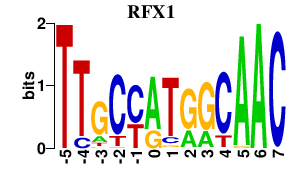
Results for RFX1
Z-value: 0.80
Transcription factors associated with RFX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX1
|
S000004166 | Major transcriptional repressor of DNA-damage-regulated genes |
Activity-expression correlation:
Activity profile of RFX1 motif
Sorted Z-values of RFX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YFR055W | 8.44 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YFR056C | 7.25 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YDR044W | 4.62 |
HEM13
|
Coproporphyrinogen III oxidase, an oxygen requiring enzyme that catalyzes the sixth step in the heme biosynthetic pathway; localizes to the mitochondrial inner membrane; transcription is repressed by oxygen and heme (via Rox1p and Hap1p) |
|
| YHR141C | 3.65 |
RPL42B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl42Ap and has similarity to rat L44; required for propagation of the killer toxin-encoding M1 double-stranded RNA satellite of the L-A double-stranded RNA virus |
|
| YNR001W-A | 3.41 |
Dubious open reading frame unlikely to encode a functional protein; identified by homology |
||
| YGR108W | 3.29 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YDL085C-A | 3.12 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YOR063W | 3.04 |
RPL3
|
Protein component of the large (60S) ribosomal subunit, has similarity to E. coli L3 and rat L3 ribosomal proteins; involved in the replication and maintenance of killer double stranded RNA virus |
|
| YOR315W | 3.01 |
SFG1
|
Nuclear protein, putative transcription factor required for growth of superficial pseudohyphae (which do not invade the agar substrate) but not for invasive pseudohyphal growth; may act together with Phd1p; potential Cdc28p substrate |
|
| YOL086C | 2.90 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YGL097W | 2.80 |
SRM1
|
Nucleotide exchange factor for Gsp1p, localizes to the nucleus, required for nucleocytoplasmic trafficking of macromolecules; suppressor of the pheromone response pathway; potentially phosphorylated by Cdc28p |
|
| YDR345C | 2.78 |
HXT3
|
Low affinity glucose transporter of the major facilitator superfamily, expression is induced in low or high glucose conditions |
|
| YNR067C | 2.70 |
DSE4
|
Daughter cell-specific secreted protein with similarity to glucanases, degrades cell wall from the daughter side causing daughter to separate from mother |
|
| YBR210W | 2.66 |
ERV15
|
Protein involved in export of proteins from the endoplasmic reticulum, has similarity to Erv14p |
|
| YIL069C | 2.64 |
RPS24B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps24Ap and has similarity to rat S24 ribosomal protein |
|
| YER137C | 2.43 |
Putative protein of unknown function |
||
| YHR136C | 2.40 |
SPL2
|
Protein with similarity to cyclin-dependent kinase inhibitors, overproduction suppresses a plc1 null mutation; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YIL029C | 2.35 |
Putative protein of unknown function; deletion confers sensitivity to 4-(N-(S-glutathionylacetyl)amino) phenylarsenoxide (GSAO) |
||
| YLR154C | 2.34 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YJR063W | 2.33 |
RPA12
|
RNA polymerase I subunit A12.2; contains two zinc binding domains, and the N terminal domain is responsible for anchoring to the RNA pol I complex |
|
| YER124C | 2.29 |
DSE1
|
Daughter cell-specific protein, may participate in pathways regulating cell wall metabolism; deletion affects cell separation after division and sensitivity to drugs targeted against the cell wall |
|
| YJL136C | 2.23 |
RPS21B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps21Ap and has similarity to rat S21 ribosomal protein |
|
| YGR180C | 2.16 |
RNR4
|
Ribonucleotide-diphosphate reductase (RNR), small subunit; the RNR complex catalyzes the rate-limiting step in dNTP synthesis and is regulated by DNA replication and DNA damage checkpoint pathways via localization of the small subunits |
|
| YDR279W | 2.15 |
RNH202
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YMR106C | 2.13 |
YKU80
|
Subunit of the telomeric Ku complex (Yku70p-Yku80p), involved in telomere length maintenance, structure and telomere position effect; relocates to sites of double-strand cleavage to promote nonhomologous end joining during DSB repair |
|
| YGL031C | 2.12 |
RPL24A
|
Ribosomal protein L30 of the large (60S) ribosomal subunit, nearly identical to Rpl24Bp and has similarity to rat L24 ribosomal protein; not essential for translation but may be required for normal translation rate |
|
| YER043C | 2.03 |
SAH1
|
S-adenosyl-L-homocysteine hydrolase, catabolizes S-adenosyl-L-homocysteine which is formed after donation of the activated methyl group of S-adenosyl-L-methionine (AdoMet) to an acceptor |
|
| YNL030W | 2.00 |
HHF2
|
One of two identical histone H4 proteins (see also HHF1); core histone required for chromatin assembly and chromosome function; contributes to telomeric silencing; N-terminal domain involved in maintaining genomic integrity |
|
| YML052W | 1.98 |
SUR7
|
Putative integral membrane protein; component of eisosomes; associated with endocytosis, along with Pil1p and Lsp1p; sporulation and plasma membrane sphingolipid content are altered in mutants |
|
| YGL030W | 1.92 |
RPL30
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L30 ribosomal protein; involved in pre-rRNA processing in the nucleolus; autoregulates splicing of its transcript |
|
| YIR021W | 1.90 |
MRS1
|
Protein required for the splicing of two mitochondrial group I introns (BI3 in COB and AI5beta in COX1); forms a splicing complex, containing four subunits of Mrs1p and two subunits of the BI3-encoded maturase, that binds to the BI3 RNA |
|
| YHL028W | 1.80 |
WSC4
|
ER membrane protein involved in the translocation of soluble secretory proteins and insertion of membrane proteins into the ER membrane; may also have a role in the stress response but has only partial functional overlap with WSC1-3 |
|
| YFR054C | 1.79 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR154W-B | 1.74 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR154W-A | 1.74 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR328W | 1.74 |
NMA1
|
Nicotinic acid mononucleotide adenylyltransferase, involved in pathways of NAD biosynthesis, including the de novo, NAD(+) salvage, and nicotinamide riboside salvage pathways |
|
| YIL114C | 1.73 |
POR2
|
Putative mitochondrial porin (voltage-dependent anion channel), related to Por1p but not required for mitochondrial membrane permeability or mitochondrial osmotic stability |
|
| YMR119W | 1.72 |
ASI1
|
Putative integral membrane E3 ubiquitin ligase; acts with Asi2p and Asi3p to ensure the fidelity of SPS-sensor signalling by maintaining the dormant repressed state of gene expression in the absence of inducing signals |
|
| YKR075C | 1.69 |
Protein of unknown function; similar to YOR062Cp and Reg1p; expression regulated by glucose and Rgt1p; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YKL120W | 1.69 |
OAC1
|
Mitochondrial inner membrane transporter, transports oxaloacetate, sulfate, and thiosulfate; member of the mitochondrial carrier family |
|
| YER070W | 1.68 |
RNR1
|
One of two large regulatory subunits of ribonucleotide-diphosphate reductase; the RNR complex catalyzes rate-limiting step in dNTP synthesis, regulated by DNA replication and DNA damage checkpoint pathways via localization of small subunits |
|
| YER011W | 1.66 |
TIR1
|
Cell wall mannoprotein of the Srp1p/Tip1p family of serine-alanine-rich proteins; expression is downregulated at acidic pH and induced by cold shock and anaerobiosis; abundance is increased in cells cultured without shaking |
|
| YML026C | 1.63 |
RPS18B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps18Ap and has similarity to E. coli S13 and rat S18 ribosomal proteins |
|
| YKR094C | 1.62 |
RPL40B
|
Fusion protein, identical to Rpl40Ap, that is cleaved to yield ubiquitin and a ribosomal protein of the large (60S) ribosomal subunit with similarity to rat L40; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes |
|
| YGR155W | 1.58 |
CYS4
|
Cystathionine beta-synthase, catalyzes the synthesis of cystathionine from serine and homocysteine, the first committed step in cysteine biosynthesis |
|
| YMR177W | 1.56 |
MMT1
|
Putative metal transporter involved in mitochondrial iron accumulation; closely related to Mmt2p |
|
| YMR011W | 1.51 |
HXT2
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YKR038C | 1.51 |
KAE1
|
Putative glycoprotease proposed to be in transcription as a component of the EKC protein complex with Bud32p, Cgi121p, Pcc1p, and Gon7p; also identified as a component of the KEOPS protein complex |
|
| YDL007W | 1.46 |
RPT2
|
One of six ATPases of the 19S regulatory particle of the 26S proteasome involved in the degradation of ubiquitinated substrates; required for normal peptide hydrolysis by the core 20S particle |
|
| YIL118W | 1.46 |
RHO3
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins involved in the establishment of cell polarity; GTPase activity positively regulated by the GTPase activating protein (GAP) Rgd1p |
|
| YMR006C | 1.45 |
PLB2
|
Phospholipase B (lysophospholipase) involved in phospholipid metabolism; displays transacylase activity in vitro; overproduction confers resistance to lysophosphatidylcholine |
|
| YOR234C | 1.44 |
RPL33B
|
Ribosomal protein L37 of the large (60S) ribosomal subunit, nearly identical to Rpl33Ap and has similarity to rat L35a; rpl33b null mutant exhibits normal growth while rpl33a rpl33b double null mutant is inviable |
|
| YLR286C | 1.43 |
CTS1
|
Endochitinase, required for cell separation after mitosis; transcriptional activation during late G and early M cell cycle phases is mediated by transcription factor Ace2p |
|
| YPR170W-B | 1.39 |
Putative protein of unknown function, conserved in fungi; partially overlaps the dubious genes YPR169W-A, YPR170W-A and YRP170C |
||
| YOR280C | 1.37 |
FSH3
|
Serine hydrolase; sequence is similar to Fsh1p and Fsh2p |
|
| YBR158W | 1.37 |
AMN1
|
Protein required for daughter cell separation, multiple mitotic checkpoints, and chromosome stability; contains 12 degenerate leucine-rich repeat motifs; expression is induced by the Mitotic Exit Network (MEN) |
|
| YJL200C | 1.37 |
ACO2
|
Putative mitochondrial aconitase isozyme; similarity to Aco1p, an aconitase required for the TCA cycle; expression induced during growth on glucose, by amino acid starvation via Gcn4p, and repressed on ethanol |
|
| YGR040W | 1.36 |
KSS1
|
Mitogen-activated protein kinase (MAPK) involved in signal transduction pathways that control filamentous growth and pheromone response; the KSS1 gene is nonfunctional in S288C strains and functional in W303 strains |
|
| YHR201C | 1.35 |
PPX1
|
Exopolyphosphatase, hydrolyzes inorganic polyphosphate (poly P) into Pi residues; located in the cytosol, plasma membrane, and mitochondrial matrix |
|
| YDR112W | 1.35 |
IRC2
|
Dubious open reading frame, unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR111C; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YGL039W | 1.33 |
Oxidoreductase, catalyzes NADPH-dependent reduction of the bicyclic diketone bicyclo[2.2.2]octane-2,6-dione (BCO2,6D) to the chiral ketoalcohol (1R,4S,6S)-6-hydroxybicyclo[2.2.2]octane-2-one (BCO2one6ol) |
||
| YDR033W | 1.33 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YDL229W | 1.33 |
SSB1
|
Cytoplasmic ATPase that is a ribosome-associated molecular chaperone, functions with J-protein partner Zuo1p; may be involved in folding of newly-made polypeptide chains; member of the HSP70 family; interacts with phosphatase subunit Reg1p |
|
| YGR181W | 1.33 |
TIM13
|
Mitochondrial intermembrane space protein, forms a complex with TIm8p that mediates import and insertion of a subset of polytopic inner membrane proteins; may prevent aggregation of incoming proteins in a chaperone-like manner |
|
| YKL096W-A | 1.32 |
CWP2
|
Covalently linked cell wall mannoprotein, major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored |
|
| YOR047C | 1.32 |
STD1
|
Protein involved in control of glucose-regulated gene expression; interacts with protein kinase Snf1p, glucose sensors Snf3p and Rgt2p, and TATA-binding protein Spt15p; acts as a regulator of the transcription factor Rgt1p |
|
| YDL148C | 1.30 |
NOP14
|
Nucleolar protein, forms a complex with Noc4p that mediates maturation and nuclear export of 40S ribosomal subunits; also present in the small subunit processome complex, which is required for processing of pre-18S rRNA |
|
| YPL177C | 1.26 |
CUP9
|
Homeodomain-containing transcriptional repressor of PTR2, which encodes a major peptide transporter; imported peptides activate ubiquitin-dependent proteolysis, resulting in degradation of Cup9p and de-repression of PTR2 transcription |
|
| YDR072C | 1.25 |
IPT1
|
Inositolphosphotransferase 1, involved in synthesis of mannose-(inositol-P)2-ceramide (M(IP)2C), which is the most abundant sphingolipid in cells, mutation confers resistance to the antifungals syringomycin E and DmAMP1 in some growth media |
|
| YJL029C | 1.24 |
VPS53
|
Component of the GARP (Golgi-associated retrograde protein) complex, Vps51p-Vps52p-Vps53p-Vps54p, which is required for the recycling of proteins from endosomes to the late Golgi; required for vacuolar protein sorting |
|
| YDL189W | 1.23 |
RBS1
|
Protein of unknown function, identified as a high copy suppressor of psk1 psk2 mutations that confer temperature-sensitivity for galactose utilization; proposed to bind single-stranded nucleic acids via its R3H domain |
|
| YER107W-A | 1.22 |
Dubious open reading frame unlikely to encode a protein, partially overlaps verified ORF GLE2/YER107C |
||
| YBL057C | 1.22 |
PTH2
|
One of two (see also PTH1) mitochondrially-localized peptidyl-tRNA hydrolases; negatively regulates the ubiquitin-proteasome pathway via interactions with ubiquitin-like ubiquitin-associated proteins; dispensable for cell growth |
|
| YPL142C | 1.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps the verified ORF RPL33A/YPL143W, a component of the large (60S) ribosomal subunit |
||
| YOL155C | 1.18 |
HPF1
|
Haze-protective mannoprotein that reduces the particle size of aggregated proteins in white wines |
|
| YOR314W | 1.18 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR170W-A | 1.17 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; identified by expression profiling and mass spectrometry |
||
| YMR083W | 1.17 |
ADH3
|
Mitochondrial alcohol dehydrogenase isozyme III; involved in the shuttling of mitochondrial NADH to the cytosol under anaerobic conditions and ethanol production |
|
| YGR090W | 1.16 |
UTP22
|
Possible U3 snoRNP protein involved in maturation of pre-18S rRNA, based on computational analysis of large-scale protein-protein interaction data |
|
| YOL085C | 1.16 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOL085W-A |
||
| YDR417C | 1.16 |
Hypothetical protein |
||
| YHL015W | 1.15 |
RPS20
|
Protein component of the small (40S) ribosomal subunit; overproduction suppresses mutations affecting RNA polymerase III-dependent transcription; has similarity to E. coli S10 and rat S20 ribosomal proteins |
|
| YER107C | 1.14 |
GLE2
|
Component of the nuclear pore complex required for polyadenylated RNA export but not for protein import, homologous to S. pombe Rae1p |
|
| YJR094W-A | 1.13 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YHR010W | 1.12 |
RPL27A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl27Bp and has similarity to rat L27 ribosomal protein |
|
| YOR313C | 1.12 |
SPS4
|
Protein whose expression is induced during sporulation; not required for sporulation; heterologous expression in E. coli induces the SOS response that senses DNA damage |
|
| YGR052W | 1.10 |
FMP48
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YLR449W | 1.09 |
FPR4
|
Peptidyl-prolyl cis-trans isomerase (PPIase) (proline isomerase) localized to the nucleus; catalyzes isomerization of proline residues in histones H3 and H4, which affects lysine methylation of those histones |
|
| YBL028C | 1.09 |
Protein of unknown function that may interact with ribosomes; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus; predicted to be involved in ribosome biogenesis |
||
| YGL034C | 1.09 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBL027W | 1.09 |
RPL19B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl19Ap and has similarity to rat L19 ribosomal protein; rpl19a and rpl19b single null mutations result in slow growth, while the double null mutation is lethal |
|
| YNL111C | 1.08 |
CYB5
|
Cytochrome b5, involved in the sterol and lipid biosynthesis pathways; acts as an electron donor to support sterol C5-6 desaturation |
|
| YBR126W-A | 1.08 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR126W-B; identified by gene-trapping, microarray analysis, and genome-wide homology searches |
||
| YDR111C | 1.08 |
ALT2
|
Putative alanine transaminase (glutamic pyruvic transaminase) |
|
| YHR137W | 1.06 |
ARO9
|
Aromatic aminotransferase II, catalyzes the first step of tryptophan, phenylalanine, and tyrosine catabolism |
|
| YDR534C | 1.05 |
FIT1
|
Mannoprotein that is incorporated into the cell wall via a glycosylphosphatidylinositol (GPI) anchor, involved in the retention of siderophore-iron in the cell wall |
|
| YJL198W | 1.05 |
PHO90
|
Low-affinity phosphate transporter; deletion of pho84, pho87, pho89, pho90, and pho91 causes synthetic lethality; transcription independent of Pi and Pho4p activity; overexpression results in vigorous growth |
|
| YHR094C | 1.05 |
HXT1
|
Low-affinity glucose transporter of the major facilitator superfamily, expression is induced by Hxk2p in the presence of glucose and repressed by Rgt1p when glucose is limiting |
|
| YGL012W | 1.03 |
ERG4
|
C-24(28) sterol reductase, catalyzes the final step in ergosterol biosynthesis; mutants are viable, but lack ergosterol |
|
| YIL078W | 1.03 |
THS1
|
Threonyl-tRNA synthetase, essential cytoplasmic protein |
|
| YKL153W | 1.02 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; transcription of both YLK153W and the overlapping essential gene GPM1 is reduced in the gcr1 null mutant |
||
| YNL289W | 1.02 |
PCL1
|
Pho85 cyclin of the Pcl1,2-like subfamily, involved in entry into the mitotic cell cycle and regulation of morphogenesis, localizes to sites of polarized cell growth |
|
| YLL034C | 1.01 |
RIX7
|
Putative ATPase of the AAA family, required for export of pre-ribosomal large subunits from the nucleus; distributed between the nucleolus, nucleoplasm, and nuclear periphery depending on growth conditions |
|
| YDR060W | 1.00 |
MAK21
|
Constituent of 66S pre-ribosomal particles, required for large (60S) ribosomal subunit biogenesis; involved in nuclear export of pre-ribosomes; required for maintenance of dsRNA virus; homolog of human CAATT-binding protein |
|
| YMR143W | 1.00 |
RPS16A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps16Bp and has similarity to E. coli S9 and rat S16 ribosomal proteins |
|
| YCR016W | 0.99 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus and nucleus; YCR016W is not an essential gene |
||
| YGL040C | 0.99 |
HEM2
|
Delta-aminolevulinate dehydratase, a homo-octameric enzyme, catalyzes the conversion of delta-aminolevulinic acid to porphobilinogen, the second step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus |
|
| YGR208W | 0.99 |
SER2
|
Phosphoserine phosphatase of the phosphoglycerate pathway, involved in serine and glycine biosynthesis, expression is regulated by the available nitrogen source |
|
| YNL031C | 0.97 |
HHT2
|
One of two identical histone H3 proteins (see also HHT1); core histone required for chromatin assembly, involved in heterochromatin-mediated telomeric and HM silencing; regulated by acetylation, methylation, and mitotic phosphorylation |
|
| YPR170C | 0.95 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORFs YPR169W-A and YPR170W-B |
||
| YLR025W | 0.94 |
SNF7
|
One of four subunits of the endosomal sorting complex required for transport III (ESCRT-III); involved in the sorting of transmembrane proteins into the multivesicular body (MVB) pathway; recruited from the cytoplasm to endosomal membranes |
|
| YIL009W | 0.94 |
FAA3
|
Long chain fatty acyl-CoA synthetase, has a preference for C16 and C18 fatty acids; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery |
|
| YMR142C | 0.94 |
RPL13B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl13Ap; not essential for viability; has similarity to rat L13 ribosomal protein |
|
| YGR050C | 0.93 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YBR078W | 0.93 |
ECM33
|
GPI-anchored protein of unknown function, has a possible role in apical bud growth; GPI-anchoring on the plasma membrane crucial to function; phosphorylated in mitochondria; similar to Sps2p and Pst1p |
|
| YHR170W | 0.92 |
NMD3
|
Protein involved in nuclear export of the large ribosomal subunit; acts as a Crm1p-dependent adapter protein for export of nascent ribosomal subunits through the nuclear pore complex |
|
| YDR418W | 0.92 |
RPL12B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl12Ap; rpl12a rpl12b double mutant exhibits slow growth and slow translation; has similarity to E. coli L11 and rat L12 ribosomal proteins |
|
| YOL127W | 0.92 |
RPL25
|
Primary rRNA-binding ribosomal protein component of the large (60S) ribosomal subunit, has similarity to E. coli L23 and rat L23a ribosomal proteins; binds to 26S rRNA via a conserved C-terminal motif |
|
| YBR209W | 0.91 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR209W is not an essential gene |
||
| YIL009C-A | 0.89 |
EST3
|
Component of the telomerase holoenzyme, involved in telomere replication |
|
| YBR121C | 0.89 |
GRS1
|
Cytoplasmic and mitochondrial glycyl-tRNA synthase that ligates glycine to the cognate anticodon bearing tRNA; transcription termination factor that may interact with the 3'-end of pre-mRNA to promote 3'-end formation |
|
| YBL056W | 0.88 |
PTC3
|
Type 2C protein phosphatase; dephosphorylates Hog1p (see also Ptc2p) to limit maximal kinase activity induced by osmotic stress; dephosphorylates T169 phosphorylated Cdc28p (see also Ptc2p); role in DNA checkpoint inactivation |
|
| YKL219W | 0.88 |
COS9
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YGL080W | 0.88 |
FMP37
|
Putative protein of unknown function; highly conserved across species and orthologous to human gene BRP44L; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YKL218C | 0.87 |
SRY1
|
3-hydroxyaspartate dehydratase, deaminates L-threo-3-hydroxyaspartate to form oxaloacetate and ammonia; required for survival in the presence of hydroxyaspartate |
|
| YDL236W | 0.86 |
PHO13
|
Alkaline phosphatase specific for p-nitrophenyl phosphate, involved in dephosphorylation of histone II-A and casein |
|
| YHR142W | 0.85 |
CHS7
|
Protein of unknown function, involved in chitin biosynthesis by regulating Chs3p export from the ER |
|
| YOR087W | 0.84 |
YVC1
|
Vacuolar cation channel, mediates release of Ca(2+) from the vacuole in response to hyperosmotic shock |
|
| YHR183W | 0.84 |
GND1
|
6-phosphogluconate dehydrogenase (decarboxylating), catalyzes an NADPH regenerating reaction in the pentose phosphate pathway; required for growth on D-glucono-delta-lactone and adaptation to oxidative stress |
|
| YNL016W | 0.82 |
PUB1
|
Poly(A)+ RNA-binding protein, abundant mRNP-component protein that binds mRNA and is required for stability of a number of mRNAs; not reported to associate with polyribosomes |
|
| YBR112C | 0.81 |
CYC8
|
General transcriptional co-repressor, acts together with Tup1p; also acts as part of a transcriptional co-activator complex that recruits the SWI/SNF and SAGA complexes to promoters |
|
| YKL110C | 0.81 |
KTI12
|
Protein that plays a role, with Elongator complex, in modification of wobble nucleosides in tRNA; involved in sensitivity to G1 arrest induced by zymocin; interacts with chromatin throughout the genome; also interacts with Cdc19p |
|
| YGR179C | 0.80 |
OKP1
|
Outer kinetochore protein, required for accurate mitotic chromosome segregation; component of the kinetochore sub-complex COMA (Ctf19p, Okp1p, Mcm21p, Ame1p) that functions as a platform for kinetochore assembly |
|
| YDR215C | 0.80 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; null mutant displays elevated sensitivity to expression of a mutant huntingtin fragment or of alpha-synuclein |
||
| YGR164W | 0.80 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHR182C-A | 0.80 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR199W | 0.78 |
CLN1
|
G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) |
|
| YBR202W | 0.78 |
MCM7
|
Component of the hexameric MCM complex, which is important for priming origins of DNA replication in G1 and becomes an active ATP-dependent helicase that promotes DNA melting and elongation when activated by Cdc7p-Dbf4p in S-phase |
|
| YGR106C | 0.78 |
VOA1
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuolar memebrane |
|
| YHR070C-A | 0.78 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene TRM5/YHR070W |
||
| YJR145C | 0.78 |
RPS4A
|
Protein component of the small (40S) ribosomal subunit; mutation affects 20S pre-rRNA processing; identical to Rps4Bp and has similarity to rat S4 ribosomal protein |
|
| YML053C | 0.76 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and the nucleus; YML053C is not an essential gene |
||
| YCR072C | 0.75 |
RSA4
|
WD-repeat protein involved in ribosome biogenesis; may interact with ribosomes; required for maturation and efficient intra-nuclear transport or pre-60S ribosomal subunits, localizes to the nucleolus |
|
| YEL020C-B | 0.75 |
Dubious open reading frame unlikely to encode a protein; partially overlaps verified gene YEL020W-A; identified by fungal homology and RT-PCR |
||
| YGR214W | 0.74 |
RPS0A
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps0Bp; required for maturation of 18S rRNA along with Rps0Bp; deletion of either RPS0 gene reduces growth rate, deletion of both genes is lethal |
|
| YEL056W | 0.72 |
HAT2
|
Subunit of the Hat1p-Hat2p histone acetyltransferase complex; required for high affinity binding of the complex to free histone H4, thereby enhancing Hat1p activity; similar to human RbAp46 and 48; has a role in telomeric silencing |
|
| YOR269W | 0.72 |
PAC1
|
Protein involved in nuclear migration, part of the dynein/dynactin pathway; targets dynein to microtubule tips, which is necessary for sliding of microtubules along bud cortex; synthetic lethal with bni1; homolog of human LIS1 |
|
| YBR126C | 0.71 |
TPS1
|
Synthase subunit of trehalose-6-phosphate synthase/phosphatase complex, which synthesizes the storage carbohydrate trehalose; also found in a monomeric form; expression is induced by the stress response and repressed by the Ras-cAMP pathway |
|
| YML119W | 0.71 |
Putative protein of unknown funtion; YML119W is not an essential gene; potential Cdc28p substrate |
||
| YMR205C | 0.71 |
PFK2
|
Beta subunit of heterooctameric phosphofructokinase involved in glycolysis, indispensable for anaerobic growth, activated by fructose-2,6-bisphosphate and AMP, mutation inhibits glucose induction of cell cycle-related genes |
|
| YDL211C | 0.71 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YER131W | 0.69 |
RPS26B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps26Ap and has similarity to rat S26 ribosomal protein |
|
| YOR164C | 0.69 |
Protein of unknown function; highly conserved across species and homologous to human gene C7orf20; interacts with Mdy2p |
||
| YBR113W | 0.68 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene CYC8 |
||
| YDR047W | 0.68 |
HEM12
|
Uroporphyrinogen decarboxylase, catalyzes the fifth step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus; activity inhibited by Cu2+, Zn2+, Fe2+, Fe3+ and sulfhydryl-specific reagents |
|
| YGR051C | 0.67 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YGR051C is not an essential gene |
||
| YNR013C | 0.66 |
PHO91
|
Low-affinity phosphate transporter of the vacuolar membrane; deletion of pho84, pho87, pho89, pho90, and pho91 causes synthetic lethality; transcription independent of Pi and Pho4p activity; overexpression results in vigorous growth |
|
| YBR032W | 0.66 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YDR535C | 0.66 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YDR535C is not an essential gene. |
||
| YDR278C | 0.65 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR218W | 0.65 |
CRM1
|
Major karyopherin, involved in export of proteins, RNAs, and ribosomal subunits from the nucleus |
|
| YAL033W | 0.65 |
POP5
|
Subunit of both RNase MRP, which cleaves pre-rRNA, and nuclear RNase P, which cleaves tRNA precursors to generate mature 5' ends |
|
| YDR410C | 0.65 |
STE14
|
Farnesyl cysteine-carboxyl methyltransferase, mediates the carboxyl methylation step during C-terminal CAAX motif processing of a-factor and RAS proteins in the endoplasmic reticulum, localizes to the ER membrane |
|
| YGL225W | 0.65 |
VRG4
|
Golgi GDP-mannose transporter; regulates Golgi function and glycosylation in Golgi |
|
| YOL103W | 0.64 |
ITR2
|
Myo-inositol transporter with strong similarity to the major myo-inositol transporter Itr1p, member of the sugar transporter superfamily; expressed constitutively |
|
| YNL043C | 0.63 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YIP3/YNL044W |
||
| YER012W | 0.62 |
PRE1
|
Beta 4 subunit of the 20S proteasome; localizes to the nucleus throughout the cell cycle |
|
| YLR400W | 0.61 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPR157W | 0.61 |
Putative protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YLR287C-A | 0.61 |
RPS30A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps30Bp and has similarity to rat S30 ribosomal protein |
|
| YDL241W | 0.61 |
Putative protein of unknown function; YDL241W is not an essential gene |
||
| YLR154W-C | 0.61 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YDR133C | 0.60 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR134C |
||
| YMR263W | 0.60 |
SAP30
|
Subunit of a histone deacetylase complex, along with Rpd3p and Sin3p, that is involved in silencing at telomeres, rDNA, and silent mating-type loci; involved in telomere maintenance |
|
| YDR188W | 0.60 |
CCT6
|
Subunit of the cytosolic chaperonin Cct ring complex, related to Tcp1p, essential protein that is required for the assembly of actin and tubulins in vivo; contains an ATP-binding motif |
|
| YIR033W | 0.59 |
MGA2
|
ER membrane protein involved in regulation of OLE1 transcription, acts with homolog Spt23p; inactive ER form dimerizes and one subunit is then activated by ubiquitin/proteasome-dependent processing followed by nuclear targeting |
|
| YGR122W | 0.59 |
Putative protein of unknown function; deletion mutants do not properly process Rim101p and have decreased resistance to rapamycin; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YJL028W | 0.59 |
Protein of unknown function; may interact with ribosomes, based on co-purification experiments |
||
| YGR234W | 0.58 |
YHB1
|
Nitric oxide oxidoreductase, flavohemoglobin involved in nitric oxide detoxification; plays a role in the oxidative and nitrosative stress responses |
|
| YGR195W | 0.58 |
SKI6
|
3'-to-5' phosphorolytic exoribonuclease that is a subunit of the exosome; required for 3' processing of the 5.8S rRNA; involved in 3' to 5' mRNA degradation and translation inhibition of non-poly(A) mRNAs |
|
| YDR073W | 0.58 |
SNF11
|
Subunit of the SWI/SNF chromatin remodeling complex involved in transcriptional regulation; interacts with a highly conserved 40-residue sequence of Snf2p |
|
| YER003C | 0.57 |
PMI40
|
Mannose-6-phosphate isomerase, catalyzes the interconversion of fructose-6-P and mannose-6-P; required for early steps in protein mannosylation |
|
| YPR171W | 0.57 |
BSP1
|
Adapter that links synaptojanins Inp52p and Inp53p to the cortical actin cytoskeleton |
|
| YIL015W | 0.57 |
BAR1
|
Aspartyl protease secreted into the periplasmic space of mating type a cells, helps cells find mating partners, cleaves and inactivates alpha factor allowing cells to recover from alpha-factor-induced cell cycle arrest |
|
| YAR002C-A | 0.57 |
ERP1
|
Protein that forms a heterotrimeric complex with Erp2p, Emp24p, and Erv25p; member, along with Emp24p and Erv25p, of the p24 family involved in ER to Golgi transport and localized to COPII-coated vesicles |
|
| YMR112C | 0.56 |
MED11
|
Subunit of the RNA polymerase II mediator complex; associates with core polymerase subunits to form the RNA polymerase II holoenzyme; essential protein |
|
| YLR321C | 0.56 |
SFH1
|
Component of the RSC chromatin remodeling complex; essential gene required for cell cycle progression and maintenance of proper ploidy; phosphorylated in the G1 phase of the cell cycle; Snf5p paralog |
|
| YGR187C | 0.56 |
HGH1
|
Nonessential protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; similar to mammalian BRP16 (Brain protein 16) |
|
| YMR099C | 0.56 |
Glucose-6-phosphate 1-epimerase (hexose-6-phosphate mutarotase), likely involved in carbohydrate metabolism; GFP-fusion protein localizes to both the nucleus and cytoplasm and is induced in response to the DNA-damaging agent MMS |
||
| YPR146C | 0.56 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR187C | 0.55 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified, essential ORF CCT6/YDR188W |
||
| YMR113W | 0.55 |
FOL3
|
Dihydrofolate synthetase, involved in folic acid biosynthesis; catalyzes the conversion of dihydropteroate to dihydrofolate in folate coenzyme biosynthesis |
|
| YHR181W | 0.54 |
SVP26
|
Integral membrane protein of the early Golgi apparatus and endoplasmic reticulum, involved in COP II vesicle transport; may also function to promote retention of proteins in the early Golgi compartment |
|
| YBL083C | 0.54 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps verified ORF ALG3 |
||
| YOR086C | 0.54 |
TCB1
|
Lipid-binding protein containing three calcium and lipid binding domains; non-tagged protein localizes to mitochondria and GFP-fusion protein localizes to the cell periphery; C-termini of Tcb1p, Tcb2p and Tcb3p interact |
|
| YGR107W | 0.53 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YIL028W | 0.53 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR008C-A | 0.52 |
Putative protein of unknown function, includes a potential transmembrane domain; deletion results in slightly lengthened telomeres |
||
| YKL179C | 0.52 |
COY1
|
Golgi membrane protein with similarity to mammalian CASP; genetic interactions with GOS1 (encoding a Golgi snare protein) suggest a role in Golgi function |
|
| YMR290W-A | 0.52 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; overlaps 5' end of essential HAS1 gene which encodes an ATP-dependent RNA helicase |
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.9 | 3.6 | GO:0046898 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 0.9 | 2.7 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.8 | 2.5 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.8 | 5.5 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.7 | 3.0 | GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000461) |
| 0.7 | 2.1 | GO:2000879 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 0.6 | 4.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.6 | 1.7 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.5 | 3.4 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 0.5 | 2.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.5 | 1.4 | GO:0006798 | polyphosphate catabolic process(GO:0006798) |
| 0.4 | 1.3 | GO:0070637 | nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) |
| 0.4 | 0.9 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.4 | 3.9 | GO:0000947 | amino acid catabolic process to alcohol via Ehrlich pathway(GO:0000947) |
| 0.4 | 2.1 | GO:0007535 | donor selection(GO:0007535) |
| 0.4 | 1.6 | GO:0019346 | transsulfuration(GO:0019346) |
| 0.4 | 3.9 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.4 | 1.5 | GO:0090338 | regulation of formin-nucleated actin cable assembly(GO:0090337) positive regulation of formin-nucleated actin cable assembly(GO:0090338) |
| 0.4 | 1.1 | GO:0031057 | negative regulation of histone modification(GO:0031057) |
| 0.4 | 1.8 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.4 | 1.1 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.3 | 5.6 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.3 | 0.9 | GO:0034059 | response to anoxia(GO:0034059) cellular response to anoxia(GO:0071454) |
| 0.3 | 0.9 | GO:0019673 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.3 | 1.5 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.3 | 0.6 | GO:2000531 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) regulation of fatty acid biosynthetic process by transcription from RNA polymerase II promoter(GO:0100070) regulation of fatty acid biosynthetic process by regulation of transcription from RNA polymerase II promoter(GO:2000531) |
| 0.3 | 0.9 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) |
| 0.3 | 2.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.3 | 0.9 | GO:0048024 | regulation of RNA splicing(GO:0043484) regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.3 | 0.6 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.3 | 0.8 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of nucleobase-containing compound transport(GO:0032239) regulation of RNA export from nucleus(GO:0046831) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.3 | 1.9 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.3 | 1.3 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.3 | 0.5 | GO:0071902 | positive regulation of protein serine/threonine kinase activity(GO:0071902) |
| 0.3 | 1.3 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.3 | 1.3 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.2 | 0.5 | GO:0090088 | regulation of peptide transport(GO:0090087) regulation of oligopeptide transport(GO:0090088) regulation of dipeptide transport(GO:0090089) |
| 0.2 | 1.9 | GO:0000376 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.2 | 7.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 2.9 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.2 | 1.1 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.2 | 0.8 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.2 | 1.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 1.0 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.2 | 1.7 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.2 | 1.8 | GO:0001402 | signal transduction involved in filamentous growth(GO:0001402) |
| 0.2 | 1.5 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.2 | 0.5 | GO:0046901 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.2 | 1.5 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.2 | 0.7 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 0.5 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.2 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 19.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 0.8 | GO:0000173 | inactivation of MAPK activity involved in osmosensory signaling pathway(GO:0000173) |
| 0.1 | 0.7 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.1 | 1.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.6 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.6 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) |
| 0.1 | 1.5 | GO:0034963 | box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) |
| 0.1 | 1.1 | GO:0034221 | cell wall chitin biosynthetic process(GO:0006038) fungal-type cell wall chitin biosynthetic process(GO:0034221) fungal-type cell wall polysaccharide biosynthetic process(GO:0051278) fungal-type cell wall polysaccharide metabolic process(GO:0071966) |
| 0.1 | 0.6 | GO:0009312 | trehalose biosynthetic process(GO:0005992) oligosaccharide biosynthetic process(GO:0009312) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 2.8 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 1.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 1.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.4 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) |
| 0.1 | 1.0 | GO:0051320 | mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.1 | 1.0 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.6 | GO:0007323 | peptide pheromone maturation(GO:0007323) |
| 0.1 | 0.3 | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) |
| 0.1 | 1.3 | GO:0001100 | negative regulation of exit from mitosis(GO:0001100) |
| 0.1 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.5 | GO:0090158 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 0.3 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.1 | 0.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 3.8 | GO:0009266 | response to temperature stimulus(GO:0009266) |
| 0.1 | 0.3 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 0.3 | GO:1900436 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.1 | 0.2 | GO:0016320 | endoplasmic reticulum membrane fusion(GO:0016320) |
| 0.1 | 0.9 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.1 | 1.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 2.8 | GO:0006694 | steroid biosynthetic process(GO:0006694) sterol biosynthetic process(GO:0016126) |
| 0.1 | 1.7 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.1 | 0.7 | GO:0031506 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) |
| 0.1 | 0.9 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.3 | GO:0072668 | obsolete tubulin complex biogenesis(GO:0072668) |
| 0.1 | 0.5 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.1 | 1.4 | GO:0006885 | regulation of pH(GO:0006885) |
| 0.1 | 0.3 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.1 | 1.7 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.2 | GO:0036213 | actomyosin contractile ring contraction(GO:0000916) actin filament-based movement(GO:0030048) contractile ring contraction(GO:0036213) actin filament-based transport(GO:0099515) |
| 0.1 | 0.2 | GO:0009187 | cyclic nucleotide metabolic process(GO:0009187) |
| 0.1 | 0.2 | GO:0000296 | spermine transport(GO:0000296) |
| 0.1 | 0.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.3 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 1.4 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.4 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.1 | 0.2 | GO:0006571 | tyrosine biosynthetic process(GO:0006571) L-phenylalanine biosynthetic process(GO:0009094) erythrose 4-phosphate/phosphoenolpyruvate family amino acid biosynthetic process(GO:1902223) |
| 0.0 | 0.5 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.9 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0046656 | folic acid metabolic process(GO:0046655) folic acid biosynthetic process(GO:0046656) |
| 0.0 | 0.2 | GO:0006797 | polyphosphate metabolic process(GO:0006797) |
| 0.0 | 0.1 | GO:0051597 | response to methylmercury(GO:0051597) cellular response to methylmercury(GO:0071406) |
| 0.0 | 0.3 | GO:0006771 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 1.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:0034517 | ribophagy(GO:0034517) |
| 0.0 | 0.1 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0090002 | protein localization to plasma membrane(GO:0072659) establishment of protein localization to plasma membrane(GO:0090002) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.2 | GO:2000278 | regulation of telomere maintenance via telomerase(GO:0032210) regulation of telomere maintenance via telomere lengthening(GO:1904356) regulation of DNA biosynthetic process(GO:2000278) |
| 0.0 | 0.1 | GO:0031684 | heterotrimeric G-protein complex cycle(GO:0031684) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0046033 | AMP biosynthetic process(GO:0006167) AMP metabolic process(GO:0046033) |
| 0.0 | 0.2 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0006779 | porphyrin-containing compound metabolic process(GO:0006778) porphyrin-containing compound biosynthetic process(GO:0006779) heme biosynthetic process(GO:0006783) tetrapyrrole metabolic process(GO:0033013) tetrapyrrole biosynthetic process(GO:0033014) heme metabolic process(GO:0042168) |
| 0.0 | 0.1 | GO:0000162 | tryptophan biosynthetic process(GO:0000162) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0048309 | endoplasmic reticulum inheritance(GO:0048309) |
| 0.0 | 2.2 | GO:0016072 | rRNA metabolic process(GO:0016072) |
| 0.0 | 0.5 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.0 | GO:0035306 | positive regulation of phosphatase activity(GO:0010922) positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0051668 | localization within membrane(GO:0051668) |
| 0.0 | 0.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.6 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.0 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.1 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.0 | GO:0019379 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.0 | 0.1 | GO:0016125 | steroid metabolic process(GO:0008202) sterol metabolic process(GO:0016125) |
| 0.0 | 0.2 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.0 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0000735 | removal of nonhomologous ends(GO:0000735) |
| 0.0 | 0.0 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.1 | GO:0043687 | C-terminal protein amino acid modification(GO:0018410) post-translational protein modification(GO:0043687) |
| 0.0 | 0.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.0 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.0 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) |
| 0.0 | 0.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.9 | 2.7 | GO:0030428 | cell septum(GO:0030428) |
| 0.6 | 1.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.5 | 2.5 | GO:0030689 | Noc complex(GO:0030689) |
| 0.4 | 1.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.4 | 1.7 | GO:0000938 | GARP complex(GO:0000938) |
| 0.3 | 2.0 | GO:0032126 | eisosome(GO:0032126) |
| 0.3 | 1.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.3 | 24.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 2.1 | GO:0005724 | nuclear heterochromatin(GO:0005720) nuclear telomeric heterochromatin(GO:0005724) telomeric heterochromatin(GO:0031933) |
| 0.2 | 1.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 2.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 3.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 12.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 1.0 | GO:0032545 | CURI complex(GO:0032545) |
| 0.2 | 0.8 | GO:0000817 | COMA complex(GO:0000817) |
| 0.2 | 2.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 2.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.2 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.2 | 1.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.6 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.1 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 0.4 | GO:0000928 | gamma-tubulin small complex, spindle pole body(GO:0000928) gamma-tubulin complex(GO:0000930) gamma-tubulin small complex(GO:0008275) |
| 0.1 | 4.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 0.7 | GO:0035339 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.1 | 0.5 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.1 | GO:0005940 | septin ring(GO:0005940) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.3 | GO:0043529 | GET complex(GO:0043529) |
| 0.1 | 4.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.3 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.1 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.5 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 0.5 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.1 | 0.8 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.8 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.1 | GO:0071261 | Ssh1 translocon complex(GO:0071261) |
| 0.0 | 0.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0016587 | Isw1 complex(GO:0016587) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.6 | GO:0070210 | Rpd3L-Expanded complex(GO:0070210) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 5.7 | GO:0005933 | cellular bud(GO:0005933) |
| 0.0 | 0.2 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 4.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.0 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.1 | GO:0048188 | histone methyltransferase complex(GO:0035097) Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0070772 | PAS complex(GO:0070772) |
| 0.0 | 1.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.0 | GO:0033254 | vacuolar transporter chaperone complex(GO:0033254) |
| 0.0 | 0.1 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0044433 | vesicle membrane(GO:0012506) cytoplasmic vesicle membrane(GO:0030659) cytoplasmic vesicle part(GO:0044433) |
| 0.0 | 0.0 | GO:0030892 | mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 8.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 1.1 | 4.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.1 | 4.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.7 | 2.9 | GO:0042973 | glucan endo-1,3-beta-D-glucosidase activity(GO:0042973) |
| 0.6 | 3.9 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.6 | 1.7 | GO:0005253 | anion channel activity(GO:0005253) voltage-gated anion channel activity(GO:0008308) porin activity(GO:0015288) wide pore channel activity(GO:0022829) |
| 0.6 | 1.7 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.5 | 1.6 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.4 | 1.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.4 | 1.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.3 | 1.7 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.3 | 0.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.3 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 0.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 0.9 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.3 | 0.8 | GO:0022832 | voltage-gated ion channel activity(GO:0005244) calcium channel activity(GO:0005262) voltage-gated channel activity(GO:0022832) |
| 0.3 | 1.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 1.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 0.9 | GO:0016854 | ammonia-lyase activity(GO:0016841) racemase and epimerase activity(GO:0016854) |
| 0.2 | 0.6 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.2 | 2.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.2 | 0.6 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.2 | 2.4 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.2 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 0.6 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) |
| 0.2 | 0.8 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 1.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.2 | 4.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 1.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.2 | 1.1 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.2 | 1.8 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.2 | 2.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 0.5 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.2 | 1.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 3.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 3.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.2 | 32.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.7 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 0.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.7 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.1 | 1.4 | GO:0015578 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.1 | 0.6 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.1 | 1.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.0 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.3 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.1 | 3.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 0.4 | GO:0005034 | osmosensor activity(GO:0005034) |
| 0.1 | 0.3 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.3 | GO:0016972 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) thiol oxidase activity(GO:0016972) |
| 0.1 | 0.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.3 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.3 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 0.9 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.1 | 2.0 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 0.2 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.1 | 1.8 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.1 | 0.2 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.1 | 1.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.9 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 3.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 0.2 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 2.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 0.6 | GO:0005496 | steroid binding(GO:0005496) sterol binding(GO:0032934) |
| 0.1 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.6 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.1 | 0.2 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.1 | 1.0 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.2 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.1 | 0.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.7 | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen(GO:0016705) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.8 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.2 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 1.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.7 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0015103 | inorganic anion transmembrane transporter activity(GO:0015103) |
| 0.0 | 0.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0032561 | GTP binding(GO:0005525) guanyl nucleotide binding(GO:0019001) guanyl ribonucleotide binding(GO:0032561) |
| 0.0 | 0.2 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 0.0 | 0.2 | GO:0008483 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0030695 | GTPase regulator activity(GO:0030695) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.1 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.0 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 7.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 1.2 | REACTOME GPCR LIGAND BINDING | Genes involved in GPCR ligand binding |
| 0.1 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 0.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |