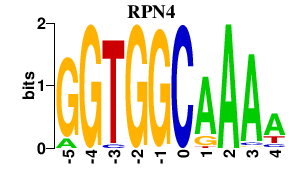
Results for RPN4
Z-value: 0.90
Transcription factors associated with RPN4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RPN4
|
S000002178 | Transcription factor that stimulates expression of proteasome genes |
Activity-expression correlation:
Activity profile of RPN4 motif
Sorted Z-values of RPN4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YJR005C-A | 2.00 |
Putative protein of unknown function, originally identified as a syntenic homolog of an Ashbya gossypii gene |
||
| YJL115W | 1.85 |
ASF1
|
Nucleosome assembly factor, involved in chromatin assembly and disassembly, anti-silencing protein that causes derepression of silent loci when overexpressed; plays a role in regulating Ty1 transposition |
|
| YBL085W | 1.71 |
BOI1
|
Protein implicated in polar growth, functionally redundant with Boi2p; interacts with bud-emergence protein Bem1p; contains an SH3 (src homology 3) domain and a PH (pleckstrin homology) domain |
|
| YJR094C | 1.51 |
IME1
|
Master regulator of meiosis that is active only during meiotic events, activates transcription of early meiotic genes through interaction with Ume6p, degraded by the 26S proteasome following phosphorylation by Ime2p |
|
| YGL035C | 1.44 |
MIG1
|
Transcription factor involved in glucose repression; sequence specific DNA binding protein containing two Cys2His2 zinc finger motifs; regulated by the SNF1 kinase and the GLC7 phosphatase |
|
| YJL116C | 1.43 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YKL218C | 1.32 |
SRY1
|
3-hydroxyaspartate dehydratase, deaminates L-threo-3-hydroxyaspartate to form oxaloacetate and ammonia; required for survival in the presence of hydroxyaspartate |
|
| YDR510W | 1.06 |
SMT3
|
Ubiquitin-like protein of the SUMO family, conjugated to lysine residues of target proteins; regulates chromatid cohesion, chromosome segregation, APC-mediated proteolysis, DNA replication and septin ring dynamics |
|
| YOL123W | 1.02 |
HRP1
|
Subunit of cleavage factor I, a five-subunit complex required for the cleavage and polyadenylation of pre-mRNA 3' ends; RRM-containing heteronuclear RNA binding protein and hnRNPA/B family member that binds to poly (A) signal sequences |
|
| YOR052C | 1.02 |
Nuclear protein of unknown function; expression induced by nitrogen limitation in a GLN3, GAT1-independent manner and by weak acid |
||
| YLL045C | 0.91 |
RPL8B
|
Ribosomal protein L4 of the large (60S) ribosomal subunit, nearly identical to Rpl8Ap and has similarity to rat L7a ribosomal protein; mutation results in decreased amounts of free 60S subunits |
|
| YDL037C | 0.91 |
BSC1
|
Protein of unconfirmed function, similar to cell surface flocculin Muc1p; ORF exhibits genomic organization compatible with a translational readthrough-dependent mode of expression |
|
| YGR286C | 0.91 |
BIO2
|
Biotin synthase, catalyzes the conversion of dethiobiotin to biotin, which is the last step of the biotin biosynthesis pathway; complements E. coli bioB mutant |
|
| YNL251C | 0.91 |
NRD1
|
RNA-binding protein that interacts with the C-terminal domain of the RNA polymerase II large subunit (Rpo21p), required for transcription termination and 3' end maturation of nonpolyadenylated RNAs |
|
| YBR114W | 0.88 |
RAD16
|
Protein that recognizes and binds damaged DNA in an ATP-dependent manner (with Rad7p) during nucleotide excision repair; subunit of Nucleotide Excision Repair Factor 4 (NEF4) and the Elongin-Cullin-Socs (ECS) ligase complex |
|
| YAL003W | 0.86 |
EFB1
|
Translation elongation factor 1 beta; stimulates nucleotide exchange to regenerate EF-1 alpha-GTP for the next elongation cycle; part of the EF-1 complex, which facilitates binding of aminoacyl-tRNA to the ribosomal A site |
|
| YJL107C | 0.85 |
Putative protein of unknown function; expression is induced by activation of the HOG1 mitogen-activated signaling pathway and this induction is Hog1p/Pbs2p dependent; YJL107C and adjacent ORF, YJL108C are merged in related fungi |
||
| YGL103W | 0.84 |
RPL28
|
Ribosomal protein of the large (60S) ribosomal subunit, has similarity to E. coli L15 and rat L27a ribosomal proteins; may have peptidyl transferase activity; can mutate to cycloheximide resistance |
|
| YMR100W | 0.82 |
MUB1
|
Protein of unknown function, deletion causes multi-budding phenotype; has similarity to Aspergillus nidulans samB gene |
|
| YER177W | 0.80 |
BMH1
|
14-3-3 protein, major isoform; controls proteome at post-transcriptional level, binds proteins and DNA, involved in regulation of many processes including exocytosis, vesicle transport, Ras/MAPK signaling, and rapamycin-sensitive signaling |
|
| YMR102C | 0.79 |
Protein of unknown function; transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in multidrug resistance; mutant shows increased resistance to azoles; YMR102C is not an essential gene |
||
| YDR224C | 0.79 |
HTB1
|
One of two nearly identical (see HTB2) histone H2B subtypes required for chromatin assembly and chromosome function; Rad6p-Bre1p-Lge1p mediated ubiquitination regulates transcriptional activation, meiotic DSB formation and H3 methylation |
|
| YFL023W | 0.75 |
BUD27
|
Protein involved in bud-site selection, nutrient signaling, and gene expression controlled by TOR kinase; diploid mutants show a random budding pattern rather than the wild-type bipolar pattern; plays a role in regulating Ty1 transposition |
|
| YBR112C | 0.73 |
CYC8
|
General transcriptional co-repressor, acts together with Tup1p; also acts as part of a transcriptional co-activator complex that recruits the SWI/SNF and SAGA complexes to promoters |
|
| YJR094W-A | 0.73 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YGR255C | 0.70 |
COQ6
|
Putative flavin-dependent monooxygenase, involved in ubiquinone (Coenzyme Q) biosynthesis; localizes to the matrix face of the mitochondrial inner membrane in a large complex with other ubiquinone biosynthetic enzymes |
|
| YBR184W | 0.69 |
Putative protein of unknown function; YBR184W is not an essential gene |
||
| YFR056C | 0.66 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YGR060W | 0.66 |
ERG25
|
C-4 methyl sterol oxidase, catalyzes the first of three steps required to remove two C-4 methyl groups from an intermediate in ergosterol biosynthesis; mutants accumulate the sterol intermediate 4,4-dimethylzymosterol |
|
| YKL216W | 0.66 |
URA1
|
Dihydroorotate dehydrogenase, catalyzes the fourth enzymatic step in the de novo biosynthesis of pyrimidines, converting dihydroorotic acid into orotic acid |
|
| YBR208C | 0.62 |
DUR1,2
|
Urea amidolyase, contains both urea carboxylase and allophanate hydrolase activities, degrades urea to CO2 and NH3; expression sensitive to nitrogen catabolite repression and induced by allophanate, an intermediate in allantoin degradation |
|
| YLR260W | 0.61 |
LCB5
|
Minor sphingoid long-chain base kinase, paralog of Lcb4p responsible for few percent of the total activity, possibly involved in synthesis of long-chain base phosphates, which function as signaling molecules |
|
| YLR448W | 0.60 |
RPL6B
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl6Ap and to rat L6 ribosomal protein; binds to 5.8S rRNA |
|
| YIL031W | 0.59 |
ULP2
|
Peptidase that deconjugates Smt3/SUMO-1 peptides from proteins, plays a role in chromosome cohesion at centromeric regions and recovery from checkpoint arrest induced by DNA damage or DNA replication defects; potential Cdc28p substrate |
|
| YHR021C | 0.58 |
RPS27B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps27Ap and has similarity to rat S27 ribosomal protein |
|
| YGR048W | 0.57 |
UFD1
|
Protein that interacts with Cdc48p and Npl4p, involved in recognition of polyubiquitinated proteins and their presentation to the 26S proteasome for degradation; involved in transporting proteins from the ER to the cytosol |
|
| YML007W | 0.56 |
YAP1
|
Basic leucine zipper (bZIP) transcription factor required for oxidative stress tolerance; activated by H2O2 through the multistep formation of disulfide bonds and transit from the cytoplasm to the nucleus; mediates resistance to cadmium |
|
| YDR414C | 0.56 |
ERD1
|
Predicted membrane protein required for the retention of lumenal endoplasmic reticulum proteins; mutants secrete the endogenous ER protein, BiP (Kar2p) |
|
| YGR140W | 0.54 |
CBF2
|
Essential kinetochore protein, component of the CBF3 multisubunit complex that binds to the CDEIII region of the centromere; Cbf2p also binds to the CDEII region possibly forming a different multimeric complex, ubiquitinated in vivo |
|
| YJL090C | 0.53 |
DPB11
|
Subunit of DNA Polymerase II Epsilon complex; has BRCT domain, required on the prereplicative complex at replication origins for loading DNA polymerases to initiate DNA synthesis, also required for S/M checkpoint control |
|
| YOL038W | 0.53 |
PRE6
|
Alpha 4 subunit of the 20S proteasome; may replace alpha 3 subunit (Pre9p) under stress conditions to create a more active proteasomal isoform; GFP-fusion protein relocates from cytosol to the mitochondrial surface upon oxidative stress |
|
| YNL323W | 0.53 |
LEM3
|
Membrane protein of the plasma membrane and ER, involved in translocation of phospholipids and alkylphosphocholine drugs across the plasma membrane |
|
| YFR055W | 0.52 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YIL142C-A | 0.51 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDL211C | 0.50 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YKL190W | 0.50 |
CNB1
|
Calcineurin B; the regulatory subunit of calcineurin, a Ca++/calmodulin-regulated type 2B protein phosphatase which regulates Crz1p (a stress-response transcription factor), the other calcineurin subunit is encoded by CNA1 and/or CMP1 |
|
| YFL022C | 0.49 |
FRS2
|
Alpha subunit of cytoplasmic phenylalanyl-tRNA synthetase, forms a tetramer with Frs1p to form active enzyme; evolutionarily distant from mitochondrial phenylalanyl-tRNA synthetase based on protein sequence, but substrate binding is similar |
|
| YNR051C | 0.49 |
BRE5
|
Ubiquitin protease cofactor, forms deubiquitination complex with Ubp3p that coregulates anterograde and retrograde transport between the endoplasmic reticulum and Golgi compartments; null is sensitive to brefeldin A |
|
| YML032C | 0.49 |
RAD52
|
Protein that stimulates strand exchange by facilitating Rad51p binding to single-stranded DNA; anneals complementary single-stranded DNA; involved in the repair of double-strand breaks in DNA during vegetative growth and meiosis |
|
| YBR106W | 0.48 |
PHO88
|
Probable membrane protein, involved in phosphate transport; pho88 pho86 double null mutant exhibits enhanced synthesis of repressible acid phosphatase at high inorganic phosphate concentrations |
|
| YDL070W | 0.47 |
BDF2
|
Protein involved in transcription initiation at TATA-containing promoters; associates with the basal transcription factor TFIID; contains two bromodomains; corresponds to the C-terminal region of mammalian TAF1; redundant with Bdf1p |
|
| YMR101C | 0.47 |
SRT1
|
Cis-prenyltransferase involved in synthesis of long-chain dolichols (19-22 isoprene units; as opposed to Rer2p which synthesizes shorter-chain dolichols); localizes to lipid bodies; transcription is induced during stationary phase |
|
| YLL027W | 0.47 |
ISA1
|
Mitochondrial matrix protein involved in biogenesis of the iron-sulfur (Fe/S) cluster of Fe/S proteins, isa1 deletion causes loss of mitochondrial DNA and respiratory deficiency; depletion reduces growth on nonfermentable carbon sources |
|
| YHL024W | 0.47 |
RIM4
|
Putative RNA-binding protein required for the expression of early and middle sporulation genes |
|
| YAL005C | 0.47 |
SSA1
|
ATPase involved in protein folding and nuclear localization signal (NLS)-directed nuclear transport; member of heat shock protein 70 (HSP70) family; forms a chaperone complex with Ydj1p; localized to the nucleus, cytoplasm, and cell wall |
|
| YML088W | 0.46 |
UFO1
|
F-box receptor protein, subunit of the Skp1-Cdc53-F-box receptor (SCF) E3 ubiquitin ligase complex; binds to phosphorylated Ho endonuclease, allowing its ubiquitylation by SCF and subsequent degradation |
|
| YAL053W | 0.46 |
FLC2
|
Putative FAD transporter; required for uptake of FAD into endoplasmic reticulum; involved in cell wall maintenance |
|
| YPL110C | 0.46 |
GDE1
|
Glycerophosphocholine (GroPCho) phosphodiesterase; hydrolyzes GroPCho to choline and glycerolphosphate, for use as a phosphate source and as a precursor for phosphocholine synthesis; may interact with ribosomes |
|
| YOR355W | 0.46 |
GDS1
|
Protein of unknown function, required for growth on glycerol as a carbon source; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YER012W | 0.46 |
PRE1
|
Beta 4 subunit of the 20S proteasome; localizes to the nucleus throughout the cell cycle |
|
| YKL118W | 0.45 |
Dubious open reading frame, unlikely to encode a protein; partially overlaps the verified gene VPH2 |
||
| YPL177C | 0.44 |
CUP9
|
Homeodomain-containing transcriptional repressor of PTR2, which encodes a major peptide transporter; imported peptides activate ubiquitin-dependent proteolysis, resulting in degradation of Cup9p and de-repression of PTR2 transcription |
|
| YJR006W | 0.44 |
POL31
|
DNA polymerase III (delta) subunit, essential for cell viability; involved in DNA replication and DNA repair |
|
| YKL054C | 0.44 |
DEF1
|
RNAPII degradation factor, forms a complex with Rad26p in chromatin, enables ubiquitination and proteolysis of RNAPII present in an elongation complex; mutant is deficient in Zip1p loading onto chromosomes during meiosis |
|
| YOR226C | 0.43 |
ISU2
|
Conserved protein of the mitochondrial matrix, required for synthesis of mitochondrial and cytosolic iron-sulfur proteins, performs a scaffolding function in mitochondria during Fe/S cluster assembly; isu1 isu2 double mutant is inviable |
|
| YGR138C | 0.43 |
TPO2
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; transcription of TPO2 is regulated by Haa1p; member of the major facilitator superfamily |
|
| YCL058C | 0.43 |
FYV5
|
Protein of unknown function, required for survival upon exposure to K1 killer toxin; involved in ion homeostasis |
|
| YIL142W | 0.43 |
CCT2
|
Subunit beta of the cytosolic chaperonin Cct ring complex, related to Tcp1p, required for the assembly of actin and tubulins in vivo |
|
| YKL110C | 0.42 |
KTI12
|
Protein that plays a role, with Elongator complex, in modification of wobble nucleosides in tRNA; involved in sensitivity to G1 arrest induced by zymocin; interacts with chromatin throughout the genome; also interacts with Cdc19p |
|
| YHR006W | 0.42 |
STP2
|
Transcription factor, activated by proteolytic processing in response to signals from the SPS sensor system for external amino acids; activates transcription of amino acid permease genes |
|
| YDL246C | 0.42 |
SOR2
|
Protein of unknown function, computational analysis of large-scale protein-protein interaction data suggests a possible role in fructose or mannose metabolism |
|
| YEL037C | 0.41 |
RAD23
|
Protein with ubiquitin-like N terminus, recognizes and binds damaged DNA (with Rad4p) during nucleotide excision repair; regulates Rad4p levels, subunit of Nuclear Excision Repair Factor 2 (NEF2); homolog of human HR23A and HR23B proteins |
|
| YOL086W-A | 0.41 |
Putative protein of unknown function; identified by homology |
||
| YAL035W | 0.41 |
FUN12
|
GTPase, required for general translation initiation by promoting Met-tRNAiMet binding to ribosomes and ribosomal subunit joining; homolog of bacterial IF2 |
|
| YIL072W | 0.40 |
HOP1
|
Meiosis-specific DNA binding protein that displays Red1p dependent localization to the unsynapsed axial-lateral elements of the synaptonemal complex; required for homologous chromosome synapsis and chiasma formation |
|
| YGR051C | 0.39 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YGR051C is not an essential gene |
||
| YDR043C | 0.39 |
NRG1
|
Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters; mediates glucose repression and negatively regulates a variety of processes including filamentous growth and alkaline pH response |
|
| YCR061W | 0.39 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YJL106W | 0.39 |
IME2
|
Serine/threonine protein kinase involved in activation of meiosis, associates with Ime1p and mediates its stability, activates Ndt80p; IME2 expression is positively regulated by Ime1p |
|
| YGR052W | 0.39 |
FMP48
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YEL033W | 0.39 |
MTC7
|
Predicted metabolic role based on network analysis derived from ChIP experiments, a large-scale deletion study and localization of transcription factor binding sites; null mutant is sensitive to temperature oscillation in a cdc13-1 mutant |
|
| YHR183W | 0.39 |
GND1
|
6-phosphogluconate dehydrogenase (decarboxylating), catalyzes an NADPH regenerating reaction in the pentose phosphate pathway; required for growth on D-glucono-delta-lactone and adaptation to oxidative stress |
|
| YHL031C | 0.39 |
GOS1
|
v-SNARE protein involved in Golgi transport, homolog of the mammalian protein GOS-28/GS28 |
|
| YKR092C | 0.38 |
SRP40
|
Nucleolar, serine-rich protein with a role in preribosome assembly or transport; may function as a chaperone of small nucleolar ribonucleoprotein particles (snoRNPs); immunologically and structurally to rat Nopp140 |
|
| YER064C | 0.38 |
Non-essential nuclear protein; null mutation has global effects on transcription |
||
| YGL122C | 0.38 |
NAB2
|
Nuclear polyadenylated RNA-binding protein; autoregulates mRNA levels; related to human hnRNPs; has nuclear localization signal sequence that binds to Kap104p; required for poly(A) tail length control and nuclear mRNA export |
|
| YBR171W | 0.38 |
SEC66
|
Non-essential subunit of Sec63 complex (Sec63p, Sec62p, Sec66p and Sec72p); with Sec61 complex, Kar2p/BiP and Lhs1p forms a channel competent for SRP-dependent and post-translational SRP-independent protein targeting and import into the ER |
|
| YOR362C | 0.37 |
PRE10
|
Alpha 7 subunit of the 20S proteasome |
|
| YBR174C | 0.37 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF YBR175W; null mutant is viable and sporulation defective |
||
| YBR009C | 0.37 |
HHF1
|
One of two identical histone H4 proteins (see also HHF2); core histone required for chromatin assembly and chromosome function; contributes to telomeric silencing; N-terminal domain involved in maintaining genomic integrity |
|
| YNL027W | 0.37 |
CRZ1
|
Transcription factor that activates transcription of genes involved in stress response; nuclear localization is positively regulated by calcineurin-mediated dephosphorylation |
|
| YBR113W | 0.37 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene CYC8 |
||
| YLR287C-A | 0.36 |
RPS30A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps30Bp and has similarity to rat S30 ribosomal protein |
|
| YOR062C | 0.36 |
Protein of unknown function; similar to YKR075Cp and Reg1p; expression regulated by glucose and Rgt1p; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YDL007W | 0.36 |
RPT2
|
One of six ATPases of the 19S regulatory particle of the 26S proteasome involved in the degradation of ubiquitinated substrates; required for normal peptide hydrolysis by the core 20S particle |
|
| YDR073W | 0.36 |
SNF11
|
Subunit of the SWI/SNF chromatin remodeling complex involved in transcriptional regulation; interacts with a highly conserved 40-residue sequence of Snf2p |
|
| YCR081C-A | 0.35 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps with SRB8/YCR081W; identified by gene-trapping, microarray expression analysis, and genome-wide homology searching |
||
| YML125C | 0.35 |
PGA3
|
Essential protein required for maturation of Gas1p and Pho8p, proposed to be involved in protein trafficking; GFP-fusion protein localizes to the endoplasmic reticulum; null mutants have a cell separation defect |
|
| YDL193W | 0.35 |
NUS1
|
Prenyltransferase, required for cell viability; involved in protein trafficking |
|
| YGR047C | 0.35 |
TFC4
|
One of six subunits of the RNA polymerase III transcription initiation factor complex (TFIIIC); part of the TauA domain of TFIIIC that binds BoxA DNA promoter sites of tRNA and similar genes; has TPR motifs; human homolog is TFIIIC-102 |
|
| YKR011C | 0.35 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus |
||
| YJL036W | 0.35 |
SNX4
|
Sorting nexin, involved in the retrieval of late-Golgi SNAREs from the post-Golgi endosome to the trans-Golgi network and in cytoplasm to vacuole transport; contains a PX domain; forms complexes with Snx41p and with Atg20p |
|
| YPR008W | 0.35 |
HAA1
|
Transcriptional activator involved in the transcription of TPO2, HSP30 and other genes encoding membrane stress proteins; involved in adaptation to weak acid stress |
|
| YER095W | 0.35 |
RAD51
|
Strand exchange protein, forms a helical filament with DNA that searches for homology; involved in the recombinational repair of double-strand breaks in DNA during vegetative growth and meiosis; homolog of Dmc1p and bacterial RecA protein |
|
| YML111W | 0.35 |
BUL2
|
Component of the Rsp5p E3-ubiquitin ligase complex, involved in intracellular amino acid permease sorting, functions in heat shock element mediated gene expression, essential for growth in stress conditions, functional homolog of BUL1 |
|
| YJL200C | 0.35 |
ACO2
|
Putative mitochondrial aconitase isozyme; similarity to Aco1p, an aconitase required for the TCA cycle; expression induced during growth on glucose, by amino acid starvation via Gcn4p, and repressed on ethanol |
|
| YOR157C | 0.34 |
PUP1
|
Endopeptidase with trypsin-like activity that cleaves after basic residues; beta 2 subunit of 20S proteasome, synthesized as a proprotein before being proteolytically processed for assembly into 20S particle; human homolog is subunit Z |
|
| YBL086C | 0.34 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery |
||
| YLL043W | 0.34 |
FPS1
|
Plasma membrane channel, member of major intrinsic protein (MIP) family; involved in efflux of glycerol and in uptake of acetic acid and the trivalent metalloids arsenite and antimonite; phosphorylated by Hog1p MAPK under acetate stress |
|
| YGL201C | 0.34 |
MCM6
|
Protein involved in DNA replication; component of the Mcm2-7 hexameric complex that binds chromatin as a part of the pre-replicative complex |
|
| YGL172W | 0.34 |
NUP49
|
Subunit of the Nsp1p-Nup57p-Nup49p-Nic96p subcomplex of the nuclear pore complex (NPC), required for nuclear export of ribosomes |
|
| YOR389W | 0.34 |
Putative protein of unknown function; expression regulated by copper levels |
||
| YHR182C-A | 0.34 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YFR003C | 0.33 |
YPI1
|
Inhibitor of the type I protein phosphatase Glc7p, which is involved in regulation of a variety of metabolic processes; overproduction causes decreased cellular content of glycogen |
|
| YGL033W | 0.33 |
HOP2
|
Meiosis-specific protein that localizes to chromosomes, preventing synapsis between nonhomologous chromosomes and ensuring synapsis between homologs; complexes with Mnd1p to promote homolog pairing and meiotic double-strand break repair |
|
| YML092C | 0.33 |
PRE8
|
Alpha 2 subunit of the 20S proteasome |
|
| YDR431W | 0.33 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR374W-A | 0.32 |
Putative protein of unknown function |
||
| YJL007C | 0.32 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJR159W | 0.32 |
SOR1
|
Sorbitol dehydrogenase; expression is induced in the presence of sorbitol |
|
| YDL028C | 0.32 |
MPS1
|
Dual-specificity kinase required for spindle pole body (SPB) duplication and spindle checkpoint function; substrates include SPB proteins Spc42p, Spc110p, and Spc98p, mitotic exit network protein Mob1p, and checkpoint protein Mad1p |
|
| YOL087C | 0.32 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; deletion mutant is sensitive to various chemicals including phenanthroline, sanguinarine, and nordihydroguaiaretic acid |
||
| YDL145C | 0.31 |
COP1
|
Alpha subunit of COPI vesicle coatomer complex, which surrounds transport vesicles in the early secretory pathway |
|
| YKL117W | 0.31 |
SBA1
|
Co-chaperone that binds to and regulates Hsp90 family chaperones; important for pp60v-src activity in yeast; homologous to the mammalian p23 proteins and like p23 can regulate telomerase activity |
|
| YBL058W | 0.31 |
SHP1
|
UBX (ubiquitin regulatory X) domain-containing protein that regulates Glc7p phosphatase activity and interacts with Cdc48p; interacts with ubiquitylated proteins in vivo and is required for degradation of a ubiquitylated model substrate |
|
| YPR102C | 0.31 |
RPL11A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl11Bp; involved in ribosomal assembly; depletion causes degradation of proteins and RNA of the 60S subunit; has similarity to E. coli L5 and rat L11 |
|
| YOR050C | 0.31 |
Hypothetical protein |
||
| YMR067C | 0.31 |
UBX4
|
UBX (ubiquitin regulatory X) domain-containing protein that interacts with Cdc48p |
|
| YLR096W | 0.30 |
KIN2
|
Serine/threonine protein kinase involved in regulation of exocytosis; localizes to the cytoplasmic face of the plasma membrane; closely related to Kin1p |
|
| YML065W | 0.30 |
ORC1
|
Largest subunit of the origin recognition complex, which directs DNA replication by binding to replication origins and is also involved in transcriptional silencing; exhibits ATPase activity |
|
| YLR154W-A | 0.30 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YMR246W | 0.29 |
FAA4
|
Long chain fatty acyl-CoA synthetase, regulates protein modification during growth in the presence of ethanol, functions to incorporate palmitic acid into phospholipids and neutral lipids |
|
| YPR041W | 0.29 |
TIF5
|
Translation initiation factor eIF-5; N-terminal domain functions as a GTPase-activating protein to mediate hydrolysis of ribosome-bound GTP; C-terminal domain is the core of ribosomal preinitiation complex formation |
|
| YOR051C | 0.29 |
Nuclear protein that inhibits replication of Brome mosaic virus in S. cerevisiae, which is a model system for studying replication of positive-strand RNA viruses in their natural hosts |
||
| YML130C | 0.28 |
ERO1
|
Thiol oxidase required for oxidative protein folding in the endoplasmic reticulum |
|
| YLR154W-B | 0.28 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YFL036W | 0.28 |
RPO41
|
Mitochondrial RNA polymerase; single subunit enzyme similar to those of T3 and T7 bacteriophages; requires a specificity subunit encoded by MTF1 for promoter recognition |
|
| YOL034W | 0.28 |
SMC5
|
Structural maintenance of chromosomes (SMC) protein; essential subunit of the Mms21-Smc5-Smc6 complex; required for growth and DNA repair; S. pombe homolog forms a heterodimer with S. pombe Rad18p that is involved in DNA repair |
|
| YKL015W | 0.28 |
PUT3
|
Transcriptional activator of proline utilization genes, constitutively binds PUT1 and PUT2 promoter sequences and undergoes a conformational change to form the active state; has a Zn(2)-Cys(6) binuclear cluster domain |
|
| YER075C | 0.27 |
PTP3
|
Phosphotyrosine-specific protein phosphatase involved in the inactivation of mitogen-activated protein kinase (MAPK) during osmolarity sensing; dephosporylates Hog1p MAPK and regulates its localization; localized to the cytoplasm |
|
| YOR146W | 0.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; open reading frame overlaps the verified gene PNO1/YOR145C |
||
| YDR363W-A | 0.27 |
SEM1
|
Component of the lid subcomplex of the regulatory subunit of the 26S proteasome; ortholog of human DSS1 |
|
| YBL041W | 0.27 |
PRE7
|
Beta 6 subunit of the 20S proteasome |
|
| YCR082W | 0.27 |
AHC2
|
Protein of unknown function, putative transcriptional regulator; proposed to be a Ada Histone acetyltransferase complex component; GFP tagged protein is localized to the cytoplasm and nucleus |
|
| YDR285W | 0.27 |
ZIP1
|
Transverse filament protein of the synaptonemal complex; required for normal levels of meiotic recombination and pairing between homologous chromosome during meiosis; potential Cdc28p substrate |
|
| YER129W | 0.27 |
SAK1
|
Upstream serine/threonine kinase for the SNF1 complex; partially redundant with Elm1p and Tos3p; members of this family have functional orthology with LKB1, a mammalian kinase associated with Peutz-Jeghers cancer-susceptibility syndrome |
|
| YBR300C | 0.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YBR301W; YBR300C is not an essential gene |
||
| YNL155W | 0.27 |
Putative protein of unknown function, contains DHHC domain, also predicted to have thiol-disulfide oxidoreductase active site |
||
| YBR010W | 0.27 |
HHT1
|
One of two identical histone H3 proteins (see also HHT2); core histone required for chromatin assembly, involved in heterochromatin-mediated telomeric and HM silencing; regulated by acetylation, methylation, and mitotic phosphorylation |
|
| YHR021W-A | 0.27 |
ECM12
|
Non-essential protein of unknown function |
|
| YEL056W | 0.27 |
HAT2
|
Subunit of the Hat1p-Hat2p histone acetyltransferase complex; required for high affinity binding of the complex to free histone H4, thereby enhancing Hat1p activity; similar to human RbAp46 and 48; has a role in telomeric silencing |
|
| YNL172W | 0.26 |
APC1
|
Largest subunit of the Anaphase-Promoting Complex/Cyclosome (APC/C), which is a ubiquitin-protein ligase required for degradation of anaphase inhibitors, including mitotic cyclins, during the metaphase/anaphase transition |
|
| YMR281W | 0.26 |
GPI12
|
ER membrane protein involved in the second step of glycosylphosphatidylinositol (GPI) anchor assembly, the de-N-acetylation of the N-acetylglucosaminylphosphatidylinositol intermediate; functional homolog of human PIG-Lp |
|
| YCL024W | 0.26 |
KCC4
|
Protein kinase of the bud neck involved in the septin checkpoint, associates with septin proteins, negatively regulates Swe1p by phosphorylation, shows structural homology to bud neck kinases Gin4p and Hsl1p |
|
| YEL032W | 0.26 |
MCM3
|
Protein involved in DNA replication; component of the Mcm2-7 hexameric complex that binds chromatin as a part of the pre-replicative complex |
|
| YIL126W | 0.26 |
STH1
|
ATPase component of the RSC chromatin remodeling complex; required for expression of early meiotic genes; essential helicase-related protein homologous to Snf2p |
|
| YGL011C | 0.26 |
SCL1
|
Alpha 1 subunit of the 20S core complex of the 26S proteasome involved in the degradation of ubiquitinated substrates; essential for growth; detected in the mitochondria |
|
| YHR027C | 0.26 |
RPN1
|
Non-ATPase base subunit of the 19S regulatory particle of the 26S proteasome; may participate in the recognition of several ligands of the proteasome; contains a leucine-rich repeat (LRR) domain, a site for protein?protein interactions |
|
| YNL313C | 0.26 |
Putative protein of unknown function; identified as interacting with Kar2p, Grs1p, and Tub3p in high-throughput TAP-tagging experiments; YNL313C is an essential gene |
||
| YJL014W | 0.26 |
CCT3
|
Subunit of the cytosolic chaperonin Cct ring complex, related to Tcp1p, required for the assembly of actin and tubulins in vivo |
|
| YNL304W | 0.26 |
YPT11
|
Rab-type small GTPase that interacts with the C-terminal tail domain of Myo2p to mediate distribution of mitochondria to daughter cells |
|
| YPR185W | 0.26 |
ATG13
|
Regulatory subunit of the Atg1p signaling complex; stimulates Atg1p kinase activity; required for vesicle formation during autophagy and the cytoplasm-to-vacuole targeting (Cvt) pathway; involved in Atg9p, Atg23p, and Atg27p cycling |
|
| YPL058C | 0.26 |
PDR12
|
Plasma membrane ATP-binding cassette (ABC) transporter, weak-acid-inducible multidrug transporter required for weak organic acid resistance; induced by sorbate and benzoate and regulated by War1p; mutants exhibit sorbate hypersensitivity |
|
| YOR145C | 0.26 |
PNO1
|
Essential nucleolar protein required for pre-18S rRNA processing, interacts with Dim1p, an 18S rRNA dimethyltransferase, and also with Nob1p, which is involved in proteasome biogenesis; contains a KH domain |
|
| YJL105W | 0.26 |
SET4
|
Protein of unknown function, contains a SET domain |
|
| YJL008C | 0.25 |
CCT8
|
Subunit of the cytosolic chaperonin Cct ring complex, related to Tcp1p, required for the assembly of actin and tubulins in vivo |
|
| YOR359W | 0.25 |
VTS1
|
Post-transcriptional gene regulator, RNA-binding protein containing a SAM domain; shows genetic interactions with Vti1p, which is a v-SNARE involved in cis-Golgi membrane traffic |
|
| YBR202W | 0.25 |
MCM7
|
Component of the hexameric MCM complex, which is important for priming origins of DNA replication in G1 and becomes an active ATP-dependent helicase that promotes DNA melting and elongation when activated by Cdc7p-Dbf4p in S-phase |
|
| YER054C | 0.25 |
GIP2
|
Putative regulatory subunit of the protein phosphatase Glc7p, involved in glycogen metabolism; contains a conserved motif (GVNK motif) that is also found in Gac1p, Pig1p, and Pig2p |
|
| YJL015C | 0.25 |
Dubious open reading frame unlikely to encode a functional protein; expression if heat-inducible; located in promoter region of essential CCT3 gene encoding a subunit of the cytosolic chaperonin Cct ring complex, overlaps ORF YJL016W |
||
| YFR051C | 0.25 |
RET2
|
Delta subunit of the coatomer complex (COPI), which coats Golgi-derived transport vesicles; involved in retrograde transport between Golgi and ER |
|
| YPR065W | 0.25 |
ROX1
|
Heme-dependent repressor of hypoxic genes; contains an HMG domain that is responsible for DNA bending activity |
|
| YEL034C-A | 0.24 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; completely overlaps HYP2/YEL034W, a verified gene that encodes eiF-5A |
||
| YLR154W-C | 0.24 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YOL086C | 0.24 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YOL159C-A | 0.24 |
Putative protein of unknown function; identified by sequence comparison with hemiascomycetous yeast species |
||
| YFL015W-A | 0.24 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR097W | 0.24 |
ASK10
|
Component of the RNA polymerase II holoenzyme, phosphorylated in response to oxidative stress; has a role in destruction of Ssn8p, which relieves repression of stress-response genes |
|
| YDR270W | 0.24 |
CCC2
|
Cu(+2)-transporting P-type ATPase, required for export of copper from the cytosol into an extracytosolic compartment; has similarity to human proteins involved in Menkes and Wilsons diseases |
|
| YPL108W | 0.24 |
Cytoplasmic protein of unknown function; non-essential gene that is induced in a GDH1 deleted strain with altered redox metabolism; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YHR203C | 0.23 |
RPS4B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps4Ap and has similarity to rat S4 ribosomal protein |
|
| YGL148W | 0.23 |
ARO2
|
Bifunctional chorismate synthase and flavin reductase, catalyzes the conversion of 5-enolpyruvylshikimate 3-phosphate (EPSP) to form chorismate, which is a precursor to aromatic amino acids |
|
| YPL205C | 0.23 |
Hypothetical protein; deletion of locus affects telomere length |
||
| YBR060C | 0.23 |
ORC2
|
Subunit of the origin recognition complex, which directs DNA replication by binding to replication origins and is also involved in transcriptional silencing; phosphorylated by Cdc28p |
|
| YPL204W | 0.23 |
HRR25
|
Protein kinase involved in regulating diverse events including vesicular trafficking, DNA repair, and chromosome segregation; binds the CTD of RNA pol II; homolog of mammalian casein kinase 1delta (CK1delta) |
|
| YBR049C | 0.23 |
REB1
|
RNA polymerase I enhancer binding protein; DNA binding protein which binds to genes transcribed by both RNA polymerase I and RNA polymerase II; required for termination of RNA polymerase I transcription |
|
| YOR315W | 0.23 |
SFG1
|
Nuclear protein, putative transcription factor required for growth of superficial pseudohyphae (which do not invade the agar substrate) but not for invasive pseudohyphal growth; may act together with Phd1p; potential Cdc28p substrate |
|
| YIR007W | 0.23 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YIR007W is a non-essential gene |
||
| YHR154W | 0.23 |
RTT107
|
Protein implicated in Mms22-dependent DNA repair during S phase, DNA damage induces phosphorylation by Mec1p at one or more SQ/TQ motifs; interacts with Mms22p and Slx4p; has four BRCT domains; has a role in regulation of Ty1 transposition |
|
| YDR279W | 0.23 |
RNH202
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YGR146C-A | 0.23 |
Putative protein of unknown function |
||
| YML031C-A | 0.23 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF NDC1/YML031W; questionable ORF from MIPS |
||
| YIL164C | 0.22 |
NIT1
|
Nitrilase, member of the nitrilase branch of the nitrilase superfamily; in closely related species and other S. cerevisiae strain backgrounds YIL164C and adjacent ORF, YIL165C, likely constitute a single ORF encoding a nitrilase gene |
|
| YPR125W | 0.22 |
YLH47
|
Mitochondrial inner membrane protein exposed to the mitochondrial matrix, associates with mitochondrial ribosomes, NOT required for respiratory growth; homolog of human Letm1, a protein implicated in Wolf-Hirschhorn syndrome |
|
| YNR054C | 0.22 |
ESF2
|
Essential nucleolar protein involved in pre-18S rRNA processing; binds to RNA and stimulates ATPase activity of Dbp8; involved in assembly of the small subunit (SSU) processome |
|
| YDL143W | 0.22 |
CCT4
|
Subunit of the cytosolic chaperonin Cct ring complex, related to Tcp1p, required for the assembly of actin and tubulins in vivo |
|
| YMR108W | 0.22 |
ILV2
|
Acetolactate synthase, catalyses the first common step in isoleucine and valine biosynthesis and is the target of several classes of inhibitors, localizes to the mitochondria; expression of the gene is under general amino acid control |
|
| YMR244C-A | 0.22 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and nucleus and is induced in response to the DNA-damaging agent MMS; YMR244C-A is not an essential gene |
||
| YFR037C | 0.22 |
RSC8
|
Component of the RSC chromatin remodeling complex; essential for viability and mitotic growth; homolog of SWI/SNF subunit Swi3p, but unlike Swi3p, does not activate transcription of reporters |
|
| YPL128C | 0.22 |
TBF1
|
Telobox-containing general regulatory factor; binds to TTAGGG repeats within subtelomeric anti-silencing regions (STARs) and possibly throughout the genome and mediates their insulating capacity by blocking silent chromatin propagation |
Network of associatons between targets according to the STRING database.
First level regulatory network of RPN4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0035950 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 0.4 | 1.5 | GO:0036170 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.4 | 1.9 | GO:2000758 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-dependent nucleosome organization(GO:0034723) positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.4 | 1.4 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.3 | 0.9 | GO:1904667 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.2 | 0.9 | GO:0046898 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 0.2 | 0.9 | GO:0071041 | antisense RNA transcript catabolic process(GO:0071041) |
| 0.2 | 1.4 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.2 | 0.6 | GO:0043419 | urea catabolic process(GO:0043419) |
| 0.2 | 2.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 0.6 | GO:0000304 | response to singlet oxygen(GO:0000304) |
| 0.2 | 0.7 | GO:0000709 | meiotic joint molecule formation(GO:0000709) |
| 0.2 | 0.5 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.2 | 0.5 | GO:0061422 | positive regulation of transcription from RNA polymerase II promoter in response to alkaline pH(GO:0061422) |
| 0.2 | 0.5 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.2 | 0.7 | GO:0048313 | Golgi inheritance(GO:0048313) |
| 0.2 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 0.5 | GO:0034517 | ribophagy(GO:0034517) |
| 0.2 | 1.8 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.2 | 1.4 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 0.4 | GO:0071469 | response to alkaline pH(GO:0010446) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.5 | GO:0000296 | spermine transport(GO:0000296) |
| 0.1 | 0.5 | GO:0015883 | FAD transport(GO:0015883) |
| 0.1 | 1.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.1 | GO:2000756 | regulation of histone acetylation(GO:0035065) regulation of protein acetylation(GO:1901983) regulation of peptidyl-lysine acetylation(GO:2000756) |
| 0.1 | 0.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.6 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.3 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) |
| 0.1 | 0.8 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.5 | GO:0071406 | response to methylmercury(GO:0051597) cellular response to methylmercury(GO:0071406) |
| 0.1 | 0.6 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.4 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.5 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.1 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.1 | GO:0019627 | urea metabolic process(GO:0019627) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) regulation of cellular amine catabolic process(GO:0033241) |
| 0.1 | 0.4 | GO:0009423 | chorismate biosynthetic process(GO:0009423) |
| 0.1 | 0.4 | GO:1903829 | retrograde protein transport, ER to cytosol(GO:0030970) positive regulation of protein localization to nucleus(GO:1900182) endoplasmic reticulum to cytosol transport(GO:1903513) positive regulation of cellular protein localization(GO:1903829) |
| 0.1 | 0.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.3 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) |
| 0.1 | 1.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.2 | GO:0034059 | response to anoxia(GO:0034059) cellular response to anoxia(GO:0071454) |
| 0.1 | 1.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.5 | GO:0044205 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.2 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.6 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 0.3 | GO:0031134 | sister chromatid biorientation(GO:0031134) |
| 0.1 | 0.4 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.1 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.2 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 0.3 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 1.4 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 0.1 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.1 | 0.2 | GO:0072363 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 0.1 | 0.5 | GO:2000278 | regulation of telomere maintenance via telomerase(GO:0032210) regulation of telomere maintenance via telomere lengthening(GO:1904356) regulation of DNA biosynthetic process(GO:2000278) |
| 0.1 | 0.5 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.1 | 0.2 | GO:0006827 | high-affinity iron ion transmembrane transport(GO:0006827) |
| 0.1 | 0.3 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 0.1 | 1.0 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.7 | GO:0042181 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) ketone biosynthetic process(GO:0042181) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.3 | GO:0032079 | positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.1 | 0.7 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 0.2 | GO:0031135 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.1 | 0.4 | GO:0001009 | transcription from RNA polymerase III type 2 promoter(GO:0001009) transcription from a RNA polymerase III hybrid type promoter(GO:0001041) |
| 0.1 | 0.5 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.2 | GO:0032443 | regulation of steroid metabolic process(GO:0019218) regulation of ergosterol biosynthetic process(GO:0032443) regulation of steroid biosynthetic process(GO:0050810) |
| 0.1 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.2 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.3 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 | 0.4 | GO:0019655 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.1 | 1.6 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.4 | GO:0045860 | positive regulation of protein kinase activity(GO:0045860) |
| 0.0 | 0.3 | GO:0071039 | nuclear polyadenylation-dependent CUT catabolic process(GO:0071039) |
| 0.0 | 0.3 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.1 | GO:1900151 | regulation of mRNA 3'-end processing(GO:0031440) regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) |
| 0.0 | 0.1 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.0 | 0.0 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.1 | GO:0070482 | response to decreased oxygen levels(GO:0036293) response to oxygen levels(GO:0070482) |
| 0.0 | 0.4 | GO:2000144 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.9 | GO:0002098 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.2 | GO:0043007 | maintenance of rDNA(GO:0043007) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.4 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.2 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.0 | 0.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0001080 | nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) nitrogen catabolite activation of transcription(GO:0090294) |
| 0.0 | 0.2 | GO:0010529 | negative regulation of transposition, RNA-mediated(GO:0010526) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.3 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.1 | GO:0001111 | promoter clearance during DNA-templated transcription(GO:0001109) promoter clearance from RNA polymerase II promoter(GO:0001111) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 3.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0000735 | removal of nonhomologous ends(GO:0000735) |
| 0.0 | 0.8 | GO:0051304 | chromosome separation(GO:0051304) |
| 0.0 | 0.3 | GO:0006037 | cell wall chitin metabolic process(GO:0006037) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) copper ion homeostasis(GO:0055070) |
| 0.0 | 0.1 | GO:0030308 | negative regulation of cell growth(GO:0030308) negative regulation of pseudohyphal growth(GO:2000221) |
| 0.0 | 0.1 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.0 | 0.0 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.2 | GO:0000376 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.0 | 0.0 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) |
| 0.0 | 0.1 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.2 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.3 | GO:0032005 | pheromone-dependent signal transduction involved in conjugation with cellular fusion(GO:0000750) signal transduction involved in conjugation with cellular fusion(GO:0032005) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.3 | GO:0000754 | adaptation of signaling pathway by response to pheromone involved in conjugation with cellular fusion(GO:0000754) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.1 | GO:0000921 | septin ring assembly(GO:0000921) |
| 0.0 | 0.1 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 1.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.0 | GO:1902749 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.0 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin structure organization(GO:0031032) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.3 | GO:0001101 | response to acid chemical(GO:0001101) |
| 0.0 | 0.1 | GO:0071035 | nuclear polyadenylation-dependent rRNA catabolic process(GO:0071035) |
| 0.0 | 0.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.2 | GO:0009228 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.0 | 0.1 | GO:0034503 | protein localization to nucleolar rDNA repeats(GO:0034503) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0006672 | ceramide metabolic process(GO:0006672) ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0042797 | tRNA transcription(GO:0009304) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0005977 | glycogen metabolic process(GO:0005977) energy reserve metabolic process(GO:0006112) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.4 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:0046219 | tryptophan biosynthetic process(GO:0000162) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.0 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.0 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.0 | 0.1 | GO:0051051 | negative regulation of transport(GO:0051051) |
| 0.0 | 0.1 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.3 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.4 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.0 | GO:0090282 | positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.0 | 0.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0034063 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) stress granule assembly(GO:0034063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.3 | 0.9 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.3 | 1.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 1.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 1.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.1 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.2 | 0.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.4 | GO:0000111 | nucleotide-excision repair factor 2 complex(GO:0000111) |
| 0.1 | 0.5 | GO:0031518 | CBF3 complex(GO:0031518) |
| 0.1 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.7 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.6 | GO:0005940 | septin ring(GO:0005940) |
| 0.1 | 0.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.4 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) Sec62/Sec63 complex(GO:0031207) |
| 0.1 | 1.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.2 | GO:0033551 | monopolin complex(GO:0033551) |
| 0.1 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.2 | GO:0035361 | Cul8-RING ubiquitin ligase complex(GO:0035361) |
| 0.1 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.2 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.1 | 4.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.0 | 0.2 | GO:0070772 | PAS complex(GO:0070772) |
| 0.0 | 0.1 | GO:0030874 | nucleolar chromatin(GO:0030874) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0043529 | GET complex(GO:0043529) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.5 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.2 | GO:0008278 | nuclear cohesin complex(GO:0000798) cohesin complex(GO:0008278) |
| 0.0 | 0.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 1.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0032126 | eisosome(GO:0032126) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.2 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0035097 | histone methyltransferase complex(GO:0035097) Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0097344 | Rix1 complex(GO:0097344) |
| 0.0 | 0.1 | GO:0031499 | TRAMP complex(GO:0031499) |
| 0.0 | 0.1 | GO:0031391 | DNA replication factor C complex(GO:0005663) Rad17 RFC-like complex(GO:0031389) Elg1 RFC-like complex(GO:0031391) |
| 0.0 | 0.1 | GO:0032545 | CURI complex(GO:0032545) |
| 0.0 | 0.1 | GO:0016587 | Isw1 complex(GO:0016587) |
| 0.0 | 0.4 | GO:0000131 | incipient cellular bud site(GO:0000131) |
| 0.0 | 0.1 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.1 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.3 | GO:0043332 | mating projection tip(GO:0043332) |
| 0.0 | 1.3 | GO:0000428 | DNA-directed RNA polymerase complex(GO:0000428) RNA polymerase complex(GO:0030880) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.0 | GO:0032806 | carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 0.1 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0070390 | transcription export complex 2(GO:0070390) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.3 | 0.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.3 | 0.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 1.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 2.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 0.9 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.2 | 0.7 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.2 | 0.5 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.2 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 0.6 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 2.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.4 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 0.5 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.1 | 0.5 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.1 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.4 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 1.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.4 | GO:0008310 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.3 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 0.9 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.1 | 0.6 | GO:0008143 | poly(A) binding(GO:0008143) poly-purine tract binding(GO:0070717) |
| 0.1 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.3 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.2 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.1 | 1.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.7 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 2.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.2 | GO:0004691 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.8 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.4 | GO:0001004 | RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) transcription factor activity, RNA polymerase III promoter sequence-specific binding, TFIIIB recruiting(GO:0001004) transcription factor activity, RNA polymerase III type 1 promoter sequence-specific binding, TFIIIB recruiting(GO:0001005) transcription factor activity, RNA polymerase III type 2 promoter sequence-specific binding, TFIIIB recruiting(GO:0001008) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.1 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.6 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 1.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.4 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.8 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0008902 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.0 | 0.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 2.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.6 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 2.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 2.1 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.9 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.3 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 5.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:0032182 | ubiquitin-like protein binding(GO:0032182) ubiquitin binding(GO:0043130) |
| 0.0 | 0.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0016872 | intramolecular lyase activity(GO:0016872) |
| 0.0 | 0.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.1 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.2 | GO:0000988 | transcription factor activity, protein binding(GO:0000988) |
| 0.0 | 0.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.0 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.1 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.0 | 0.0 | GO:0031683 | receptor binding(GO:0005102) G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.0 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0099600 | transmembrane signaling receptor activity(GO:0004888) signaling receptor activity(GO:0038023) transmembrane receptor activity(GO:0099600) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 0.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 1.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.2 | 0.5 | REACTOME SIGNALLING TO ERKS | Genes involved in Signalling to ERKs |
| 0.2 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.2 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 0.4 | REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | Genes involved in E2F mediated regulation of DNA replication |
| 0.0 | 1.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.1 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME TRANSCRIPTION COUPLED NER TC NER | Genes involved in Transcription-coupled NER (TC-NER) |
| 0.0 | 0.1 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.0 | REACTOME SIGNALING BY NOTCH | Genes involved in Signaling by NOTCH |
| 0.0 | 0.1 | REACTOME RNA POL II TRANSCRIPTION | Genes involved in RNA Polymerase II Transcription |
| 0.0 | 0.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |