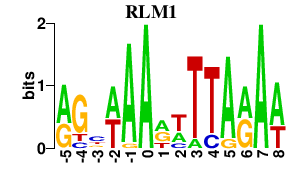
Results for RLM1
Z-value: 1.13
Transcription factors associated with RLM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RLM1
|
S000006010 | MADS-box transcription factor |
Activity-expression correlation:
Activity profile of RLM1 motif
Sorted Z-values of RLM1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YHR212C | 6.14 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR013C | 6.08 |
SEC59
|
Dolichol kinase, catalyzes the terminal step in dolichyl monophosphate (Dol-P) biosynthesis; required for viability and for normal rates of lipid intermediate synthesis and protein N-glycosylation |
|
| YHR212W-A | 5.96 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YAR053W | 5.74 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YKR097W | 5.72 |
PCK1
|
Phosphoenolpyruvate carboxykinase, key enzyme in gluconeogenesis, catalyzes early reaction in carbohydrate biosynthesis, glucose represses transcription and accelerates mRNA degradation, regulated by Mcm1p and Cat8p, located in the cytosol |
|
| YAL062W | 5.48 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YIL125W | 5.01 |
KGD1
|
Component of the mitochondrial alpha-ketoglutarate dehydrogenase complex, which catalyzes a key step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation of alpha-ketoglutarate to form succinyl-CoA |
|
| YPL171C | 4.87 |
OYE3
|
Widely conserved NADPH oxidoreductase containing flavin mononucleotide (FMN), homologous to Oye2p with slight differences in ligand binding and catalytic properties; may be involved in sterol metabolism |
|
| YAR060C | 4.64 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR120W | 4.48 |
GCY1
|
Putative NADP(+) coupled glycerol dehydrogenase, proposed to be involved in an alternative pathway for glycerol catabolism; member of the aldo-keto reductase (AKR) family |
|
| YMR107W | 4.40 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YDR342C | 4.32 |
HXT7
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels |
|
| YNL093W | 4.22 |
YPT53
|
GTPase, similar to Ypt51p and Ypt52p and to mammalian rab5; required for vacuolar protein sorting and endocytosis |
|
| YEL012W | 4.21 |
UBC8
|
Ubiquitin-conjugating enzyme that negatively regulates gluconeogenesis by mediating the glucose-induced ubiquitination of fructose-1,6-bisphosphatase (FBPase); cytoplasmic enzyme that catalyzes the ubiquitination of histones in vitro |
|
| YFR053C | 4.19 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YOR381W | 4.09 |
FRE3
|
Ferric reductase, reduces siderophore-bound iron prior to uptake by transporters; expression induced by low iron levels |
|
| YNR073C | 4.03 |
Putative mannitol dehydrogenase |
||
| YKR102W | 3.88 |
FLO10
|
Lectin-like protein with similarity to Flo1p, thought to be involved in flocculation |
|
| YDR343C | 3.80 |
HXT6
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt7p, expressed at high basal levels relative to other HXTs, repression of expression by high glucose requires SNF3 |
|
| YEL008W | 3.67 |
Hypothetical protein predicted to be involved in metabolism |
||
| YAR023C | 3.60 |
Putative integral membrane protein, member of DUP240 gene family |
||
| YCL001W-B | 3.57 |
Putative protein of unknown function; identified by homology |
||
| YEL009C | 3.43 |
GCN4
|
Transcriptional activator of amino acid biosynthetic genes in response to amino acid starvation; expression is tightly regulated at both the transcriptional and translational levels |
|
| YFL051C | 3.42 |
Putative protein of unknown function; YFL051C is not an essential gene |
||
| YHR217C | 3.21 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; located in the telomeric region TEL08R. |
||
| YKL217W | 3.18 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YIL162W | 3.11 |
SUC2
|
Invertase, sucrose hydrolyzing enzyme; a secreted, glycosylated form is regulated by glucose repression, and an intracellular, nonglycosylated enzyme is produced constitutively |
|
| YAR050W | 3.03 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YPR010C-A | 3.01 |
Putative protein of unknown function; conserved among Saccharomyces sensu stricto species |
||
| YAL054C | 3.01 |
ACS1
|
Acetyl-coA synthetase isoform which, along with Acs2p, is the nuclear source of acetyl-coA for histone acetlyation; expressed during growth on nonfermentable carbon sources and under aerobic conditions |
|
| YOR152C | 2.99 |
Putative protein of unknown function; has no similarity to any known protein; YOR152C is not an essential gene |
||
| YAR047C | 2.97 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YKL163W | 2.91 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YPL165C | 2.89 |
SET6
|
Protein of unknown function; deletion heterozygote is sensitive to compounds that target ergosterol biosynthesis, may be involved in compound availability |
|
| YGL187C | 2.86 |
COX4
|
Subunit IV of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; N-terminal 25 residues of precursor are cleaved during mitochondrial import; phosphorylated |
|
| YDR538W | 2.82 |
PAD1
|
Phenylacrylic acid decarboxylase, confers resistance to cinnamic acid, decarboxylates aromatic carboxylic acids to the corresponding vinyl derivatives; homolog of E. coli UbiX |
|
| YML089C | 2.81 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression induced by calcium shortage |
||
| YMR090W | 2.80 |
Putative protein of unknown function with similarity to DTDP-glucose 4,6-dehydratases; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YMR090W is not an essential gene |
||
| YGR070W | 2.78 |
ROM1
|
GDP/GTP exchange protein (GEP) for Rho1p; mutations are synthetically lethal with mutations in rom2, which also encodes a GEP |
|
| YEL009C-A | 2.77 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER065C | 2.71 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YDR059C | 2.63 |
UBC5
|
Ubiquitin-conjugating enzyme that mediates selective degradation of short-lived and abnormal proteins, central component of the cellular stress response; expression is heat inducible |
|
| YGR069W | 2.60 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDL210W | 2.56 |
UGA4
|
Permease that serves as a gamma-aminobutyrate (GABA) transport protein involved in the utilization of GABA as a nitrogen source; catalyzes the transport of putrescine and delta-aminolevulinic acid (ALA); localized to the vacuolar membrane |
|
| YBR200W-A | 2.48 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YDL169C | 2.48 |
UGX2
|
Protein of unknown function, transcript accumulates in response to any combination of stress conditions |
|
| YER014C-A | 2.48 |
BUD25
|
Protein involved in bud-site selection; diploid mutants display a random budding pattern instead of the wild-type bipolar pattern |
|
| YLR366W | 2.38 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR364C-A |
||
| YGR087C | 2.38 |
PDC6
|
Minor isoform of pyruvate decarboxylase, decarboxylates pyruvate to acetaldehyde, involved in amino acid catabolism; transcription is glucose- and ethanol-dependent, and is strongly induced during sulfur limitation |
|
| YER158C | 2.37 |
Protein of unknown function, has similarity to Afr1p; potentially phosphorylated by Cdc28p |
||
| YER103W | 2.35 |
SSA4
|
Heat shock protein that is highly induced upon stress; plays a role in SRP-dependent cotranslational protein-membrane targeting and translocation; member of the HSP70 family; cytoplasmic protein that concentrates in nuclei upon starvation |
|
| YJL045W | 2.33 |
Minor succinate dehydrogenase isozyme; homologous to Sdh1p, the major isozyme reponsible for the oxidation of succinate and transfer of electrons to ubiquinone; induced during the diauxic shift in a Cat8p-dependent manner |
||
| YFL063W | 2.30 |
Dubious open reading frame, based on available experimental and comparative sequence data |
||
| YGR032W | 2.28 |
GSC2
|
Catalytic subunit of 1,3-beta-glucan synthase, involved in formation of the inner layer of the spore wall; activity positively regulated by Rho1p and negatively by Smk1p; has similarity to an alternate catalytic subunit, Fks1p (Gsc1p) |
|
| YMR081C | 2.24 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YNR034W-A | 2.23 |
Putative protein of unknown function; expression is regulated by Msn2p/Msn4p |
||
| YHR071W | 2.21 |
PCL5
|
Cyclin, interacts with Pho85p cyclin-dependent kinase (Cdk), induced by Gcn4p at level of transcription, specifically required for Gcn4p degradation, may be sensor of cellular protein biosynthetic capacity |
|
| YDR540C | 2.21 |
IRC4
|
Putative protein of unknown function; null mutant displays increased levels of spontaneous Rad52p foci; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
|
| YBR076W | 2.19 |
ECM8
|
Non-essential protein of unknown function |
|
| YCR025C | 2.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YCR025C is not an essential gene |
||
| YLR047C | 2.14 |
FRE8
|
Protein with sequence similarity to iron/copper reductases, involved in iron homeostasis; deletion mutant has iron deficiency/accumulation growth defects; expression increased in the absence of copper-responsive transcription factor Mac1p |
|
| YDR119W-A | 2.13 |
Putative protein of unknown function |
||
| YER033C | 2.13 |
ZRG8
|
Protein of unknown function; authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; GFP-fusion protein is localized to the cytoplasm; transcription induced under conditions of zinc deficiency |
|
| YPR150W | 2.11 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene SUE1/YPR151C |
||
| YDR043C | 2.10 |
NRG1
|
Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters; mediates glucose repression and negatively regulates a variety of processes including filamentous growth and alkaline pH response |
|
| YJL130C | 2.06 |
URA2
|
Bifunctional carbamoylphosphate synthetase (CPSase)-aspartate transcarbamylase (ATCase), catalyzes the first two enzymatic steps in the de novo biosynthesis of pyrimidines; both activities are subject to feedback inhibition by UTP |
|
| YPL119C-A | 2.05 |
Putative protein of unknown function; identified by expression profiling and mass spectrometry |
||
| YMR014W | 2.03 |
BUD22
|
Protein involved in bud-site selection; diploid mutants display a random budding pattern instead of the wild-type bipolar pattern |
|
| YEL010W | 2.02 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR211C | 2.02 |
MGM1
|
Mitochondrial GTPase related to dynamin, present in a complex containing Ugo1p and Fzo1p; required for normal morphology of cristae and for stability of Tim11p; homolog of human OPA1 involved in autosomal dominant optic atrophy |
|
| YGR144W | 1.98 |
THI4
|
Thiazole synthase, catalyzes formation of the thiazole moiety of thiamin pyrophosphate; required for thiamine biosynthesis and for mitochondrial genome stability |
|
| YOR100C | 1.94 |
CRC1
|
Mitochondrial inner membrane carnitine transporter, required for carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria during fatty acid beta-oxidation |
|
| YBR077C | 1.93 |
SLM4
|
Component of the EGO complex, which is involved in the regulation of microautophagy, and of the GSE complex, which is required for proper sorting of amino acid permease Gap1p; gene exhibits synthetic genetic interaction with MSS4 |
|
| YGR045C | 1.92 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR023W | 1.90 |
MTL1
|
Protein with both structural and functional similarity to Mid2p, which is a plasma membrane sensor required for cell integrity signaling during pheromone-induced morphogenesis; suppresses rgd1 null mutations |
|
| YEL070W | 1.90 |
DSF1
|
Deletion suppressor of mpt5 mutation |
|
| YAL063C | 1.89 |
FLO9
|
Lectin-like protein with similarity to Flo1p, thought to be expressed and involved in flocculation |
|
| YIL155C | 1.87 |
GUT2
|
Mitochondrial glycerol-3-phosphate dehydrogenase; expression is repressed by both glucose and cAMP and derepressed by non-fermentable carbon sources in a Snf1p, Rsf1p, Hap2/3/4/5 complex dependent manner |
|
| YCR091W | 1.86 |
KIN82
|
Putative serine/threonine protein kinase, most similar to cyclic nucleotide-dependent protein kinase subfamily and the protein kinase C subfamily |
|
| YBR182C | 1.85 |
SMP1
|
Putative transcription factor involved in regulating the response to osmotic stress; member of the MADS-box family of transcription factors |
|
| YGL146C | 1.81 |
Putative protein of unknown function, contains two putative transmembrane spans, shows no significant homology to any other known protein sequence, YGL146C is not an essential gene |
||
| YHR138C | 1.79 |
Putative protein of unknown function; has similarity to Pbi2p; double null mutant lacking Pbi2p and Yhr138p exhibits highly fragmented vacuoles |
||
| YHR218W | 1.74 |
Helicase-like protein encoded within the telomeric Y' element |
||
| YLL053C | 1.74 |
Putative protein; in the Sigma 1278B strain background YLL053C is contiguous with AQY2 which encodes an aquaporin |
||
| YJR150C | 1.71 |
DAN1
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YLR004C | 1.70 |
THI73
|
Putative plasma membrane permease proposed to be involved in carboxylic acid uptake and repressed by thiamine; substrate of Dbf2p/Mob1p kinase; transcription is altered if mitochondrial dysfunction occurs |
|
| YML090W | 1.69 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YML089C; exhibits growth defect on a non-fermentable (respiratory) carbon source |
||
| YDR536W | 1.65 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YGR022C | 1.64 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF MTL1/YGR023W |
||
| YHR211W | 1.64 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YPR027C | 1.63 |
Putative protein of unknown function |
||
| YFL011W | 1.61 |
HXT10
|
Putative hexose transporter, expressed at low levels and expression is repressed by glucose |
|
| YCR040W | 1.60 |
MATALPHA1
|
Transcriptional co-activator involved in regulation of mating-type-specific gene expression; targets the transcription factor Mcm1p to the promoters of alpha-specific genes; one of two genes encoded by the MATalpha mating type cassette |
|
| YOL082W | 1.59 |
ATG19
|
Receptor protein for the cytoplasm-to-vacuole targeting (Cvt) pathway; recognizes cargo proteins aminopeptidase I (Lap4p) and alpha-mannosidase (Ams1p) and delivers them to the preautophagosomal structure for packaging into Cvt vesicles |
|
| YJL116C | 1.59 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YEL007W | 1.58 |
Putative protein with sequence similarity to S. pombe gti1+ (gluconate transport inducer 1) |
||
| YMR104C | 1.58 |
YPK2
|
Protein kinase with similarityto serine/threonine protein kinase Ypk1p; functionally redundant with YPK1 at the genetic level; participates in a signaling pathway required for optimal cell wall integrity; homolog of mammalian kinase SGK |
|
| YGL081W | 1.55 |
Putative protein of unknown function; non-essential gene; interacts genetically with CHS5, a gene involved in chitin biosynthesis |
||
| YKL004W | 1.55 |
AUR1
|
Phosphatidylinositol:ceramide phosphoinositol transferase (IPC synthase), required for sphingolipid synthesis; can mutate to confer aureobasidin A resistance |
|
| YLL052C | 1.54 |
AQY2
|
Water channel that mediates the transport of water across cell membranes, only expressed in proliferating cells, controlled by osmotic signals, may be involved in freeze tolerance; disrupted by a stop codon in many S. cerevisiae strains |
|
| YDL194W | 1.53 |
SNF3
|
Plasma membrane glucose sensor that regulates glucose transport; has 12 predicted transmembrane segments; long cytoplasmic C-terminal tail is required for low glucose induction of hexose transporter genes HXT2 and HXT4 |
|
| YNL333W | 1.52 |
SNZ2
|
Member of a stationary phase-induced gene family; transcription of SNZ2 is induced prior to diauxic shift, and also in the absence of thiamin in a Thi2p-dependent manner; forms a coregulated gene pair with SNO2; interacts with Thi11p |
|
| YMR206W | 1.52 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YPR015C | 1.49 |
Putative protein of unknown function |
||
| YOL154W | 1.45 |
ZPS1
|
Putative GPI-anchored protein; transcription is induced under low-zinc conditions, as mediated by the Zap1p transcription factor, and at alkaline pH |
|
| YBR212W | 1.44 |
NGR1
|
RNA binding protein that negatively regulates growth rate; interacts with the 3' UTR of the mitochondrial porin (POR1) mRNA and enhances its degradation; overexpression impairs mitochondrial function; expressed in stationary phase |
|
| YGR088W | 1.42 |
CTT1
|
Cytosolic catalase T, has a role in protection from oxidative damage by hydrogen peroxide |
|
| YFL064C | 1.42 |
Putative protein of unknown function |
||
| YOR235W | 1.41 |
IRC13
|
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YNL334C | 1.41 |
SNO2
|
Protein of unknown function, nearly identical to Sno3p; expression is induced before the diauxic shift and also in the absence of thiamin |
|
| YPR202W | 1.41 |
Putative protein of unknown function with similarity to telomere-encoded helicases; YPR202W is not an essential gene; transcript is predicted to be spliced but there is no evidence that it is spliced in vivo |
||
| YLR365W | 1.40 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps dubious gene YLR364C-A; YLR365W is not an essential gene |
||
| YAR027W | 1.38 |
UIP3
|
Putative integral membrane protein of unknown function; interacts with Ulp1p at the nuclear periphery; member of DUP240 gene family |
|
| YHR009C | 1.37 |
Putative protein of unknown function; not an essential gene |
||
| YJL161W | 1.36 |
FMP33
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YML091C | 1.36 |
RPM2
|
Protein subunit of mitochondrial RNase P, has roles in nuclear transcription, cytoplasmic and mitochondrial RNA processing, and mitochondrial translation; distributed to mitochondria, cytoplasmic processing bodies, and the nucleus |
|
| YLR377C | 1.35 |
FBP1
|
Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway, required for glucose metabolism |
|
| YOL060C | 1.32 |
MAM3
|
Protein required for normal mitochondrial morphology, has similarity to hemolysins |
|
| YAL039C | 1.30 |
CYC3
|
Cytochrome c heme lyase (holocytochrome c synthase), attaches heme to apo-cytochrome c (Cyc1p or Cyc7p) in the mitochondrial intermembrane space; human ortholog may have a role in microphthalmia with linear skin defects (MLS) |
|
| YCR007C | 1.30 |
Putative integral membrane protein, member of DUP240 gene family; YCR007C is not an essential gene |
||
| YPL216W | 1.29 |
Putative protein of unknown function; YPL216W is not an essential gene |
||
| YKL044W | 1.28 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPR196W | 1.28 |
Putative maltose activator |
||
| YPR078C | 1.27 |
Putative protein of unknown function; possible role in DNA metabolism and/or in genome stability; expression is heat-inducible |
||
| YPL089C | 1.27 |
RLM1
|
MADS-box transcription factor, component of the protein kinase C-mediated MAP kinase pathway involved in the maintenance of cell integrity; phosphorylated and activated by the MAP-kinase Slt2p |
|
| YDR461C-A | 1.26 |
Putative protein of unknown function |
||
| YOR011W-A | 1.26 |
Putative protein of unknown function |
||
| YDR014W-A | 1.25 |
HED1
|
Meiosis-specific protein that down-regulates Rad51p-mediated mitotic recombination when the meiotic recombination machinery is impaired; early meiotic gene, transcribed specifically during meiotic prophase |
|
| YER150W | 1.24 |
SPI1
|
GPI-anchored cell wall protein involved in weak acid resistance; basal expression requires Msn2p/Msn4p; expression is induced under conditions of stress and during the diauxic shift; similar to Sed1p |
|
| YLL030C | 1.21 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YKL005C | 1.20 |
BYE1
|
Negative regulator of transcription elongation, contains a TFIIS-like domain and a PHD finger, multicopy suppressor of temperature-sensitive ess1 mutations, probably binds RNA polymerase II large subunit |
|
| YCL038C | 1.20 |
ATG22
|
Vacuolar integral membrane protein required for efflux of amino acids during autophagic body breakdown in the vacuole; null mutation causes a gradual loss of viability during starvation |
|
| YEL049W | 1.20 |
PAU2
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YFL062W | 1.19 |
COS4
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YHR092C | 1.18 |
HXT4
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YDR313C | 1.18 |
PIB1
|
RING-type ubiquitin ligase of the endosomal and vacuolar membranes, binds phosphatidylinositol(3)-phosphate; contains a FYVE finger domain |
|
| YMR137C | 1.17 |
PSO2
|
Required for a post-incision step in the repair of DNA single and double-strand breaks that result from interstrand crosslinks produced by a variety of mono- and bi-functional psoralen derivatives; induced by UV-irradiation |
|
| YER024W | 1.16 |
YAT2
|
Carnitine acetyltransferase; has similarity to Yat1p, which is a carnitine acetyltransferase associated with the mitochondrial outer membrane |
|
| YDL244W | 1.15 |
THI13
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YGR065C | 1.15 |
VHT1
|
High-affinity plasma membrane H+-biotin (vitamin H) symporter; mutation results in fatty acid auxotrophy; 12 transmembrane domain containing major facilitator subfamily member; mRNA levels negatively regulated by iron deprivation and biotin |
|
| YCL065W | 1.14 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps HMLALPHA1 |
||
| YDR542W | 1.13 |
PAU10
|
Hypothetical protein |
|
| YNL202W | 1.12 |
SPS19
|
Peroxisomal 2,4-dienoyl-CoA reductase, auxiliary enzyme of fatty acid beta-oxidation; homodimeric enzyme required for growth and sporulation on petroselineate medium; expression induced during late sporulation and in the presence of oleate |
|
| YCL054W | 1.12 |
SPB1
|
AdoMet-dependent methyltransferase involved in rRNA processing and 60S ribosomal subunit maturation; methylates G2922 in the tRNA docking site of the large subunit rRNA and in the absence of snR52, U2921; suppressor of PAB1 mutants |
|
| YDL180W | 1.12 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YFL012W | 1.12 |
Putative protein of unknown function; transcribed during sporulation; null mutant exhibits increased resistance to rapamycin |
||
| YGR041W | 1.11 |
BUD9
|
Protein involved in bud-site selection; diploid mutants display a unipolar budding pattern instead of the wild-type bipolar pattern, and bud at the distal pole |
|
| YBR292C | 1.11 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR292C is not an essential gene |
||
| YNL144C | 1.10 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YNL144C is not an essential gene |
||
| YNL326C | 1.10 |
PFA3
|
Palmitoyltransferase for Vac8p, required for vacuolar membrane fusion; contains an Asp-His-His-Cys-cysteine rich (DHHC-CRD) domain; autoacylates; required for vacuolar integrity under stress conditions |
|
| YIL059C | 1.09 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YIL060W |
||
| YPL088W | 1.09 |
Putative aryl alcohol dehydrogenase; transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in multidrug resistance |
||
| YCR039C | 1.06 |
MATALPHA2
|
Homeobox-domain protein that, with Mcm1p, represses a-specific genes in haploids; acts with A1p to repress transcription of haploid-specific genes in diploids; one of two genes encoded by the MATalpha mating type cassette |
|
| YDR223W | 1.05 |
CRF1
|
Transcriptional corepressor involved in the regulation of ribosomal protein gene transcription via the TOR signaling pathway and protein kinase A, phosphorylated by activated Yak1p which promotes accumulation of Crf1p in the nucleus |
|
| YGR066C | 1.05 |
Putative protein of unknown function |
||
| YHR002W | 1.02 |
LEU5
|
Mitochondrial carrier protein involved in the accumulation of CoA in the mitochondrial matrix; homolog of human Graves disease protein; does not encode an isozyme of Leu4p, as first hypothesized |
|
| YER163C | 1.02 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YEL030C-A | 1.02 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YBR296C | 1.00 |
PHO89
|
Na+/Pi cotransporter, active in early growth phase; similar to phosphate transporters of Neurospora crassa; transcription regulated by inorganic phosphate concentrations and Pho4p |
|
| YGR039W | 1.00 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the Autonomously Replicating Sequence ARS722 |
||
| YLR189C | 0.98 |
ATG26
|
UDP-glucose:sterol glucosyltransferase, conserved enzyme involved in synthesis of sterol glucoside membrane lipids; in contrast to ATG26 from P. pastoris, S. cerevisiae ATG26 is not involved in autophagy |
|
| YOR289W | 0.96 |
Putative protein of unknown function; transcription induced by the unfolded protein response; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and the nucleus |
||
| YER167W | 0.96 |
BCK2
|
Protein rich in serine and threonine residues involved in protein kinase C signaling pathway, which controls cell integrity; overproduction suppresses pkc1 mutations |
|
| YHR048W | 0.96 |
YHK8
|
Presumed antiporter of the DHA1 family of multidrug resistance transporters; contains 12 predicted transmembrane spans; expression of gene is up-regulated in cells exhibiting reduced susceptibility to azoles |
|
| YEL072W | 0.95 |
RMD6
|
Protein required for sporulation |
|
| YLR023C | 0.95 |
IZH3
|
Membrane protein involved in zinc metabolism, member of the four-protein IZH family, expression induced by zinc deficiency; deletion reduces sensitivity to elevated zinc and shortens lag phase, overexpression reduces Zap1p activity |
|
| YOR348C | 0.94 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YLL029W | 0.94 |
FRA1
|
Protein involved in negative regulation of transcription of iron regulon; forms an iron independent complex with Fra2p, Grx3p, and Grx4p; cytosolic; mutant fails to repress transcription of iron regulon and is defective in spore formation |
|
| YIL134C-A | 0.94 |
Putative protein of unknown function, identified by fungal homology and RT-PCR |
||
| YDR387C | 0.94 |
Putative transporter, member of the sugar porter family; YDR387C is not an essential gene |
||
| YLR327C | 0.93 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YKL109W | 0.93 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YJL037W | 0.92 |
IRC18
|
Putative protein of unknown function; expression induced in respiratory-deficient cells and in carbon-limited chemostat cultures; similar to adjacent ORF, YJL038C; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YOR156C | 0.92 |
NFI1
|
SUMO ligase, catalyzes the covalent attachment of SUMO (Smt3p) to proteins; involved in maintenance of proper telomere length |
|
| YGL255W | 0.92 |
ZRT1
|
High-affinity zinc transporter of the plasma membrane, responsible for the majority of zinc uptake; transcription is induced under low-zinc conditions by the Zap1p transcription factor |
|
| YNL005C | 0.91 |
MRP7
|
Mitochondrial ribosomal protein of the large subunit |
|
| YER187W | 0.91 |
Putative protein of unknown function; induced in respiratory-deficient cells |
||
| YLR356W | 0.91 |
Putative protein of unknown function with similarity to SCM4; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR356W is not an essential gene |
||
| YER101C | 0.90 |
AST2
|
Protein that may have a role in targeting of plasma membrane [H+]ATPase (Pma1p) to the plasma membrane, as suggested by analysis of genetic interactions |
|
| YOR339C | 0.90 |
UBC11
|
Ubiquitin-conjugating enzyme most similar in sequence to Xenopus ubiquitin-conjugating enzyme E2-C, but not a true functional homolog of this E2; unlike E2-C, not required for the degradation of mitotic cyclin Clb2 |
|
| YBR045C | 0.90 |
GIP1
|
Meiosis-specific regulatory subunit of the Glc7p protein phosphatase, regulates spore wall formation and septin organization, required for expression of some late meiotic genes and for normal localization of Glc7p |
|
| YHR096C | 0.90 |
HXT5
|
Hexose transporter with moderate affinity for glucose, induced in the presence of non-fermentable carbon sources, induced by a decrease in growth rate, contains an extended N-terminal domain relative to other HXTs |
|
| YJR120W | 0.90 |
Protein of unknown function; essential for growth under anaerobic conditions; mutation causes decreased expression of ATP2, impaired respiration, defective sterol uptake, and altered levels/localization of ABC transporters Aus1p and Pdr11p |
||
| YLR331C | 0.89 |
JIP3
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 98% of ORF overlaps the verified gene MID2 |
|
| YOL159C | 0.89 |
Soluble protein of unknown function; deletion mutants are viable and have elevated levels of Ty1 retrotransposition and Ty1 cDNA |
||
| YER066W | 0.89 |
Putative protein of unknown function; YER066W is not an essential gene |
||
| YDR528W | 0.88 |
HLR1
|
Protein involved in regulation of cell wall composition and integrity and response to osmotic stress; overproduction suppresses a lysis sensitive PKC mutation; similar to Lre1p, which functions antagonistically to protein kinase A |
|
| YGL219C | 0.88 |
MDM34
|
Mitochondrial outer membrane protein, required for normal mitochondrial morphology and inheritance; localizes to dots on the mitochondrial surface near mtDNA nucleoids; interacts genetically with MDM31 and MDM32 |
|
| YNL142W | 0.88 |
MEP2
|
Ammonium permease involved in regulation of pseudohyphal growth; belongs to a ubiquitous family of cytoplasmic membrane proteins that transport only ammonium (NH4+); expression is under the nitrogen catabolite repression regulation |
|
| YHR210C | 0.88 |
Putative protein of unknown function; non-essential gene; highly expressed under anaeorbic conditions; sequence similarity to aldose 1-epimerases such as GAL10 |
||
| YOL118C | 0.88 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YOL075C | 0.87 |
Putative ABC transporter |
||
| YHL041W | 0.87 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YCL066W | 0.87 |
HMLALPHA1
|
Silenced copy of ALPHA1 at HML, encoding a transcriptional coactivator involved in the regulation of mating-type alpha-specific gene expression |
|
| YOR292C | 0.87 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YOR292C is not an essential gene |
||
| YCR073W-A | 0.86 |
SOL2
|
Protein with a possible role in tRNA export; shows similarity to 6-phosphogluconolactonase non-catalytic domains but does not exhibit this enzymatic activity; homologous to Sol1p, Sol3p, and Sol4p |
|
| YGR121C | 0.86 |
MEP1
|
Ammonium permease; belongs to a ubiquitous family of cytoplasmic membrane proteins that transport only ammonium (NH4+); expression is under the nitrogen catabolite repression regulation |
|
| YER004W | 0.86 |
FMP52
|
Protein of unknown function, localized to the mitochondrial outer membrane; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YLR332W | 0.85 |
MID2
|
O-glycosylated plasma membrane protein that acts as a sensor for cell wall integrity signaling and activates the pathway; interacts with Rom2p, a guanine nucleotide exchange factor for Rho1p, and with cell integrity pathway protein Zeo1p |
|
| YHL024W | 0.85 |
RIM4
|
Putative RNA-binding protein required for the expression of early and middle sporulation genes |
Network of associatons between targets according to the STRING database.
First level regulatory network of RLM1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0015755 | fructose transport(GO:0015755) |
| 1.7 | 10.4 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 1.6 | 4.9 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 1.5 | 4.6 | GO:0042843 | D-xylose catabolic process(GO:0042843) |
| 1.5 | 5.9 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 1.1 | 4.2 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.9 | 3.8 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.9 | 2.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.8 | 2.5 | GO:0045895 | positive regulation of mating-type specific transcription, DNA-templated(GO:0045895) |
| 0.8 | 2.3 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.8 | 3.1 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.6 | 0.6 | GO:0071361 | positive regulation of transcription from RNA polymerase II promoter in response to ethanol(GO:0061410) cellular response to ethanol(GO:0071361) cellular response to alcohol(GO:0097306) |
| 0.6 | 2.4 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.6 | 1.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.6 | 1.7 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.6 | 2.3 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.6 | 3.9 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.6 | 1.7 | GO:0045894 | regulation of mating-type specific transcription from RNA polymerase II promoter(GO:0001196) negative regulation of mating-type specific transcription from RNA polymerase II promoter(GO:0001198) negative regulation of mating-type specific transcription, DNA-templated(GO:0045894) |
| 0.5 | 2.6 | GO:0019405 | alditol catabolic process(GO:0019405) glycerol catabolic process(GO:0019563) |
| 0.5 | 4.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.5 | 1.9 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.5 | 1.4 | GO:0042744 | hydrogen peroxide metabolic process(GO:0042743) hydrogen peroxide catabolic process(GO:0042744) |
| 0.5 | 2.8 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.5 | 4.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 1.4 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.5 | 8.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.4 | 3.6 | GO:0005987 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.4 | 2.2 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.4 | 2.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.4 | 2.1 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.4 | 1.6 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.4 | 1.2 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.4 | 1.1 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.4 | 7.6 | GO:0015749 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.4 | 0.7 | GO:0043419 | urea catabolic process(GO:0043419) |
| 0.3 | 1.4 | GO:0046352 | disaccharide catabolic process(GO:0046352) |
| 0.3 | 3.0 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.3 | 7.5 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.3 | 1.3 | GO:0016577 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 0.3 | 1.6 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.3 | 0.9 | GO:0034395 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.3 | 0.9 | GO:0015691 | cadmium ion transport(GO:0015691) |
| 0.3 | 1.8 | GO:0061393 | positive regulation of transcription from RNA polymerase II promoter in response to osmotic stress(GO:0061393) |
| 0.3 | 0.6 | GO:2000219 | positive regulation of invasive growth in response to glucose limitation by positive regulation of transcription from RNA polymerase II promoter(GO:0036095) positive regulation of invasive growth in response to glucose limitation(GO:2000219) |
| 0.3 | 1.7 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.2 | 1.2 | GO:0032048 | cardiolipin metabolic process(GO:0032048) |
| 0.2 | 0.5 | GO:0000304 | response to singlet oxygen(GO:0000304) |
| 0.2 | 0.5 | GO:0009415 | response to water(GO:0009415) cellular response to water stimulus(GO:0071462) |
| 0.2 | 1.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 1.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.2 | 1.2 | GO:0034487 | amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 0.2 | 1.0 | GO:0006171 | cAMP biosynthetic process(GO:0006171) cyclic nucleotide biosynthetic process(GO:0009190) cyclic purine nucleotide metabolic process(GO:0052652) |
| 0.2 | 1.0 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.2 | 0.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 5.5 | GO:0006915 | apoptotic process(GO:0006915) cell death(GO:0008219) programmed cell death(GO:0012501) |
| 0.2 | 0.7 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.2 | 0.9 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.2 | 0.9 | GO:0032373 | positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) positive regulation of sterol transport(GO:0032373) positive regulation of transmembrane transport(GO:0034764) regulation of sterol import by regulation of transcription from RNA polymerase II promoter(GO:0035968) positive regulation of sterol import by positive regulation of transcription from RNA polymerase II promoter(GO:0035969) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.2 | 4.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 0.9 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.2 | 0.4 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 1.3 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) manganese ion homeostasis(GO:0055071) |
| 0.2 | 2.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 1.0 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.2 | 0.2 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) |
| 0.2 | 0.6 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.2 | 1.7 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 1.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.2 | 0.4 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.2 | 3.8 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.2 | 0.7 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.2 | 1.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 0.7 | GO:0070054 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) IRE1-mediated unfolded protein response(GO:0036498) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) |
| 0.2 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.7 | GO:0051346 | negative regulation of hydrolase activity(GO:0051346) |
| 0.2 | 1.0 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.2 | 1.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.2 | 0.7 | GO:2000284 | positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.2 | 0.7 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.2 | 0.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.2 | 2.5 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.2 | 1.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.2 | 0.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.2 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 0.9 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.9 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.7 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.1 | 1.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.4 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.1 | 0.4 | GO:0045117 | azole transport(GO:0045117) |
| 0.1 | 1.2 | GO:0043487 | regulation of RNA stability(GO:0043487) regulation of mRNA stability(GO:0043488) |
| 0.1 | 0.7 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.1 | 0.7 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.7 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.1 | 0.1 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) regulation of maintenance of sister chromatid cohesion(GO:0034091) |
| 0.1 | 0.4 | GO:1904951 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 0.1 | 1.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.5 | GO:0046464 | triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.1 | 0.5 | GO:0031135 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.1 | 1.7 | GO:0001101 | response to acid chemical(GO:0001101) |
| 0.1 | 0.7 | GO:0010388 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.1 | 0.8 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.4 | GO:0046021 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) |
| 0.1 | 0.2 | GO:0001178 | regulation of transcriptional start site selection at RNA polymerase II promoter(GO:0001178) |
| 0.1 | 1.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 1.7 | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress(GO:0036003) |
| 0.1 | 2.5 | GO:0097034 | mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.5 | GO:0051126 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.1 | 0.3 | GO:0071850 | mitotic cell cycle arrest in response to pheromone(GO:0000751) mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 3.4 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.1 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.5 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.1 | GO:0045991 | carbon catabolite activation of transcription(GO:0045991) |
| 0.1 | 0.3 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.1 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.6 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.2 | GO:0000272 | polysaccharide catabolic process(GO:0000272) |
| 0.1 | 0.1 | GO:0019627 | urea metabolic process(GO:0019627) |
| 0.1 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.3 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.1 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 1.9 | GO:0010324 | membrane invagination(GO:0010324) lysosomal microautophagy(GO:0016237) single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.5 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.4 | GO:0001080 | nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) nitrogen catabolite activation of transcription(GO:0090294) |
| 0.1 | 1.0 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 | 0.8 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.2 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.1 | 0.9 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.1 | 0.5 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.1 | 0.3 | GO:0015856 | cytosine transport(GO:0015856) purine-containing compound transmembrane transport(GO:0072530) |
| 0.1 | 0.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.3 | GO:0006641 | neutral lipid metabolic process(GO:0006638) acylglycerol metabolic process(GO:0006639) triglyceride metabolic process(GO:0006641) |
| 0.1 | 0.8 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.1 | 1.3 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 0.1 | 0.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.5 | GO:0043461 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) proton-transporting ATP synthase complex assembly(GO:0043461) |
| 0.1 | 0.6 | GO:0000114 | obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.1 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.2 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.1 | 0.1 | GO:0070726 | cell wall assembly(GO:0070726) |
| 0.1 | 0.3 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.1 | 0.4 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.1 | 0.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.4 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 0.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.3 | GO:0006078 | (1->6)-beta-D-glucan biosynthetic process(GO:0006078) |
| 0.1 | 0.3 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.1 | 0.2 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 1.4 | GO:0032258 | CVT pathway(GO:0032258) |
| 0.1 | 0.3 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.2 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.1 | 0.4 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 0.8 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.1 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of metal ion(GO:0051238) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.3 | GO:0034982 | protein processing involved in protein targeting to mitochondrion(GO:0006627) mitochondrial protein processing(GO:0034982) |
| 0.1 | 0.4 | GO:0010529 | negative regulation of transposition, RNA-mediated(GO:0010526) negative regulation of transposition(GO:0010529) |
| 0.1 | 0.3 | GO:0046033 | AMP biosynthetic process(GO:0006167) AMP metabolic process(GO:0046033) |
| 0.1 | 0.5 | GO:0031578 | mitotic spindle orientation checkpoint(GO:0031578) |
| 0.1 | 0.6 | GO:0019740 | nitrogen utilization(GO:0019740) |
| 0.1 | 0.2 | GO:0042762 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of sulfur metabolic process(GO:0042762) |
| 0.1 | 0.1 | GO:0006077 | (1->6)-beta-D-glucan metabolic process(GO:0006077) |
| 0.1 | 0.4 | GO:0055074 | cellular calcium ion homeostasis(GO:0006874) calcium ion homeostasis(GO:0055074) |
| 0.1 | 2.3 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) allantoin catabolic process(GO:0000256) cellular amide catabolic process(GO:0043605) |
| 0.1 | 0.2 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) negative regulation of developmental process(GO:0051093) |
| 0.1 | 0.1 | GO:0016320 | endoplasmic reticulum membrane fusion(GO:0016320) |
| 0.1 | 0.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.1 | 0.2 | GO:0034755 | high-affinity iron ion transmembrane transport(GO:0006827) iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.0 | GO:0000416 | positive regulation of histone H3-K36 methylation(GO:0000416) histone H3-K36 trimethylation(GO:0097198) regulation of histone H3-K36 trimethylation(GO:2001253) positive regulation of histone H3-K36 trimethylation(GO:2001255) |
| 0.0 | 0.0 | GO:0009730 | detection of chemical stimulus(GO:0009593) detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) detection of stimulus(GO:0051606) |
| 0.0 | 0.4 | GO:0000755 | cytogamy(GO:0000755) |
| 0.0 | 0.8 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.2 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0051382 | centromere complex assembly(GO:0034508) kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.0 | 0.1 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.0 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.0 | 0.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.7 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.2 | GO:0060237 | regulation of fungal-type cell wall organization(GO:0060237) |
| 0.0 | 0.3 | GO:0010038 | response to metal ion(GO:0010038) |
| 0.0 | 0.3 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0000388 | spliceosome conformational change to release U4 (or U4atac) and U1 (or U11)(GO:0000388) |
| 0.0 | 0.3 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.0 | GO:0031111 | negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.1 | GO:2000002 | regulation of cell cycle checkpoint(GO:1901976) negative regulation of cell cycle checkpoint(GO:1901977) regulation of DNA damage checkpoint(GO:2000001) negative regulation of DNA damage checkpoint(GO:2000002) negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.0 | 0.1 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) regulation of membrane lipid metabolic process(GO:1905038) |
| 0.0 | 0.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.6 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.0 | 0.3 | GO:0040001 | establishment of mitotic spindle orientation(GO:0000132) establishment of mitotic spindle localization(GO:0040001) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.2 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.6 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.1 | GO:0072417 | cellular response to biotic stimulus(GO:0071216) response to cell cycle checkpoint signaling(GO:0072396) response to mitotic cell cycle checkpoint signaling(GO:0072414) response to spindle checkpoint signaling(GO:0072417) response to mitotic spindle checkpoint signaling(GO:0072476) response to mitotic cell cycle spindle assembly checkpoint signaling(GO:0072479) response to spindle assembly checkpoint signaling(GO:0072485) negative regulation of protein import into nucleus during spindle assembly checkpoint(GO:1901925) |
| 0.0 | 0.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.0 | 0.6 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.0 | 0.2 | GO:0046020 | negative regulation of transcription by pheromones(GO:0045996) negative regulation of transcription from RNA polymerase II promoter by pheromones(GO:0046020) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0035753 | maintenance of DNA trinucleotide repeats(GO:0035753) |
| 0.0 | 0.0 | GO:0050685 | positive regulation of mRNA processing(GO:0050685) |
| 0.0 | 0.0 | GO:0015851 | nucleobase transport(GO:0015851) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.2 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) |
| 0.0 | 0.1 | GO:0000161 | MAPK cascade involved in osmosensory signaling pathway(GO:0000161) |
| 0.0 | 0.0 | GO:0044275 | cellular carbohydrate catabolic process(GO:0044275) |
| 0.0 | 0.0 | GO:0030308 | negative regulation of cell growth(GO:0030308) negative regulation of pseudohyphal growth(GO:2000221) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.0 | GO:0043270 | positive regulation of ion transport(GO:0043270) |
| 0.0 | 0.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.0 | GO:0071454 | response to anoxia(GO:0034059) cellular response to anoxia(GO:0071454) |
| 0.0 | 0.5 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.0 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.0 | 0.1 | GO:0000753 | cell morphogenesis involved in conjugation with cellular fusion(GO:0000753) |
| 0.0 | 0.5 | GO:0000282 | cellular bud site selection(GO:0000282) mitotic cytokinesis, site selection(GO:1902408) |
| 0.0 | 0.0 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.0 | GO:0031494 | regulation of cell fate commitment(GO:0010453) positive regulation of cell fate commitment(GO:0010455) regulation of mating type switching(GO:0031494) positive regulation of mating type switching(GO:0031496) positive regulation of cell differentiation(GO:0045597) positive regulation of developmental process(GO:0051094) |
| 0.0 | 0.1 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 1.1 | GO:0030435 | sporulation resulting in formation of a cellular spore(GO:0030435) |
| 0.0 | 0.1 | GO:0010647 | positive regulation of cell communication(GO:0010647) |
| 0.0 | 0.1 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 0.1 | GO:0090522 | vesicle tethering involved in exocytosis(GO:0090522) |
| 0.0 | 0.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0009353 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial oxoglutarate dehydrogenase complex(GO:0009353) dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.8 | 2.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.6 | 2.4 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.6 | 1.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.4 | 1.3 | GO:0005756 | mitochondrial proton-transporting ATP synthase, central stalk(GO:0005756) proton-transporting ATP synthase, central stalk(GO:0045269) |
| 0.4 | 1.2 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.3 | 3.1 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.3 | 1.7 | GO:0034448 | EGO complex(GO:0034448) |
| 0.3 | 1.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 0.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 0.8 | GO:0033309 | SBF transcription complex(GO:0033309) |
| 0.2 | 0.9 | GO:0035361 | Cul8-RING ubiquitin ligase complex(GO:0035361) |
| 0.2 | 0.9 | GO:0032865 | ERMES complex(GO:0032865) ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 5.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 0.4 | GO:0045265 | mitochondrial proton-transporting ATP synthase, stator stalk(GO:0000274) proton-transporting ATP synthase, stator stalk(GO:0045265) |
| 0.1 | 0.3 | GO:0097002 | mitochondrial inner boundary membrane(GO:0097002) |
| 0.1 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.7 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.6 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.1 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.7 | GO:0072379 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 0.1 | 1.3 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.1 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 2.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.5 | GO:0016587 | Isw1 complex(GO:0016587) |
| 0.1 | 0.7 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.1 | 0.4 | GO:0033551 | monopolin complex(GO:0033551) |
| 0.1 | 0.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 2.4 | GO:0031226 | integral component of plasma membrane(GO:0005887) intrinsic component of plasma membrane(GO:0031226) |
| 0.1 | 0.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.6 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) core TFIIH complex(GO:0000439) |
| 0.1 | 4.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.1 | 0.3 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.3 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.1 | 0.5 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.9 | GO:0005628 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.1 | 0.6 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.1 | 0.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 3.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 1.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.3 | GO:0001400 | mating projection base(GO:0001400) |
| 0.1 | 2.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.9 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 1.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0070211 | Snt2C complex(GO:0070211) |
| 0.0 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0070274 | RES complex(GO:0070274) |
| 0.0 | 0.3 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.1 | GO:0070823 | HDA1 complex(GO:0070823) |
| 0.0 | 5.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 1.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.3 | GO:0098852 | fungal-type vacuole membrane(GO:0000329) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0033698 | Rpd3L complex(GO:0033698) |
| 0.0 | 18.7 | GO:0031224 | intrinsic component of membrane(GO:0031224) |
| 0.0 | 0.1 | GO:0000818 | MIS12/MIND type complex(GO:0000444) nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.2 | GO:0005619 | ascospore wall(GO:0005619) spore wall(GO:0031160) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0030139 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle(GO:0030139) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0044455 | mitochondrial membrane part(GO:0044455) |
| 0.0 | 0.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0032806 | carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 0.1 | GO:0032126 | eisosome(GO:0032126) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.4 | GO:0005537 | mannose binding(GO:0005537) |
| 1.5 | 7.7 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 1.3 | 5.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 1.3 | 5.2 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 1.2 | 4.7 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 1.2 | 4.6 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 1.1 | 3.4 | GO:0004575 | beta-fructofuranosidase activity(GO:0004564) sucrose alpha-glucosidase activity(GO:0004575) |
| 1.0 | 4.2 | GO:0004396 | hexokinase activity(GO:0004396) |
| 1.0 | 3.0 | GO:0016880 | AMP binding(GO:0016208) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.8 | 2.4 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.8 | 6.9 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.7 | 2.1 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.7 | 2.0 | GO:0005536 | glucose binding(GO:0005536) |
| 0.6 | 2.5 | GO:0000772 | mating pheromone activity(GO:0000772) pheromone activity(GO:0005186) |
| 0.6 | 1.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.6 | 2.3 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 0.5 | 2.2 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 0.5 | 3.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.5 | 3.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.4 | 0.4 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.4 | 4.4 | GO:0015578 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.4 | 7.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.4 | 1.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.4 | 1.8 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.4 | 1.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.4 | 1.4 | GO:0022821 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.3 | 3.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 4.1 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.3 | 1.7 | GO:0019903 | phosphatase binding(GO:0019902) protein phosphatase binding(GO:0019903) |
| 0.3 | 1.0 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.3 | 7.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.3 | 1.6 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.3 | 2.6 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.3 | 1.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.3 | 1.5 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.3 | 0.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 3.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.3 | 1.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.3 | 2.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 0.8 | GO:0008972 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.3 | 0.8 | GO:0008137 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 1.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.2 | 1.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 0.9 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 2.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 1.0 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.2 | 0.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 1.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.2 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.7 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 0.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) flap endonuclease activity(GO:0048256) |
| 0.2 | 0.9 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.2 | 0.5 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.2 | 0.6 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.2 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 0.7 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.6 | GO:0060089 | receptor activity(GO:0004872) transmembrane signaling receptor activity(GO:0004888) signaling receptor activity(GO:0038023) molecular transducer activity(GO:0060089) transmembrane receptor activity(GO:0099600) |
| 0.1 | 0.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 1.1 | GO:0034979 | NAD-dependent histone deacetylase activity(GO:0017136) NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.1 | 2.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.4 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 1.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.5 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 1.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.0 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.3 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.1 | 0.6 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 1.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.2 | GO:0008106 | aldo-keto reductase (NADP) activity(GO:0004033) alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 0.3 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.6 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 1.2 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 0.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.6 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.1 | 0.7 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 2.5 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.1 | 0.1 | GO:0015151 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.1 | 0.8 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 3.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.3 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.1 | 0.5 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 0.3 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 0.1 | 1.0 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.1 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.2 | GO:0004601 | peroxidase activity(GO:0004601) oxidoreductase activity, acting on peroxide as acceptor(GO:0016684) |
| 0.1 | 1.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.2 | GO:0015288 | anion channel activity(GO:0005253) voltage-gated anion channel activity(GO:0008308) porin activity(GO:0015288) wide pore channel activity(GO:0022829) |
| 0.1 | 1.6 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.1 | 0.7 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.1 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.3 | GO:0015343 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.1 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 1.2 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.1 | 0.3 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 0.1 | 0.3 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.1 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.2 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.2 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0031406 | amino acid binding(GO:0016597) carboxylic acid binding(GO:0031406) organic acid binding(GO:0043177) |
| 0.0 | 0.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.2 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.0 | 0.5 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:0034061 | DNA polymerase activity(GO:0034061) |
| 0.0 | 0.5 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 3.8 | GO:0016773 | phosphotransferase activity, alcohol group as acceptor(GO:0016773) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.3 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.0 | 0.0 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.0 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.3 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.1 | GO:0052866 | phosphatidylinositol phosphate phosphatase activity(GO:0052866) |
| 0.0 | 0.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 0.0 | 0.0 | GO:0016894 | endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 3'-phosphomonoesters(GO:0016894) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.5 | 2.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.4 | 1.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 124.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.7 | 2.0 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.3 | 0.8 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.2 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 0.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 0.6 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.1 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 0.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 0.2 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 124.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.2 | REACTOME TCA CYCLE AND RESPIRATORY ELECTRON TRANSPORT | Genes involved in The citric acid (TCA) cycle and respiratory electron transport |
| 0.1 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.1 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |