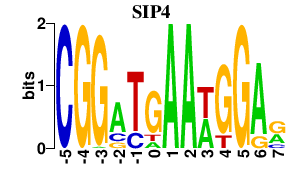
Results for SIP4
Z-value: 2.84
Transcription factors associated with SIP4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIP4
|
S000003625 | Transcriptional activator that regulates gluconeogenic genes |
Activity-expression correlation:
Activity profile of SIP4 motif
Sorted Z-values of SIP4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YLR307C-A | 82.73 |
Putative protein of unknown function |
||
| YJR095W | 62.11 |
SFC1
|
Mitochondrial succinate-fumarate transporter, transports succinate into and fumarate out of the mitochondrion; required for ethanol and acetate utilization |
|
| YKR097W | 51.78 |
PCK1
|
Phosphoenolpyruvate carboxykinase, key enzyme in gluconeogenesis, catalyzes early reaction in carbohydrate biosynthesis, glucose represses transcription and accelerates mRNA degradation, regulated by Mcm1p and Cat8p, located in the cytosol |
|
| YNL117W | 48.99 |
MLS1
|
Malate synthase, enzyme of the glyoxylate cycle, involved in utilization of non-fermentable carbon sources; expression is subject to carbon catabolite repression; localizes in peroxisomes during growth in oleic acid medium |
|
| YER065C | 41.77 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YPL223C | 26.06 |
GRE1
|
Hydrophilin of unknown function; stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YKL217W | 25.54 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YHR096C | 25.50 |
HXT5
|
Hexose transporter with moderate affinity for glucose, induced in the presence of non-fermentable carbon sources, induced by a decrease in growth rate, contains an extended N-terminal domain relative to other HXTs |
|
| YOR100C | 23.34 |
CRC1
|
Mitochondrial inner membrane carnitine transporter, required for carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria during fatty acid beta-oxidation |
|
| YDR536W | 22.26 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YCR005C | 21.09 |
CIT2
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate, peroxisomal isozyme involved in glyoxylate cycle; expression is controlled by Rtg1p and Rtg2p transcription factors |
|
| YAR035W | 20.30 |
YAT1
|
Outer mitochondrial carnitine acetyltransferase, minor ethanol-inducible enzyme involved in transport of activated acyl groups from the cytoplasm into the mitochondrial matrix; phosphorylated |
|
| YML042W | 17.98 |
CAT2
|
Carnitine acetyl-CoA transferase present in both mitochondria and peroxisomes, transfers activated acetyl groups to carnitine to form acetylcarnitine which can be shuttled across membranes |
|
| YPL054W | 15.29 |
LEE1
|
Zinc-finger protein of unknown function |
|
| YER024W | 14.37 |
YAT2
|
Carnitine acetyltransferase; has similarity to Yat1p, which is a carnitine acetyltransferase associated with the mitochondrial outer membrane |
|
| YAL054C | 13.22 |
ACS1
|
Acetyl-coA synthetase isoform which, along with Acs2p, is the nuclear source of acetyl-coA for histone acetlyation; expressed during growth on nonfermentable carbon sources and under aerobic conditions |
|
| YJL045W | 12.81 |
Minor succinate dehydrogenase isozyme; homologous to Sdh1p, the major isozyme reponsible for the oxidation of succinate and transfer of electrons to ubiquinone; induced during the diauxic shift in a Cat8p-dependent manner |
||
| YBL015W | 12.12 |
ACH1
|
Acetyl-coA hydrolase, primarily localized to mitochondria; phosphorylated; required for acetate utilization and for diploid pseudohyphal growth |
|
| YLR308W | 11.95 |
CDA2
|
Chitin deacetylase, together with Cda1p involved in the biosynthesis ascospore wall component, chitosan; required for proper rigidity of the ascospore wall |
|
| YLR377C | 11.94 |
FBP1
|
Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway, required for glucose metabolism |
|
| YBR296C | 11.63 |
PHO89
|
Na+/Pi cotransporter, active in early growth phase; similar to phosphate transporters of Neurospora crassa; transcription regulated by inorganic phosphate concentrations and Pho4p |
|
| YGR067C | 11.58 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YBR147W | 11.40 |
RTC2
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays fluconazole resistance and suppresses cdc13-1 temperature sensitivity |
|
| YPL222W | 10.88 |
FMP40
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YJL089W | 10.09 |
SIP4
|
C6 zinc cluster transcriptional activator that binds to the carbon source-responsive element (CSRE) of gluconeogenic genes; involved in the positive regulation of gluconeogenesis; regulated by Snf1p protein kinase; localized to the nucleus |
|
| YBR051W | 9.43 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partiallly overlaps the REG2/YBR050C regulatory subunit of the Glc7p type-1 protein phosphatase |
||
| YBR050C | 9.00 |
REG2
|
Regulatory subunit of the Glc7p type-1 protein phosphatase; involved with Reg1p, Glc7p, and Snf1p in regulation of glucose-repressible genes, also involved in glucose-induced proteolysis of maltose permease |
|
| YFL030W | 8.93 |
AGX1
|
Alanine:glyoxylate aminotransferase (AGT), catalyzes the synthesis of glycine from glyoxylate, which is one of three pathways for glycine biosynthesis in yeast; has similarity to mammalian and plant alanine:glyoxylate aminotransferases |
|
| YER066W | 8.91 |
Putative protein of unknown function; YER066W is not an essential gene |
||
| YAR053W | 8.89 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR186C-A | 8.84 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YKL163W | 8.82 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YDR542W | 8.66 |
PAU10
|
Hypothetical protein |
|
| YFL024C | 8.31 |
EPL1
|
Component of NuA4, which is an essential histone H4/H2A acetyltransferase complex; homologous to Drosophila Enhancer of Polycomb |
|
| YBR230C | 7.70 |
OM14
|
Integral mitochondrial outer membrane protein; abundance is decreased in cells grown in glucose relative to other carbon sources; appears to contain 3 alpha-helical transmembrane segments; ORF encodes a 97-basepair intron |
|
| YHR211W | 7.34 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YLR004C | 7.33 |
THI73
|
Putative plasma membrane permease proposed to be involved in carboxylic acid uptake and repressed by thiamine; substrate of Dbf2p/Mob1p kinase; transcription is altered if mitochondrial dysfunction occurs |
|
| YNL194C | 7.25 |
Integral membrane protein localized to eisosomes; sporulation and plasma membrane sphingolipid content are altered in mutants; has homologs SUR7 and FMP45; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YPL021W | 7.20 |
ECM23
|
Non-essential protein of unconfirmed function; affects pre-rRNA processing, may act as a negative regulator of the transcription of genes involved in pseudohyphal growth; homologous to Srd1p |
|
| YLR296W | 7.11 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YJR146W | 6.98 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene HMS2 |
||
| YOR338W | 6.56 |
Putative protein of unknown function; YOR338W transcription is regulated by Azf1p and its transcript is a specific target of the G protein effector Scp160p; identified as being required for sporulation in a high-throughput mutant screen |
||
| YER088C | 6.53 |
DOT6
|
Protein of unknown function, involved in telomeric gene silencing and filamentation |
|
| YPR026W | 6.49 |
ATH1
|
Acid trehalase required for utilization of extracellular trehalose |
|
| YMR017W | 6.44 |
SPO20
|
Meiosis-specific subunit of the t-SNARE complex, required for prospore membrane formation during sporulation; similar to but not functionally redundant with Sec9p; SNAP-25 homolog |
|
| YNR064C | 6.33 |
Epoxide hydrolase, member of the alpha/beta hydrolase fold family; may have a role in detoxification of epoxides |
||
| YAR060C | 6.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR033W | 5.99 |
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm |
||
| YMR191W | 5.96 |
SPG5
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YOL126C | 5.95 |
MDH2
|
Cytoplasmic malate dehydrogenase, one of three isozymes that catalyze interconversion of malate and oxaloacetate; involved in the glyoxylate cycle and gluconeogenesis during growth on two-carbon compounds; interacts with Pck1p and Fbp1 |
|
| YMR135C | 5.85 |
GID8
|
Protein of unknown function, involved in proteasome-dependent catabolite inactivation of fructose-1,6-bisphosphatase; contains LisH and CTLH domains, like Vid30p; dosage-dependent regulator of START |
|
| YMR175W | 5.78 |
SIP18
|
Protein of unknown function whose expression is induced by osmotic stress |
|
| YOR343C | 5.69 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YLR295C | 5.58 |
ATP14
|
Subunit h of the F0 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YIL146C | 5.56 |
ECM37
|
Non-essential protein of unknown function |
|
| YIR039C | 5.51 |
YPS6
|
Putative GPI-anchored aspartic protease |
|
| YGL096W | 5.44 |
TOS8
|
Homeodomain-containing transcription factor; SBF regulated target gene that in turn regulates expression of genes involved in G1/S phase events such as bud site selection, bud emergence and cell cycle progression; similarity to Cup9p |
|
| YEL059W | 5.37 |
Dubious open reading frame unlikely to encode a functional protein |
||
| YGR043C | 5.30 |
NQM1
|
Protein of unknown function; transcription is repressed by Mot1p and induced by alpha-factor and during diauxic shift; null mutant non-quiescent cells exhibit reduced reproductive capacity |
|
| YGR065C | 5.30 |
VHT1
|
High-affinity plasma membrane H+-biotin (vitamin H) symporter; mutation results in fatty acid auxotrophy; 12 transmembrane domain containing major facilitator subfamily member; mRNA levels negatively regulated by iron deprivation and biotin |
|
| YPR009W | 5.29 |
SUT2
|
Putative transcription factor; multicopy suppressor of mutations that cause low activity of the cAMP/protein kinase A pathway; highly similar to Sut1p |
|
| YML131W | 5.29 |
Putative protein of unknown function with similarity to oxidoreductases; HOG1 and SKO1-dependent mRNA expression is induced after osmotic shock; GFP-fusion protein localizes to the cytoplasm and is induced by the DNA-damaging agent MMS |
||
| YMR081C | 5.19 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YIL066C | 4.91 |
RNR3
|
One of two large regulatory subunits of ribonucleotide-diphosphate reductase; the RNR complex catalyzes rate-limiting step in dNTP synthesis, regulated by DNA replication and DNA damage checkpoint pathways via localization of small subunits |
|
| YEL024W | 4.88 |
RIP1
|
Ubiquinol-cytochrome-c reductase, a Rieske iron-sulfur protein of the mitochondrial cytochrome bc1 complex; transfers electrons from ubiquinol to cytochrome c1 during respiration |
|
| YLR111W | 4.87 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPL134C | 4.69 |
ODC1
|
Mitochondrial inner membrane transporter, exports 2-oxoadipate and 2-oxoglutarate from the mitochondrial matrix to the cytosol for lysine and glutamate biosynthesis and lysine catabolism; suppresses, in multicopy, an fmc1 null mutation |
|
| YFL055W | 4.66 |
AGP3
|
Low-affinity amino acid permease, may act to supply the cell with amino acids as nitrogen source in nitrogen-poor conditions; transcription is induced under conditions of sulfur limitation; plays a role in regulating Ty1 transposition |
|
| YPR153W | 4.60 |
Putative protein of unknown function |
||
| YPL036W | 4.53 |
PMA2
|
Plasma membrane H+-ATPase, isoform of Pma1p, involved in pumping protons out of the cell; regulator of cytoplasmic pH and plasma membrane potential |
|
| YPR036W-A | 4.48 |
Protein of unknown function; transcription is regulated by Pdr1p |
||
| YIL045W | 4.48 |
PIG2
|
Putative type-1 protein phosphatase targeting subunit that tethers Glc7p type-1 protein phosphatase to Gsy2p glycogen synthase |
|
| YPL061W | 4.43 |
ALD6
|
Cytosolic aldehyde dehydrogenase, activated by Mg2+ and utilizes NADP+ as the preferred coenzyme; required for conversion of acetaldehyde to acetate; constitutively expressed; locates to the mitochondrial outer surface upon oxidative stress |
|
| YDR541C | 4.40 |
Putative dihydrokaempferol 4-reductase |
||
| YMR103C | 4.39 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGL104C | 4.36 |
VPS73
|
Mitochondrial protein; mutation affects vacuolar protein sorting; putative transporter; member of the sugar porter family |
|
| YGR289C | 4.26 |
MAL11
|
Inducible high-affinity maltose transporter (alpha-glucoside transporter); encoded in the MAL1 complex locus; member of the 12 transmembrane domain superfamily of sugar transporters; broad substrate specificity that includes maltotriose |
|
| YJR152W | 4.21 |
DAL5
|
Allantoin permease; ureidosuccinate permease; expression is constitutive but sensitive to nitrogen catabolite repression |
|
| YFR033C | 4.20 |
QCR6
|
Subunit 6 of the ubiquinol cytochrome-c reductase complex, which is a component of the mitochondrial inner membrane electron transport chain; highly acidic protein; required for maturation of cytochrome c1 |
|
| YGL208W | 4.15 |
SIP2
|
One of three beta subunits of the Snf1 serine/threonine protein kinase complex involved in the response to glucose starvation; null mutants exhibit accelerated aging; N-myristoylprotein localized to the cytoplasm and the plasma membrane |
|
| YDR034C | 4.14 |
LYS14
|
Transcriptional activator involved in regulation of genes of the lysine biosynthesis pathway; requires 2-aminoadipate semialdehyde as co-inducer |
|
| YER158C | 4.04 |
Protein of unknown function, has similarity to Afr1p; potentially phosphorylated by Cdc28p |
||
| YHR095W | 4.04 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJL163C | 4.03 |
Putative protein of unknown function |
||
| YHR086W | 3.98 |
NAM8
|
RNA binding protein, component of the U1 snRNP protein; mutants are defective in meiotic recombination and in formation of viable spores, involved in the formation of DSBs through meiosis-specific splicing of MER2 pre-mRNA |
|
| YPR078C | 3.94 |
Putative protein of unknown function; possible role in DNA metabolism and/or in genome stability; expression is heat-inducible |
||
| YKR011C | 3.87 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus |
||
| YDR342C | 3.87 |
HXT7
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels |
|
| YPL206C | 3.80 |
PGC1
|
Phosphatidyl Glycerol phospholipase C; regulates the phosphatidylglycerol (PG) content via a phospholipase C-type degradation mechanism; contains glycerophosphodiester phosphodiesterase motifs |
|
| YBR200W-A | 3.78 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YOR348C | 3.71 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YDL244W | 3.71 |
THI13
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YJR008W | 3.64 |
Putative protein of unknown function; expression repressed by inosine and choline in an Opi1p-dependent manner; expression induced by mild heat-stress on a non-fermentable carbon source. |
||
| YGR290W | 3.57 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; putative HLH protein; partially overlaps the verified ORF MAL11/YGR289C (a high-affinity maltose transporter) |
||
| YLR235C | 3.51 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene TOP3 |
||
| YBR212W | 3.51 |
NGR1
|
RNA binding protein that negatively regulates growth rate; interacts with the 3' UTR of the mitochondrial porin (POR1) mRNA and enhances its degradation; overexpression impairs mitochondrial function; expressed in stationary phase |
|
| YIL086C | 3.47 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBR230W-A | 3.44 |
Putative protein of unknown function |
||
| YLR173W | 3.44 |
Putative protein of unknown function |
||
| YDR223W | 3.40 |
CRF1
|
Transcriptional corepressor involved in the regulation of ribosomal protein gene transcription via the TOR signaling pathway and protein kinase A, phosphorylated by activated Yak1p which promotes accumulation of Crf1p in the nucleus |
|
| YNL055C | 3.38 |
POR1
|
Mitochondrial porin (voltage-dependent anion channel), outer membrane protein required for the maintenance of mitochondrial osmotic stability and mitochondrial membrane permeability; phosphorylated |
|
| YLL019C | 3.37 |
KNS1
|
Nonessential putative protein kinase of unknown cellular role; member of the LAMMER family of protein kinases, which are serine/threonine kinases also capable of phosphorylating tyrosine residues |
|
| YPL150W | 3.32 |
Putative protein kinase of unknown cellular role |
||
| YML129C | 3.31 |
COX14
|
Mitochondrial membrane protein, involved in translational regulation of Cox1p and assembly of cytochrome c oxidase (complex IV); associates with complex IV assembly intermediates and complex III/complex IV supercomplexes |
|
| YKL044W | 3.31 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR236C | 3.31 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNL115C | 3.30 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YNL115C is not an essential gene |
||
| YML058W | 3.22 |
SML1
|
Ribonucleotide reductase inhibitor involved in regulating dNTP production; regulated by Mec1p and Rad53p during DNA damage and S phase |
|
| YDR042C | 3.19 |
Putative protein of unknown function; expression is increased in ssu72-ts69 mutant |
||
| YLR346C | 3.18 |
Putative protein of unknown function found in mitochondria; expression is regulated by transcription factors involved in pleiotropic drug resistance, Pdr1p and Yrr1p; YLR346C is not an essential gene |
||
| YGL177W | 3.18 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHR210C | 3.18 |
Putative protein of unknown function; non-essential gene; highly expressed under anaeorbic conditions; sequence similarity to aldose 1-epimerases such as GAL10 |
||
| YDR275W | 3.17 |
BSC2
|
Protein of unknown function, ORF exhibits genomic organization compatible with a translational readthrough-dependent mode of expression |
|
| YJR061W | 3.16 |
Putative protein of unknown function; non-essential gene with similarity to Mnn4, a putative membrane protein involved in glycosylation; transcription repressed by Rm101p |
||
| YDR171W | 3.09 |
HSP42
|
Small heat shock protein (sHSP) with chaperone activity; forms barrel-shaped oligomers that suppress unfolded protein aggregation; involved in cytoskeleton reorganization after heat shock |
|
| YDR205W | 3.04 |
MSC2
|
Member of the cation diffusion facilitator family, localizes to the endoplasmic reticulum and nucleus; mutations affect the cellular distribution of zinc and also confer defects in meiotic recombination between homologous chromatids |
|
| YLR297W | 3.01 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YLR297W is not an essential gene; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YGL192W | 2.97 |
IME4
|
Probable mRNA N6-adenosine methyltransferase that is required for IME1 transcript accumulation and for sporulation; expression is induced in starved MATa/MAT alpha diploid cells |
|
| YGL193C | 2.95 |
Haploid-specific gene repressed by a1-alpha2, turned off in sir3 null strains, absence enhances the sensitivity of rad52-327 cells to campothecin almost 100-fold |
||
| YPR120C | 2.89 |
CLB5
|
B-type cyclin involved in DNA replication during S phase; activates Cdc28p to promote initiation of DNA synthesis; functions in formation of mitotic spindles along with Clb3p and Clb4p; most abundant during late G1 phase |
|
| YJL100W | 2.89 |
LSB6
|
Type II phosphatidylinositol 4-kinase that binds Las17p, which is a homolog of human Wiskott-Aldrich Syndrome protein involved in actin patch assembly and actin polymerization |
|
| YJR155W | 2.87 |
AAD10
|
Putative aryl-alcohol dehydrogenase with similarity to P. chrysosporium aryl-alcohol dehydrogenase; mutational analysis has not yet revealed a physiological role |
|
| YBR292C | 2.86 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR292C is not an essential gene |
||
| YBR045C | 2.85 |
GIP1
|
Meiosis-specific regulatory subunit of the Glc7p protein phosphatase, regulates spore wall formation and septin organization, required for expression of some late meiotic genes and for normal localization of Glc7p |
|
| YBR056W-A | 2.84 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR056C-B |
||
| YDR216W | 2.83 |
ADR1
|
Carbon source-responsive zinc-finger transcription factor, required for transcription of the glucose-repressed gene ADH2, of peroxisomal protein genes, and of genes required for ethanol, glycerol, and fatty acid utilization |
|
| YIL055C | 2.82 |
Putative protein of unknown function |
||
| YNL104C | 2.81 |
LEU4
|
Alpha-isopropylmalate synthase (2-isopropylmalate synthase); the main isozyme responsible for the first step in the leucine biosynthesis pathway |
|
| YCR007C | 2.71 |
Putative integral membrane protein, member of DUP240 gene family; YCR007C is not an essential gene |
||
| YNL332W | 2.70 |
THI12
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YDL246C | 2.69 |
SOR2
|
Protein of unknown function, computational analysis of large-scale protein-protein interaction data suggests a possible role in fructose or mannose metabolism |
|
| YMR206W | 2.65 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YKL093W | 2.63 |
MBR1
|
Protein involved in mitochondrial functions and stress response; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants |
|
| YLR237W | 2.62 |
THI7
|
Plasma membrane transporter responsible for the uptake of thiamine, member of the major facilitator superfamily of transporters; mutation of human ortholog causes thiamine-responsive megaloblastic anemia |
|
| YIL087C | 2.62 |
LRC2
|
Putative protein of unknown function; protein is detected in purified mitochondria in high-throughput studies; null mutant displays decreased mitochondrial genome loss (petite formation) and severe growth defect in minimal glycerol media |
|
| YPR145C-A | 2.59 |
Putative protein of unknown function |
||
| YDR258C | 2.59 |
HSP78
|
Oligomeric mitochondrial matrix chaperone that cooperates with Ssc1p in mitochondrial thermotolerance after heat shock; prevents the aggregation of misfolded matrix proteins; component of the mitochondrial proteolysis system |
|
| YDR256C | 2.59 |
CTA1
|
Catalase A, breaks down hydrogen peroxide in the peroxisomal matrix formed by acyl-CoA oxidase (Pox1p) during fatty acid beta-oxidation |
|
| YLL029W | 2.58 |
FRA1
|
Protein involved in negative regulation of transcription of iron regulon; forms an iron independent complex with Fra2p, Grx3p, and Grx4p; cytosolic; mutant fails to repress transcription of iron regulon and is defective in spore formation |
|
| YHR139C | 2.54 |
SPS100
|
Protein required for spore wall maturation; expressed during sporulation; may be a component of the spore wall |
|
| YFL023W | 2.53 |
BUD27
|
Protein involved in bud-site selection, nutrient signaling, and gene expression controlled by TOR kinase; diploid mutants show a random budding pattern rather than the wild-type bipolar pattern; plays a role in regulating Ty1 transposition |
|
| YGR087C | 2.52 |
PDC6
|
Minor isoform of pyruvate decarboxylase, decarboxylates pyruvate to acetaldehyde, involved in amino acid catabolism; transcription is glucose- and ethanol-dependent, and is strongly induced during sulfur limitation |
|
| YPL183W-A | 2.46 |
RTC6
|
Homolog of the prokaryotic ribosomal protein L36, likely to be a mitochondrial ribosomal protein coded in the nuclear genome; null mutation suppresses cdc13-1 temperature sensitivity |
|
| YER053C | 2.46 |
PIC2
|
Mitochondrial phosphate carrier, imports inorganic phosphate into mitochondria; functionally redundant with Mir1p but less abundant than Mir1p under normal conditions; expression is induced at high temperature |
|
| YGR292W | 2.45 |
MAL12
|
Maltase (alpha-D-glucosidase), inducible protein involved in maltose catabolism; encoded in the MAL1 complex locus |
|
| YBR201C-A | 2.44 |
Putative protein of unknown function |
||
| YDL243C | 2.37 |
AAD4
|
Putative aryl-alcohol dehydrogenase with similarity to P. chrysosporium aryl-alcohol dehydrogenase, involved in the oxidative stress response |
|
| YJR160C | 2.34 |
MPH3
|
Alpha-glucoside permease, transports maltose, maltotriose, alpha-methylglucoside, and turanose; identical to Mph2p; encoded in a subtelomeric position in a region likely to have undergone duplication |
|
| YJR147W | 2.33 |
HMS2
|
Protein with similarity to heat shock transcription factors; overexpression suppresses the pseudohyphal filamentation defect of a diploid mep1 mep2 homozygous null mutant |
|
| YIL160C | 2.33 |
POT1
|
3-ketoacyl-CoA thiolase with broad chain length specificity, cleaves 3-ketoacyl-CoA into acyl-CoA and acetyl-CoA during beta-oxidation of fatty acids |
|
| YGL139W | 2.32 |
FLC3
|
Putative FAD transporter, similar to Flc1p and Flc2p; localized to the ER |
|
| YPR127W | 2.31 |
Putative protein of unknown function; expression is activated by transcription factor YRM1/YOR172W; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and the nucleus |
||
| YHR071W | 2.31 |
PCL5
|
Cyclin, interacts with Pho85p cyclin-dependent kinase (Cdk), induced by Gcn4p at level of transcription, specifically required for Gcn4p degradation, may be sensor of cellular protein biosynthetic capacity |
|
| YPL151C | 2.28 |
PRP46
|
Splicing factor that is found in the Cef1p subcomplex of the spliceosome |
|
| YMR174C | 2.27 |
PAI3
|
Cytoplasmic proteinase A (Pep4p) inhibitor, dependent on Pbs2p and Hog1p protein kinases for osmotic induction; intrinsically unstructured, N-terminal half becomes ordered in the active site of proteinase A upon contact |
|
| YPR027C | 2.26 |
Putative protein of unknown function |
||
| YDR186C | 2.25 |
Putative protein of unknown function; may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YNL103W | 2.25 |
MET4
|
Leucine-zipper transcriptional activator, responsible for the regulation of the sulfur amino acid pathway, requires different combinations of the auxiliary factors Cbf1p, Met28p, Met31p and Met32p |
|
| YHR009C | 2.25 |
Putative protein of unknown function; not an essential gene |
||
| YAL005C | 2.22 |
SSA1
|
ATPase involved in protein folding and nuclear localization signal (NLS)-directed nuclear transport; member of heat shock protein 70 (HSP70) family; forms a chaperone complex with Ydj1p; localized to the nucleus, cytoplasm, and cell wall |
|
| YHR048W | 2.22 |
YHK8
|
Presumed antiporter of the DHA1 family of multidrug resistance transporters; contains 12 predicted transmembrane spans; expression of gene is up-regulated in cells exhibiting reduced susceptibility to azoles |
|
| YDL204W | 2.22 |
RTN2
|
Protein of unknown function; has similarity to mammalian reticulon proteins; member of the RTNLA (reticulon-like A) subfamily |
|
| YNL015W | 2.21 |
PBI2
|
Cytosolic inhibitor of vacuolar proteinase B, required for efficient vacuole inheritance; with thioredoxin forms protein complex LMA1, which assists in priming SNARE molecules and promotes vacuole fusion |
|
| YIL143C | 2.17 |
SSL2
|
Component of the holoenzyme form of RNA polymerase transcription factor TFIIH, has DNA-dependent ATPase/helicase activity and is required, with Rad3p, for unwinding promoter DNA; involved in DNA repair; homolog of human ERCC3 |
|
| YKL043W | 2.16 |
PHD1
|
Transcriptional activator that enhances pseudohyphal growth; regulates expression of FLO11, an adhesin required for pseudohyphal filament formation; similar to StuA, an A. nidulans developmental regulator; potential Cdc28p substrate |
|
| YBR054W | 2.12 |
YRO2
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in a phosphorylated state in highly purified mitochondria in high-throughput studies; transcriptionally regulated by Haa1p |
|
| YKL109W | 2.12 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YKL221W | 2.08 |
MCH2
|
Protein with similarity to mammalian monocarboxylate permeases, which are involved in transport of monocarboxylic acids across the plasma membrane; mutant is not deficient in monocarboxylate transport |
|
| YER167W | 2.07 |
BCK2
|
Protein rich in serine and threonine residues involved in protein kinase C signaling pathway, which controls cell integrity; overproduction suppresses pkc1 mutations |
|
| YGR184C | 2.06 |
UBR1
|
Ubiquitin-protein ligase (E3) that interacts with Rad6p/Ubc2p to ubiquitinate substrates of the N-end rule pathway; binds to the Rpn2p, Rpt1p, and Rpt6p proteins of the 19S particle of the 26S proteasome |
|
| YDR379W | 2.03 |
RGA2
|
GTPase-activating protein for the polarity-establishment protein Cdc42p; implicated in control of septin organization, pheromone response, and haploid invasive growth; regulated by Pho85p and Cdc28p |
|
| YGL140C | 2.03 |
Putative protein of unknown function; non-essential gene; contains multiple predicted transmembrane domains |
||
| YLR140W | 2.03 |
Dubious open reading frame unlikely to encode a functional protein; overlaps essential RRN5 gene which encodes a member of the UAF transcription factor involved in transcription of rDNA by RNA polymerase I |
||
| YNL243W | 2.02 |
SLA2
|
Transmembrane actin-binding protein involved in membrane cytoskeleton assembly and cell polarization; adaptor protein that links actin to clathrin and endocytosis; present in the actin cortical patch of the emerging bud tip; dimer in vivo |
|
| YFL041W | 2.02 |
FET5
|
Multicopper oxidase, integral membrane protein with similarity to Fet3p; may have a role in iron transport |
|
| YPL024W | 2.01 |
RMI1
|
Involved in response to DNA damage; null mutants have increased rates of recombination and delayed S phase; interacts physically and genetically with Sgs1p (RecQ family member) and Top3p (topoisomerase III) |
|
| YNL005C | 2.01 |
MRP7
|
Mitochondrial ribosomal protein of the large subunit |
|
| YLR122C | 1.98 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR123C |
||
| YHR180W | 1.98 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOL156W | 1.97 |
HXT11
|
Putative hexose transporter that is nearly identical to Hxt9p, has similarity to major facilitator superfamily (MFS) transporters and is involved in pleiotropic drug resistance |
|
| YNR002C | 1.96 |
ATO2
|
Putative transmembrane protein involved in export of ammonia, a starvation signal that promotes cell death in aging colonies; phosphorylated in mitochondria; member of the TC 9.B.33 YaaH family; homolog of Ady2p and Y. lipolytica Gpr1p |
|
| YDL245C | 1.94 |
HXT15
|
Protein of unknown function with similarity to hexose transporter family members, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YOL157C | 1.93 |
Putative protein of unknown function |
||
| YER101C | 1.91 |
AST2
|
Protein that may have a role in targeting of plasma membrane [H+]ATPase (Pma1p) to the plasma membrane, as suggested by analysis of genetic interactions |
|
| YPL058C | 1.89 |
PDR12
|
Plasma membrane ATP-binding cassette (ABC) transporter, weak-acid-inducible multidrug transporter required for weak organic acid resistance; induced by sorbate and benzoate and regulated by War1p; mutants exhibit sorbate hypersensitivity |
|
| YPL026C | 1.89 |
SKS1
|
Putative serine/threonine protein kinase; involved in the adaptation to low concentrations of glucose independent of the SNF3 regulated pathway |
|
| YNL250W | 1.88 |
RAD50
|
Subunit of MRX complex, with Mre11p and Xrs2p, involved in processing double-strand DNA breaks in vegetative cells, initiation of meiotic DSBs, telomere maintenance, and nonhomologous end joining |
|
| YNL308C | 1.88 |
KRI1
|
Essential nucleolar protein required for 40S ribosome biogenesis; physically and functionally interacts with Krr1p |
|
| YGL255W | 1.88 |
ZRT1
|
High-affinity zinc transporter of the plasma membrane, responsible for the majority of zinc uptake; transcription is induced under low-zinc conditions by the Zap1p transcription factor |
|
| YPL262W | 1.84 |
FUM1
|
Fumarase, converts fumaric acid to L-malic acid in the TCA cycle; cytosolic and mitochondrial localization determined by the N-terminal mitochondrial targeting sequence and protein conformation; phosphorylated in mitochondria |
|
| YMR172W | 1.84 |
HOT1
|
Transcription factor required for the transient induction of glycerol biosynthetic genes GPD1 and GPP2 in response to high osmolarity; targets Hog1p to osmostress responsive promoters; has similarity to Msn1p and Gcr1p |
|
| YMR272C | 1.83 |
SCS7
|
Sphingolipid alpha-hydroxylase, functions in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids, has both cytochrome b5-like and hydroxylase/desaturase domains, not essential for growth |
|
| YFL011W | 1.82 |
HXT10
|
Putative hexose transporter, expressed at low levels and expression is repressed by glucose |
|
| YDR406W | 1.81 |
PDR15
|
Plasma membrane ATP binding cassette (ABC) transporter, multidrug transporter and general stress response factor implicated in cellular detoxification; regulated by Pdr1p, Pdr3p and Pdr8p; promoter contains a PDR responsive element |
|
| YKR042W | 1.81 |
UTH1
|
Mitochondrial outer membrane and cell wall localized SUN family member required for mitochondrial autophagy; involved in the oxidative stress response, life span during starvation, mitochondrial biogenesis, and cell death |
|
| YIL136W | 1.78 |
OM45
|
Protein of unknown function, major constituent of the mitochondrial outer membrane; located on the outer (cytosolic) face of the outer membrane |
|
| YOR316C | 1.77 |
COT1
|
Vacuolar transporter that mediates zinc transport into the vacuole; overexpression confers resistance to cobalt and rhodium |
|
| YKL159C | 1.77 |
RCN1
|
Protein involved in calcineurin regulation during calcium signaling; has similarity to H. sapiens DSCR1 which is found in the Down Syndrome candidate region |
|
| YBR241C | 1.76 |
Putative transporter, member of the sugar porter family; green fluorescent protein (GFP)-fusion protein localizes to the vacuolar membrane; YBR241C is not an essential gene |
||
| YEL070W | 1.76 |
DSF1
|
Deletion suppressor of mpt5 mutation |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIP4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.7 | 62.1 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 17.6 | 52.7 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 12.2 | 109.5 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 6.4 | 25.5 | GO:0006848 | pyruvate transport(GO:0006848) |
| 5.0 | 10.1 | GO:0061414 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 4.5 | 22.5 | GO:0015793 | glycerol transport(GO:0015793) |
| 3.9 | 27.4 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 3.0 | 8.9 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 2.8 | 2.8 | GO:0034310 | ethanol catabolic process(GO:0006068) primary alcohol catabolic process(GO:0034310) |
| 2.6 | 75.3 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 2.3 | 11.4 | GO:0034487 | amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 2.2 | 11.0 | GO:0006026 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 2.2 | 6.5 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 1.5 | 10.8 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 1.5 | 4.5 | GO:0052547 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 1.4 | 4.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 1.4 | 8.3 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 1.4 | 5.5 | GO:0009415 | response to water(GO:0009415) cellular response to water stimulus(GO:0071462) |
| 1.3 | 4.0 | GO:0050685 | regulation of RNA splicing(GO:0043484) regulation of mRNA splicing, via spliceosome(GO:0048024) positive regulation of mRNA processing(GO:0050685) |
| 1.3 | 5.3 | GO:0072367 | positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) positive regulation of sterol transport(GO:0032373) positive regulation of transmembrane transport(GO:0034764) regulation of sterol import by regulation of transcription from RNA polymerase II promoter(GO:0035968) positive regulation of sterol import by positive regulation of transcription from RNA polymerase II promoter(GO:0035969) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 1.3 | 9.0 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 1.2 | 9.2 | GO:0051180 | vitamin transport(GO:0051180) |
| 1.1 | 4.5 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 1.1 | 3.4 | GO:0034395 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 1.1 | 10.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 1.1 | 11.7 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 1.1 | 4.3 | GO:0016241 | regulation of macroautophagy(GO:0016241) negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.9 | 5.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.9 | 3.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.9 | 2.6 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.8 | 3.4 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.8 | 9.2 | GO:0015758 | glucose transport(GO:0015758) |
| 0.8 | 4.1 | GO:2000284 | positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.8 | 3.1 | GO:0043388 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.8 | 3.0 | GO:0032071 | regulation of deoxyribonuclease activity(GO:0032070) regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.7 | 6.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.7 | 2.9 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.7 | 1.3 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.6 | 0.6 | GO:0051099 | positive regulation of binding(GO:0051099) |
| 0.6 | 0.6 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.6 | 2.5 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.6 | 4.9 | GO:0070873 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glycogen metabolic process(GO:0070873) |
| 0.6 | 4.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.6 | 1.8 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.5 | 2.2 | GO:0001109 | promoter clearance during DNA-templated transcription(GO:0001109) promoter clearance from RNA polymerase II promoter(GO:0001111) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.5 | 0.5 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.5 | 1.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.5 | 3.2 | GO:0071472 | cellular response to salt stress(GO:0071472) |
| 0.5 | 2.6 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.5 | 3.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.5 | 2.1 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.5 | 1.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.5 | 2.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.5 | 6.0 | GO:0008645 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.5 | 1.4 | GO:0032874 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.5 | 6.9 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 0.5 | 2.7 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.4 | 2.2 | GO:0043954 | cellular component maintenance(GO:0043954) |
| 0.4 | 1.7 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.4 | 5.4 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.4 | 0.4 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.4 | 2.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.4 | 1.2 | GO:0031292 | gene conversion at mating-type locus, DNA double-strand break processing(GO:0031292) |
| 0.4 | 3.9 | GO:0015893 | drug transport(GO:0015893) |
| 0.4 | 5.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.4 | 6.4 | GO:0042724 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.4 | 1.5 | GO:0072417 | cellular response to biotic stimulus(GO:0071216) response to cell cycle checkpoint signaling(GO:0072396) response to mitotic cell cycle checkpoint signaling(GO:0072414) response to spindle checkpoint signaling(GO:0072417) response to mitotic spindle checkpoint signaling(GO:0072476) response to mitotic cell cycle spindle assembly checkpoint signaling(GO:0072479) response to spindle assembly checkpoint signaling(GO:0072485) negative regulation of protein import into nucleus during spindle assembly checkpoint(GO:1901925) |
| 0.4 | 0.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.4 | 1.8 | GO:0001308 | negative regulation of chromatin silencing involved in replicative cell aging(GO:0001308) |
| 0.4 | 2.9 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.4 | 1.1 | GO:0072530 | cytosine transport(GO:0015856) purine-containing compound transmembrane transport(GO:0072530) |
| 0.4 | 1.4 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) aspartate family amino acid catabolic process(GO:0009068) |
| 0.4 | 3.5 | GO:0043487 | regulation of RNA stability(GO:0043487) regulation of mRNA stability(GO:0043488) |
| 0.4 | 4.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.3 | 0.7 | GO:0034755 | high-affinity iron ion transmembrane transport(GO:0006827) iron ion transmembrane transport(GO:0034755) |
| 0.3 | 4.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.3 | 3.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.3 | 2.7 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.3 | 1.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.3 | 3.0 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.3 | 0.7 | GO:0000729 | meiotic DNA double-strand break processing(GO:0000706) DNA double-strand break processing(GO:0000729) |
| 0.3 | 1.6 | GO:0000173 | inactivation of MAPK activity involved in osmosensory signaling pathway(GO:0000173) |
| 0.3 | 1.0 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.3 | 1.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.3 | 1.2 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.3 | 1.8 | GO:0007119 | budding cell isotropic bud growth(GO:0007119) |
| 0.3 | 0.9 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.3 | 0.3 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.3 | 3.2 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.3 | 0.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.3 | 1.2 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) |
| 0.3 | 0.8 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 2.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.3 | 4.3 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.3 | 0.8 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.3 | 2.4 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.3 | 0.8 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.2 | 0.2 | GO:0001178 | regulation of transcriptional start site selection at RNA polymerase II promoter(GO:0001178) |
| 0.2 | 0.9 | GO:0046503 | glycerolipid catabolic process(GO:0046503) |
| 0.2 | 0.9 | GO:0016577 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 0.2 | 11.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.2 | 1.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.2 | 0.6 | GO:0018195 | peptidyl-arginine modification(GO:0018195) peptidyl-arginine methylation(GO:0018216) |
| 0.2 | 0.8 | GO:0060194 | antisense RNA transcription(GO:0009300) regulation of antisense RNA transcription(GO:0060194) negative regulation of antisense RNA transcription(GO:0060195) |
| 0.2 | 1.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.2 | 1.2 | GO:0044205 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.2 | 2.2 | GO:0000436 | carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.2 | 0.6 | GO:0044273 | sulfur compound catabolic process(GO:0044273) |
| 0.2 | 1.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 6.4 | GO:0000902 | cell morphogenesis(GO:0000902) |
| 0.2 | 1.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) meiotic sister chromatid cohesion(GO:0051177) |
| 0.2 | 0.7 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 1.6 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.2 | 0.5 | GO:0055078 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.2 | 1.0 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.2 | 1.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 0.9 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.2 | 0.7 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.2 | 0.5 | GO:0019627 | putrescine transport(GO:0015847) urea metabolic process(GO:0019627) urea catabolic process(GO:0043419) |
| 0.2 | 0.8 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.2 | 0.5 | GO:1902533 | positive regulation of intracellular signal transduction(GO:1902533) |
| 0.2 | 0.8 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 4.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 0.8 | GO:0009208 | CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) |
| 0.1 | 0.9 | GO:0051322 | anaphase(GO:0051322) |
| 0.1 | 0.4 | GO:0051222 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 0.1 | 0.7 | GO:0001881 | receptor recycling(GO:0001881) protein import into peroxisome matrix, receptor recycling(GO:0016562) receptor metabolic process(GO:0043112) |
| 0.1 | 2.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.4 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.1 | 1.5 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 1.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 1.2 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.1 | 0.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.5 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.1 | 1.1 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.1 | 1.1 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 5.6 | GO:0007124 | pseudohyphal growth(GO:0007124) |
| 0.1 | 1.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 1.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 4.4 | GO:0015849 | organic acid transport(GO:0015849) |
| 0.1 | 0.7 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.1 | 0.6 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 1.0 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.1 | 0.5 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.6 | GO:0001041 | transcription from RNA polymerase III type 2 promoter(GO:0001009) transcription from a RNA polymerase III hybrid type promoter(GO:0001041) |
| 0.1 | 0.6 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.1 | 1.4 | GO:0023057 | negative regulation of signal transduction(GO:0009968) negative regulation of cell communication(GO:0010648) negative regulation of signaling(GO:0023057) |
| 0.1 | 0.2 | GO:0090527 | actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.1 | 0.9 | GO:0007129 | synapsis(GO:0007129) |
| 0.1 | 0.4 | GO:0015883 | FAD transport(GO:0015883) |
| 0.1 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 2.8 | GO:0070590 | ascospore wall assembly(GO:0030476) spore wall assembly(GO:0042244) spore wall biogenesis(GO:0070590) ascospore wall biogenesis(GO:0070591) fungal-type cell wall assembly(GO:0071940) |
| 0.1 | 0.3 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 | 0.8 | GO:0000147 | actin cortical patch assembly(GO:0000147) actin cortical patch organization(GO:0044396) |
| 0.1 | 1.5 | GO:0031503 | protein complex localization(GO:0031503) |
| 0.1 | 0.5 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.1 | 0.3 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 0.1 | GO:0016320 | endoplasmic reticulum membrane fusion(GO:0016320) |
| 0.1 | 0.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 2.2 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.1 | 0.7 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.1 | 0.8 | GO:0042493 | response to drug(GO:0042493) |
| 0.1 | 0.7 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.1 | 0.6 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 0.3 | GO:0006452 | translational frameshifting(GO:0006452) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 4.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0009847 | spore germination(GO:0009847) |
| 0.0 | 0.3 | GO:0010388 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 | 0.2 | GO:0034354 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) cyclic-nucleotide-mediated signaling(GO:0019935) |
| 0.0 | 1.3 | GO:0044843 | G1/S transition of mitotic cell cycle(GO:0000082) cell cycle G1/S phase transition(GO:0044843) |
| 0.0 | 0.5 | GO:0000001 | mitochondrion inheritance(GO:0000001) mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.3 | GO:0006056 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0009607 | response to biotic stimulus(GO:0009607) |
| 0.0 | 1.6 | GO:0072666 | protein targeting to vacuole(GO:0006623) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 0.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.9 | GO:0061025 | membrane fusion(GO:0061025) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.8 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0051274 | (1->6)-beta-D-glucan biosynthetic process(GO:0006078) beta-glucan biosynthetic process(GO:0051274) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 52.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 3.8 | 11.4 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 2.4 | 12.0 | GO:0044426 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 1.6 | 12.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 1.4 | 8.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.1 | 3.4 | GO:0046930 | pore complex(GO:0046930) |
| 1.1 | 3.2 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 1.0 | 9.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 1.0 | 6.8 | GO:0034657 | GID complex(GO:0034657) |
| 0.8 | 4.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.8 | 148.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.7 | 9.9 | GO:0031307 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.6 | 1.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.6 | 1.8 | GO:0032177 | split septin rings(GO:0032176) cellular bud neck split septin rings(GO:0032177) |
| 0.6 | 1.2 | GO:0032126 | eisosome(GO:0032126) |
| 0.5 | 3.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.5 | 2.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.5 | 7.5 | GO:0033698 | Rpd3L complex(GO:0033698) |
| 0.5 | 3.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.5 | 3.2 | GO:0000439 | nucleotide-excision repair factor 3 complex(GO:0000112) core TFIIH complex(GO:0000439) |
| 0.4 | 1.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.4 | 23.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.4 | 1.2 | GO:0098562 | side of membrane(GO:0098552) cytoplasmic side of membrane(GO:0098562) |
| 0.4 | 1.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 3.0 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.3 | 1.4 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.3 | 6.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.3 | 1.4 | GO:0070210 | Rpd3L-Expanded complex(GO:0070210) |
| 0.3 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 2.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 4.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 2.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 2.5 | GO:0005619 | ascospore wall(GO:0005619) |
| 0.2 | 1.3 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.2 | 2.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.2 | 1.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 0.7 | GO:0033309 | SBF transcription complex(GO:0033309) |
| 0.2 | 59.4 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.2 | 1.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.9 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 5.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.4 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 1.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.6 | GO:0000796 | transcription factor TFIIIC complex(GO:0000127) condensin complex(GO:0000796) nuclear condensin complex(GO:0000799) |
| 0.1 | 0.3 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.1 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.2 | GO:0030874 | nucleolar chromatin(GO:0030874) |
| 0.1 | 2.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 1.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.9 | GO:0000131 | incipient cellular bud site(GO:0000131) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0034990 | mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) |
| 0.0 | 0.2 | GO:0090544 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.0 | 0.6 | GO:0061645 | actin cortical patch(GO:0030479) endocytic patch(GO:0061645) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.8 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.4 | GO:0031984 | organelle subcompartment(GO:0031984) |
| 0.0 | 0.0 | GO:0097196 | Shu complex(GO:0097196) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.6 | 52.7 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 15.5 | 62.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 10.4 | 52.1 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 7.0 | 21.1 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 6.0 | 41.8 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 5.8 | 46.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 5.0 | 15.0 | GO:0016208 | AMP binding(GO:0016208) |
| 4.0 | 12.1 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 3.0 | 9.1 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 2.8 | 53.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 2.4 | 11.9 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 2.2 | 8.8 | GO:0005537 | mannose binding(GO:0005537) |
| 2.2 | 6.5 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 1.5 | 12.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 1.5 | 4.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 1.5 | 17.4 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 1.4 | 8.5 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 1.3 | 3.8 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 1.2 | 3.6 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 1.2 | 2.3 | GO:0042947 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 1.2 | 4.7 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 1.1 | 9.1 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.1 | 3.4 | GO:0008308 | anion channel activity(GO:0005253) voltage-gated anion channel activity(GO:0008308) porin activity(GO:0015288) wide pore channel activity(GO:0022829) |
| 1.0 | 14.3 | GO:0015578 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 1.0 | 4.9 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 1.0 | 17.6 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.9 | 1.8 | GO:0019903 | phosphatase binding(GO:0019902) protein phosphatase binding(GO:0019903) |
| 0.9 | 1.7 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.8 | 6.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.8 | 4.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.8 | 3.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.8 | 2.4 | GO:0004575 | beta-fructofuranosidase activity(GO:0004564) sucrose alpha-glucosidase activity(GO:0004575) |
| 0.7 | 3.7 | GO:0052742 | phosphatidylinositol kinase activity(GO:0052742) |
| 0.7 | 13.8 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.7 | 2.8 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.7 | 2.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.7 | 7.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.6 | 1.9 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.6 | 1.9 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.6 | 6.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.5 | 1.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.5 | 5.5 | GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism(GO:0015662) |
| 0.5 | 0.5 | GO:0015489 | putrescine transmembrane transporter activity(GO:0015489) spermidine transmembrane transporter activity(GO:0015606) |
| 0.5 | 1.5 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.5 | 8.5 | GO:0008483 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.5 | 5.6 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.4 | 2.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.4 | 21.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.4 | 2.3 | GO:0004030 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.4 | 0.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.4 | 1.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.4 | 2.9 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.4 | 1.4 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.3 | 1.4 | GO:0016725 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 2.0 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.3 | 1.7 | GO:0016830 | carbon-carbon lyase activity(GO:0016830) |
| 0.3 | 1.9 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.3 | 8.3 | GO:0061733 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.3 | 0.9 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.3 | 2.1 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.3 | 1.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 1.4 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.3 | 6.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.3 | 1.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.2 | 7.2 | GO:0005342 | organic acid transmembrane transporter activity(GO:0005342) |
| 0.2 | 0.9 | GO:0015343 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.2 | 1.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.2 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 0.6 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.2 | 1.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.2 | 0.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.2 | 0.9 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.2 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 2.9 | GO:0016684 | peroxidase activity(GO:0004601) oxidoreductase activity, acting on peroxide as acceptor(GO:0016684) |
| 0.2 | 38.9 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.2 | 0.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.2 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 1.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.4 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) |
| 0.1 | 0.7 | GO:0017108 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) 5'-flap endonuclease activity(GO:0017108) flap endonuclease activity(GO:0048256) |
| 0.1 | 0.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.1 | GO:0016722 | oxidoreductase activity, oxidizing metal ions(GO:0016722) |
| 0.1 | 0.7 | GO:0042124 | glucanosyltransferase activity(GO:0042123) 1,3-beta-glucanosyltransferase activity(GO:0042124) |
| 0.1 | 0.7 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 1.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.5 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.1 | 0.4 | GO:0005034 | osmosensor activity(GO:0005034) |
| 0.1 | 0.6 | GO:0001153 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) transcription factor activity, RNA polymerase III promoter sequence-specific binding, TFIIIB recruiting(GO:0001004) transcription factor activity, RNA polymerase III type 1 promoter sequence-specific binding, TFIIIB recruiting(GO:0001005) transcription factor activity, RNA polymerase III type 2 promoter sequence-specific binding, TFIIIB recruiting(GO:0001008) RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) transcription factor activity, RNA polymerase III transcription factor recruiting(GO:0001153) 5S rDNA binding(GO:0080084) |
| 0.1 | 1.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.3 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 0.4 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.1 | 0.5 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.1 | 2.4 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.8 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 0.9 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 1.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 0.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 1.2 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.1 | 1.5 | GO:0019900 | kinase binding(GO:0019900) |
| 0.1 | 0.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 1.3 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.1 | 0.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.2 | GO:0015038 | peptide disulfide oxidoreductase activity(GO:0015037) glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.1 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 1.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.3 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 3.1 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.7 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.1 | GO:0017171 | serine hydrolase activity(GO:0017171) |
| 0.0 | 0.3 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.2 | GO:0003968 | RNA polymerase II activity(GO:0001055) RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 1.6 | GO:0032403 | protein complex binding(GO:0032403) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) second spliceosomal transesterification activity(GO:0000386) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.3 | 51.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.7 | 4.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.7 | 2.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.6 | 1.9 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.3 | 2.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 0.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 0.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 199.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 69.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 1.3 | 3.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.7 | 1.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.7 | 2.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.5 | 2.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 1.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 0.3 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.1 | 2.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 0.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 197.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |