071819) |
| 0.0 | 0.0 | GO:0000131 | incipient cellular bud site(GO:0000131) |
| 0.0 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.6 | GO:0043332 | mating projection tip(GO:0043332) |
| 0.0 | 0.1 | GO:0000417 | HIR complex(GO:0000417) |
| 0.0 | 0.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0030659 | vesicle membrane(GO:0012506) cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.0 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.6 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.5 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.0 | GO:0033597 | mitotic checkpoint complex(GO:0033597) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.5 | 1.6 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 1.6 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.5 | 1.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.5 | 2.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.4 | 1.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.4 | 2.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.3 | 1.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.3 | 1.1 | GO:0016854 | ammonia-lyase activity(GO:0016841) racemase and epimerase activity(GO:0016854) |
| 0.3 | 0.8 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 1.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.3 | 0.8 | GO:0001187 | transcription factor activity, RNA polymerase I CORE element sequence-specific binding(GO:0001169) transcription factor activity, RNA polymerase I CORE element binding transcription factor recruiting(GO:0001187) |
| 0.2 | 1.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 1.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 3.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 0.7 | GO:0043325 | phosphatidylinositol-5-phosphate binding(GO:0010314) phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 2.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 3.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 0.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 38.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 0.8 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.2 | 1.8 | GO:0043178 | alcohol binding(GO:0043178) |
| 0.2 | 0.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.2 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.4 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.1 | 0.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.1 | 0.6 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 0.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.1 | 0.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.8 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 0.4 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.1 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.4 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.3 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 1.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.3 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.6 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.3 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.1 | 0.7 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.1 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.2 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.1 | 0.5 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.1 | 1.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.4 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.1 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.1 | 0.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 0.1 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 1.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.3 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 2.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.6 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.2 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.7 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 0.6 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.5 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 1.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0004691 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.2 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0070717 | poly(A) binding(GO:0008143) poly-purine tract binding(GO:0070717) |
| 0.0 | 1.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.4 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0042124 | glucanosyltransferase activity(GO:0042123) 1,3-beta-glucanosyltransferase activity(GO:0042124) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0004529 | exodeoxyribonuclease activity(GO:0004529) |
| 0.0 | 1.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.0 | 0.1 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.0 | 1.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 0.2 | GO:0001016 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 1 promoter DNA binding(GO:0001030) transcription factor activity, RNA polymerase III transcription factor recruiting(GO:0001153) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 3.8 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0019207 | kinase regulator activity(GO:0019207) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.3 | 1.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 0.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 0.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 0.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 0.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.0 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | REACTOME SIGNALLING TO ERKS | Genes involved in Signalling to ERKs |
| 0.3 | 2.3 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.2 | 3.9 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.2 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 0.8 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.2 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 0.6 | REACTOME LAGGING STRAND SYNTHESIS | Genes involved in Lagging Strand Synthesis |
| 0.1 | 0.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 0.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 0.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME METABOLISM OF MRNA | Genes involved in Metabolism of mRNA |
| 0.0 | 0.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME AXON GUIDANCE | Genes involved in Axon guidance |
| 0.0 | 0.1 | REACTOME APOPTOSIS | Genes involved in Apoptosis |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |