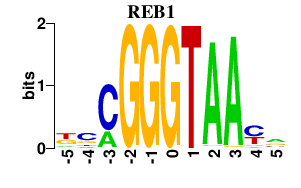
Results for REB1
Z-value: 0.92
Transcription factors associated with REB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
REB1
|
S000000253 | RNA polymerase I enhancer binding protein |
Activity-expression correlation:
Activity profile of REB1 motif
Sorted Z-values of REB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YLR154W-A | 5.41 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR154W-B | 4.83 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR154W-C | 4.04 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YNR013C | 3.55 |
PHO91
|
Low-affinity phosphate transporter of the vacuolar membrane; deletion of pho84, pho87, pho89, pho90, and pho91 causes synthetic lethality; transcription independent of Pi and Pho4p activity; overexpression results in vigorous growth |
|
| YGR242W | 3.08 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF YAP1802/YGR241C |
||
| YFL015W-A | 2.95 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR241C | 2.58 |
YAP1802
|
Protein involved in clathrin cage assembly; binds Pan1p and clathrin; homologous to Yap1801p, member of the AP180 protein family |
|
| YLR154C | 2.56 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YCR031C | 2.55 |
RPS14A
|
Ribosomal protein 59 of the small subunit, required for ribosome assembly and 20S pre-rRNA processing; mutations confer cryptopleurine resistance; nearly identical to Rps14Bp and similar to E. coli S11 and rat S14 ribosomal proteins |
|
| YFL015C | 2.48 |
Dubious open reading frame unlikely to encode a protein; partially overlaps dubious ORF YFL015W-A; YFL015C is not an essential gene |
||
| YER131W | 2.31 |
RPS26B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps26Ap and has similarity to rat S26 ribosomal protein |
|
| YLR162W-A | 2.26 |
Putative protein of unknown function identified by fungal homology comparisons and RT-PCR |
||
| YGR265W | 2.25 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF MES1/YGR264C, which encodes methionyl-tRNA synthetase |
||
| YOL005C | 2.13 |
RPB11
|
RNA polymerase II subunit B12.5; part of central core; similar to Rpc19p and bacterial alpha subunit |
|
| YOL086C | 2.04 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YML053C | 2.03 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and the nucleus; YML053C is not an essential gene |
||
| YGR264C | 2.02 |
MES1
|
Methionyl-tRNA synthetase, forms a complex with glutamyl-tRNA synthetase (Gus1p) and Arc1p, which increases the catalytic efficiency of both tRNA synthetases; also has a role in nuclear export of tRNAs |
|
| YMR290W-A | 2.01 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; overlaps 5' end of essential HAS1 gene which encodes an ATP-dependent RNA helicase |
||
| YJL107C | 1.98 |
Putative protein of unknown function; expression is induced by activation of the HOG1 mitogen-activated signaling pathway and this induction is Hog1p/Pbs2p dependent; YJL107C and adjacent ORF, YJL108C are merged in related fungi |
||
| YDL047W | 1.96 |
SIT4
|
Type 2A-related serine-threonine phosphatase that functions in the G1/S transition of the mitotic cycle; cytoplasmic and nuclear protein that modulates functions mediated by Pkc1p including cell wall and actin cytoskeleton organization |
|
| YMR183C | 1.95 |
SSO2
|
Plasma membrane t-SNARE involved in fusion of secretory vesicles at the plasma membrane; syntaxin homolog that is functionally redundant with Sso1p |
|
| YLL008W | 1.95 |
DRS1
|
Nucleolar DEAD-box protein required for ribosome assembly and function, including synthesis of 60S ribosomal subunits; constituent of 66S pre-ribosomal particles |
|
| YHR023W | 1.92 |
MYO1
|
Type II myosin heavy chain, required for wild-type cytokinesis and cell separation; localizes to the actomyosin ring; binds to myosin light chains Mlc1p and Mlc2p through its IQ1 and IQ2 motifs respectively |
|
| YDR098C | 1.85 |
GRX3
|
Hydroperoxide and superoxide-radical responsive glutathione-dependent oxidoreductase; monothiol glutaredoxin subfamily member along with Grx4p and Grx5p; protects cells from oxidative damage |
|
| YIL118W | 1.84 |
RHO3
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins involved in the establishment of cell polarity; GTPase activity positively regulated by the GTPase activating protein (GAP) Rgd1p |
|
| YKL218C | 1.82 |
SRY1
|
3-hydroxyaspartate dehydratase, deaminates L-threo-3-hydroxyaspartate to form oxaloacetate and ammonia; required for survival in the presence of hydroxyaspartate |
|
| YOL139C | 1.82 |
CDC33
|
Cytoplasmic mRNA cap binding protein; the eIF4E-cap complex is responsible for mediating cap-dependent mRNA translation via interactions with the translation initiation factor eIF4G (Tif4631p or Tif4632p) |
|
| YPL250W-A | 1.79 |
Identified by fungal homology and RT-PCR |
||
| YPR029C | 1.76 |
APL4
|
Gamma-adaptin, large subunit of the clathrin-associated protein (AP-1) complex; binds clathrin; involved in vesicle mediated transport |
|
| YMR290C | 1.74 |
HAS1
|
ATP-dependent RNA helicase; localizes to both the nuclear periphery and nucleolus; highly enriched in nuclear pore complex fractions; constituent of 66S pre-ribosomal particles |
|
| YDR146C | 1.73 |
SWI5
|
Transcription factor that activates transcription of genes expressed at the M/G1 phase boundary and in G1 phase; localization to the nucleus occurs during G1 and appears to be regulated by phosphorylation by Cdc28p kinase |
|
| YLR150W | 1.68 |
STM1
|
Protein that binds G4 quadruplex and purine motif triplex nucleic acid; acts with Cdc13p to maintain telomere structure; interacts with ribosomes and subtelomeric Y' DNA; multicopy suppressor of tom1 and pop2 mutations |
|
| YOL123W | 1.64 |
HRP1
|
Subunit of cleavage factor I, a five-subunit complex required for the cleavage and polyadenylation of pre-mRNA 3' ends; RRM-containing heteronuclear RNA binding protein and hnRNPA/B family member that binds to poly (A) signal sequences |
|
| YOR226C | 1.61 |
ISU2
|
Conserved protein of the mitochondrial matrix, required for synthesis of mitochondrial and cytosolic iron-sulfur proteins, performs a scaffolding function in mitochondria during Fe/S cluster assembly; isu1 isu2 double mutant is inviable |
|
| YPR119W | 1.58 |
CLB2
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YPL037C | 1.53 |
EGD1
|
Subunit beta1 of the nascent polypeptide-associated complex (NAC) involved in protein targeting, associated with cytoplasmic ribosomes; enhances DNA binding of the Gal4p activator; homolog of human BTF3b |
|
| YPR102C | 1.51 |
RPL11A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl11Bp; involved in ribosomal assembly; depletion causes degradation of proteins and RNA of the 60S subunit; has similarity to E. coli L5 and rat L11 |
|
| YFL016C | 1.50 |
MDJ1
|
Co-chaperone that stimulates the ATPase activity of the HSP70 protein Ssc1p; involved in protein folding/refolding in the mitochodrial matrix; required for proteolysis of misfolded proteins; member of the HSP40 (DnaJ) family of chaperones |
|
| YML043C | 1.49 |
RRN11
|
Protein required for rDNA transcription by RNA polymerase I, component of the core factor (CF) of rDNA transcription factor, which also contains Rrn6p and Rrn7p |
|
| YOR101W | 1.47 |
RAS1
|
GTPase involved in G-protein signaling in the adenylate cyclase activating pathway, plays a role in cell proliferation; localized to the plasma membrane; homolog of mammalian RAS proto-oncogenes |
|
| YOL124C | 1.47 |
TRM11
|
Catalytic subunit of an adoMet-dependent tRNA methyltransferase complex (Trm11p-Trm112p), required for the methylation of the guanosine nucleotide at position 10 (m2G10) in tRNAs; contains a THUMP domain and a methyltransferase domain |
|
| YJL106W | 1.46 |
IME2
|
Serine/threonine protein kinase involved in activation of meiosis, associates with Ime1p and mediates its stability, activates Ndt80p; IME2 expression is positively regulated by Ime1p |
|
| YFL022C | 1.45 |
FRS2
|
Alpha subunit of cytoplasmic phenylalanyl-tRNA synthetase, forms a tetramer with Frs1p to form active enzyme; evolutionarily distant from mitochondrial phenylalanyl-tRNA synthetase based on protein sequence, but substrate binding is similar |
|
| YGL097W | 1.37 |
SRM1
|
Nucleotide exchange factor for Gsp1p, localizes to the nucleus, required for nucleocytoplasmic trafficking of macromolecules; suppressor of the pheromone response pathway; potentially phosphorylated by Cdc28p |
|
| YER146W | 1.35 |
LSM5
|
Lsm (Like Sm) protein; part of heteroheptameric complexes (Lsm2p-7p and either Lsm1p or 8p): cytoplasmic Lsm1p complex involved in mRNA decay; nuclear Lsm8p complex part of U6 snRNP and possibly involved in processing tRNA, snoRNA, and rRNA |
|
| YNL118C | 1.33 |
DCP2
|
Catalytic subunit of the Dcp1p-Dcp2p decapping enzyme complex, which removes the 5' cap structure from mRNAs prior to their degradation; member of the Nudix hydrolase family |
|
| YPR043W | 1.30 |
RPL43A
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Bp and has similarity to rat L37a ribosomal protein; null mutation confers a dominant lethal phenotype |
|
| YLR355C | 1.29 |
ILV5
|
Acetohydroxyacid reductoisomerase, mitochondrial protein involved in branched-chain amino acid biosynthesis, also required for maintenance of wild-type mitochondrial DNA and found in mitochondrial nucleoids |
|
| YMR123W | 1.27 |
PKR1
|
V-ATPase assembly factor, functions with other V-ATPase assembly factors in the ER to efficiently assemble the V-ATPase membrane sector (V0); overproduction confers resistance to Pichia farinosa killer toxin |
|
| YDR060W | 1.26 |
MAK21
|
Constituent of 66S pre-ribosomal particles, required for large (60S) ribosomal subunit biogenesis; involved in nuclear export of pre-ribosomes; required for maintenance of dsRNA virus; homolog of human CAATT-binding protein |
|
| YOR157C | 1.26 |
PUP1
|
Endopeptidase with trypsin-like activity that cleaves after basic residues; beta 2 subunit of 20S proteasome, synthesized as a proprotein before being proteolytically processed for assembly into 20S particle; human homolog is subunit Z |
|
| YBL085W | 1.26 |
BOI1
|
Protein implicated in polar growth, functionally redundant with Boi2p; interacts with bud-emergence protein Bem1p; contains an SH3 (src homology 3) domain and a PH (pleckstrin homology) domain |
|
| YMR184W | 1.24 |
ADD37
|
Protein of unknown function involved in ER-associated protein degradation; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and is induced in response to the DNA-damaging agent MMS; YMR184W is not an essential gene |
|
| YHL015W | 1.20 |
RPS20
|
Protein component of the small (40S) ribosomal subunit; overproduction suppresses mutations affecting RNA polymerase III-dependent transcription; has similarity to E. coli S10 and rat S20 ribosomal proteins |
|
| YKR075C | 1.18 |
Protein of unknown function; similar to YOR062Cp and Reg1p; expression regulated by glucose and Rgt1p; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YDR041W | 1.15 |
RSM10
|
Mitochondrial ribosomal protein of the small subunit, has similarity to E. coli S10 ribosomal protein; essential for viability, unlike most other mitoribosomal proteins |
|
| YKL063C | 1.14 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the Golgi |
||
| YNL113W | 1.13 |
RPC19
|
RNA polymerase subunit, common to RNA polymerases I and III |
|
| YPL075W | 1.11 |
GCR1
|
Transcriptional activator of genes involved in glycolysis; DNA-binding protein that interacts and functions with the transcriptional activator Gcr2p |
|
| YOR318C | 1.09 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; transcript is predicted to be spliced but there is no evidence that it is spliced in vivo |
||
| YOR047C | 1.09 |
STD1
|
Protein involved in control of glucose-regulated gene expression; interacts with protein kinase Snf1p, glucose sensors Snf3p and Rgt2p, and TATA-binding protein Spt15p; acts as a regulator of the transcription factor Rgt1p |
|
| YIL053W | 1.08 |
RHR2
|
Constitutively expressed isoform of DL-glycerol-3-phosphatase; involved in glycerol biosynthesis, induced in response to both anaerobic and, along with the Hor2p/Gpp2p isoform, osmotic stress |
|
| YFR051C | 1.05 |
RET2
|
Delta subunit of the coatomer complex (COPI), which coats Golgi-derived transport vesicles; involved in retrograde transport between Golgi and ER |
|
| YKR094C | 1.05 |
RPL40B
|
Fusion protein, identical to Rpl40Ap, that is cleaved to yield ubiquitin and a ribosomal protein of the large (60S) ribosomal subunit with similarity to rat L40; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes |
|
| YDL048C | 1.04 |
STP4
|
Protein containing a Kruppel-type zinc-finger domain; has similarity to Stp1p, Stp2p, and Stp3p |
|
| YDR040C | 1.04 |
ENA1
|
P-type ATPase sodium pump, involved in Na+ and Li+ efflux to allow salt tolerance |
|
| YGR108W | 1.03 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YMR001C-A | 1.02 |
Putative protein of unknown function |
||
| YGR145W | 1.02 |
ENP2
|
Essential nucleolar protein of unknown function; contains WD repeats, interacts with Mpp10p and Bfr2p, and has homology to Spb1p |
|
| YEL053W-A | 1.01 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YEL054C |
||
| YKL110C | 1.01 |
KTI12
|
Protein that plays a role, with Elongator complex, in modification of wobble nucleosides in tRNA; involved in sensitivity to G1 arrest induced by zymocin; interacts with chromatin throughout the genome; also interacts with Cdc19p |
|
| YLR333C | 1.00 |
RPS25B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps25Ap and has similarity to rat S25 ribosomal protein |
|
| YBR049C | 0.99 |
REB1
|
RNA polymerase I enhancer binding protein; DNA binding protein which binds to genes transcribed by both RNA polymerase I and RNA polymerase II; required for termination of RNA polymerase I transcription |
|
| YOR369C | 0.98 |
RPS12
|
Protein component of the small (40S) ribosomal subunit; has similarity to rat ribosomal protein S12 |
|
| YGR195W | 0.97 |
SKI6
|
3'-to-5' phosphorolytic exoribonuclease that is a subunit of the exosome; required for 3' processing of the 5.8S rRNA; involved in 3' to 5' mRNA degradation and translation inhibition of non-poly(A) mRNAs |
|
| YBL077W | 0.95 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene ILS1/YBL076C |
||
| YKL122C | 0.94 |
SRP21
|
Subunit of the signal recognition particle (SRP), which functions in protein targeting to the endoplasmic reticulum membrane; not found in mammalian SRP; forms a pre-SRP structure in the nucleolus that is translocated to the cytoplasm |
|
| YJL200C | 0.93 |
ACO2
|
Putative mitochondrial aconitase isozyme; similarity to Aco1p, an aconitase required for the TCA cycle; expression induced during growth on glucose, by amino acid starvation via Gcn4p, and repressed on ethanol |
|
| YOR299W | 0.92 |
BUD7
|
Member of the ChAPs family of proteins (Chs5p-Arf1p-binding proteins: Bch1p, Bch2p, Bud7p, Chs6p), that forms the exomer complex with Chs5p to mediate export of specific cargo proteins, including Chs3p, from the Golgi to the plasma membrane |
|
| YDL085C-A | 0.92 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YNL065W | 0.92 |
AQR1
|
Plasma membrane multidrug transporter of the major facilitator superfamily, confers resistance to short-chain monocarboxylic acids and quinidine; involved in the excretion of excess amino acids |
|
| YGR245C | 0.92 |
SDA1
|
Highly conserved nuclear protein required for actin cytoskeleton organization and passage through Start, plays a critical role in G1 events, binds Nap1p, also involved in 60S ribosome biogenesis |
|
| YML121W | 0.91 |
GTR1
|
Cytoplasmic GTP binding protein and negative regulator of the Ran/Tc4 GTPase cycle; component of GSE complex, which is required for sorting of Gap1p; involved in phosphate transport and telomeric silencing; similar to human RagA and RagB |
|
| YGL070C | 0.91 |
RPB9
|
RNA polymerase II subunit B12.6; contacts DNA; mutations affect transcription start site; involved in telomere maintenance |
|
| YIL056W | 0.88 |
VHR1
|
Transcriptional activator, required for the vitamin H-responsive element (VHRE) mediated induction of VHT1 (Vitamin H transporter) and BIO5 (biotin biosynthesis intermediate transporter) in response to low biotin concentrations |
|
| YLL021W | 0.88 |
SPA2
|
Component of the polarisome, which functions in actin cytoskeletal organization during polarized growth; acts as a scaffold for Mkk1p and Mpk1p cell wall integrity signaling components; potential Cdc28p substrate |
|
| YLR196W | 0.87 |
PWP1
|
Protein with WD-40 repeats involved in rRNA processing; associates with trans-acting ribosome biogenesis factors; similar to beta-transducin superfamily |
|
| YLR249W | 0.87 |
YEF3
|
Translational elongation factor 3, stimulates the binding of aminoacyl-tRNA (AA-tRNA) to ribosomes by releasing EF-1 alpha from the ribosomal complex; contains two ABC cassettes; binds and hydrolyses ATP |
|
| YBR002C | 0.87 |
RER2
|
Cis-prenyltransferase involved in dolichol synthesis; participates in endoplasmic reticulum (ER) protein sorting |
|
| YOL012C | 0.87 |
HTZ1
|
Histone variant H2AZ, exchanged for histone H2A in nucleosomes by the SWR1 complex; involved in transcriptional regulation through prevention of the spread of silent heterochromatin |
|
| YMR016C | 0.86 |
SOK2
|
Nuclear protein that plays a regulatory role in the cyclic AMP (cAMP)-dependent protein kinase (PKA) signal transduction pathway; negatively regulates pseudohyphal differentiation; homologous to several transcription factors |
|
| YER130C | 0.86 |
Hypothetical protein |
||
| YNL111C | 0.86 |
CYB5
|
Cytochrome b5, involved in the sterol and lipid biosynthesis pathways; acts as an electron donor to support sterol C5-6 desaturation |
|
| YIL148W | 0.85 |
RPL40A
|
Fusion protein, identical to Rpl40Bp, that is cleaved to yield ubiquitin and a ribosomal protein of the large (60S) ribosomal subunit with similarity to rat L40; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes |
|
| YPL055C | 0.85 |
LGE1
|
Protein of unknown function; null mutant forms abnormally large cells, and homozygous diploid null mutant displays delayed premeiotic DNA synthesis and reduced efficiency of meiotic nuclear division |
|
| YFL014W | 0.85 |
HSP12
|
Plasma membrane localized protein that protects membranes from desiccation; induced by heat shock, oxidative stress, osmostress, stationary phase entry, glucose depletion, oleate and alcohol; regulated by the HOG and Ras-Pka pathways |
|
| YOR025W | 0.85 |
HST3
|
Member of the Sir2 family of NAD(+)-dependent protein deacetylases; involved along with Hst4p in telomeric silencing, cell cycle progression, radiation resistance, genomic stability and short-chain fatty acid metabolism |
|
| YER019C-A | 0.85 |
SBH2
|
Ssh1p-Sss1p-Sbh2p complex component, involved in protein translocation into the endoplasmic reticulum; homologous to Sbh1p |
|
| YLR401C | 0.84 |
DUS3
|
Dihydrouridine synthase, member of a widespread family of conserved proteins including Smm1p, Dus1p, and Dus4p; contains a consensus oleate response element (ORE) in its promoter region |
|
| YLR358C | 0.84 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF RSC2/YLR357W |
||
| YER156C | 0.84 |
Putative protein of unknown function; identified as interacting with Hsp82p in a high-throughput two-hybrid screen; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YDL173W | 0.84 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YDL173W is not an essential gene |
||
| YJL198W | 0.83 |
PHO90
|
Low-affinity phosphate transporter; deletion of pho84, pho87, pho89, pho90, and pho91 causes synthetic lethality; transcription independent of Pi and Pho4p activity; overexpression results in vigorous growth |
|
| YOR046C | 0.83 |
DBP5
|
Cytoplasmic ATP-dependent RNA helicase of the DEAD-box family involved in mRNA export from the nucleus; involved in translation termination |
|
| YLR254C | 0.83 |
NDL1
|
Homolog of nuclear distribution factor NudE, NUDEL; interacts with Pac1p and regulates dynein targeting to microtubule plus ends |
|
| YJR076C | 0.83 |
CDC11
|
Component of the septin ring of the mother-bud neck that is required for cytokinesis; septins recruit proteins to the neck and can act as a barrier to diffusion at the membrane, and they comprise the 10nm filaments seen with EM |
|
| YDR156W | 0.83 |
RPA14
|
RNA polymerase I subunit A14 |
|
| YDR144C | 0.83 |
MKC7
|
GPI-anchored aspartyl protease (yapsin) involved in protein processing; shares functions with Yap3p and Kex2p |
|
| YDL031W | 0.82 |
DBP10
|
Putative ATP-dependent RNA helicase of the DEAD-box protein family, constituent of 66S pre-ribosomal particles; essential protein involved in ribosome biogenesis |
|
| YBR017C | 0.82 |
KAP104
|
Transportin, cytosolic karyopherin beta 2 involved in delivery of heterogeneous nuclear ribonucleoproteins to the nucleoplasm, binds rg-nuclear localization signals on Nab2p and Hrp1p, plays a role in cell-cycle progression |
|
| YHR127W | 0.82 |
Protein of unknown function; localizes to the nucleus |
||
| YPL160W | 0.82 |
CDC60
|
Cytosolic leucyl tRNA synthetase, ligates leucine to the appropriate tRNA |
|
| YIR012W | 0.82 |
SQT1
|
Essential protein involved in a late step of 60S ribosomal subunit assembly or modification; contains multiple WD repeats; interacts with Qsr1p in a two-hybrid assay |
|
| YML119W | 0.80 |
Putative protein of unknown funtion; YML119W is not an essential gene; potential Cdc28p substrate |
||
| YDR147W | 0.80 |
EKI1
|
Ethanolamine kinase, primarily responsible for phosphatidylethanolamine synthesis via the CDP-ethanolamine pathway; exhibits some choline kinase activity, thus contributing to phosphatidylcholine synthesis via the CDP-choline pathway |
|
| YPL238C | 0.80 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; partially overlaps 5' end of the verified essential gene SUI3/YPL237W |
||
| YJL194W | 0.80 |
CDC6
|
Essential ATP-binding protein required for DNA replication, component of the pre-replicative complex (pre-RC) which requires ORC to associate with chromatin and is in turn required for Mcm2-7p DNA association; homologous to S. pombe Cdc18p |
|
| YDL148C | 0.80 |
NOP14
|
Nucleolar protein, forms a complex with Noc4p that mediates maturation and nuclear export of 40S ribosomal subunits; also present in the small subunit processome complex, which is required for processing of pre-18S rRNA |
|
| YFR055W | 0.79 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YOR271C | 0.79 |
FSF1
|
Putative protein, predicted to be an alpha-isopropylmalate carrier; belongs to the sideroblastic-associated protein family; non-tagged protein is detected in purified mitochondria; likely to play a role in iron homeostasis |
|
| YGR280C | 0.78 |
PXR1
|
Essential protein involved in rRNA and snoRNA maturation; competes with TLC1 RNA for binding to Est2p, suggesting a role in negative regulation of telomerase; human homolog inhibits telomerase; contains a G-patch RNA interacting domain |
|
| YLL012W | 0.77 |
YEH1
|
Steryl ester hydrolase, one of three gene products (Yeh1p, Yeh2p, Tgl1p) responsible for steryl ester hydrolase activity and involved in sterol homeostasis; localized to lipid particle membranes |
|
| YAL025C | 0.77 |
MAK16
|
Essential nuclear protein, constituent of 66S pre-ribosomal particles; required for maturation of 25S and 5.8S rRNAs; required for maintenance of M1 satellite double-stranded RNA of the L-A virus |
|
| YPL212C | 0.77 |
PUS1
|
tRNA:pseudouridine synthase, introduces pseudouridines at positions 26-28, 34-36, 65, and 67 of tRNA; nuclear protein that appears to be involved in tRNA export; also acts on U2 snRNA |
|
| YKL118W | 0.77 |
Dubious open reading frame, unlikely to encode a protein; partially overlaps the verified gene VPH2 |
||
| YMR214W | 0.76 |
SCJ1
|
One of several homologs of bacterial chaperone DnaJ, located in the ER lumen where it cooperates with Kar2p to mediate maturation of proteins |
|
| YPR163C | 0.76 |
TIF3
|
Translation initiation factor eIF-4B, has RNA annealing activity; contains an RNA recognition motif and binds to single-stranded RNA |
|
| YOR073W-A | 0.76 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially/completely overlaps the verified ORF CDC21/YOR074C; identified by RT-PCR |
||
| YOL085C | 0.75 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOL085W-A |
||
| YNL301C | 0.74 |
RPL18B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl18Ap and has similarity to rat L18 ribosomal protein |
|
| YER159C | 0.74 |
BUR6
|
Subunit of a heterodimeric NC2 transcription regulator complex with Ncb2p; complex binds to TBP and can repress transcription by preventing preinitiation complex assembly or stimulate activated transcription; homologous to human NC2alpha |
|
| YMR121C | 0.74 |
RPL15B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl15Ap and has similarity to rat L15 ribosomal protein; binds to 5.8 S rRNA |
|
| YKL182W | 0.73 |
FAS1
|
Beta subunit of fatty acid synthetase, which catalyzes the synthesis of long-chain saturated fatty acids; contains acetyltransacylase, dehydratase, enoyl reductase, malonyl transacylase, and palmitoyl transacylase activities |
|
| YIL009W | 0.73 |
FAA3
|
Long chain fatty acyl-CoA synthetase, has a preference for C16 and C18 fatty acids; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery |
|
| YOR361C | 0.72 |
PRT1
|
Subunit of the core complex of translation initiation factor 3(eIF3), essential for translation; part of a subcomplex (Prt1p-Rpg1p-Nip1p) that stimulates binding of mRNA and tRNA(i)Met to ribosomes |
|
| YDL213C | 0.72 |
NOP6
|
Putative RNA-binding protein implicated in ribosome biogenesis; contains an RNA recognition motif (RRM) and has similarity to hydrophilins; NOP6 may be a fungal-specific gene as no homologs have been yet identified in higher eukaryotes |
|
| YGR078C | 0.72 |
PAC10
|
Part of the heteromeric co-chaperone GimC/prefoldin complex, which promotes efficient protein folding |
|
| YLR406C | 0.71 |
RPL31B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl31Ap and has similarity to rat L31 ribosomal protein; associates with the karyopherin Sxm1p |
|
| YKL072W | 0.71 |
STB6
|
Protein that binds Sin3p in a two-hybrid assay |
|
| YHR148W | 0.70 |
IMP3
|
Component of the SSU processome, which is required for pre-18S rRNA processing, essential protein that interacts with Mpp10p and mediates interactions of Imp4p and Mpp10p with U3 snoRNA |
|
| YDR299W | 0.70 |
BFR2
|
Essential protein possibly involved in secretion; multicopy suppressor of sensitivity to Brefeldin A |
|
| YER064C | 0.70 |
Non-essential nuclear protein; null mutation has global effects on transcription |
||
| YGL179C | 0.69 |
TOS3
|
Protein kinase, related to and functionally redundant with Elm1p and Sak1p for the phosphorylation and activation of Snf1p; functionally orthologous to LKB1, a mammalian kinase associated with Peutz-Jeghers cancer-susceptibility syndrome |
|
| YJL115W | 0.69 |
ASF1
|
Nucleosome assembly factor, involved in chromatin assembly and disassembly, anti-silencing protein that causes derepression of silent loci when overexpressed; plays a role in regulating Ty1 transposition |
|
| YOR287C | 0.69 |
Putative protein of unknown function; may play a role in the ribosome and rRNA biosynthesis based on expression profiles and mutant phenotype |
||
| YDL192W | 0.69 |
ARF1
|
ADP-ribosylation factor, GTPase of the Ras superfamily involved in regulation of coated vesicle formation in intracellular trafficking within the Golgi; functionally interchangeable with Arf2p |
|
| YBL032W | 0.69 |
HEK2
|
RNA binding protein with similarity to hnRNP-K that localizes to the cytoplasm and to subtelomeric DNA; required for the proper localization of ASH1 mRNA; involved in the regulation of telomere position effect and telomere length |
|
| YDL208W | 0.68 |
NHP2
|
Nuclear protein related to mammalian high mobility group (HMG) proteins, essential for function of H/ACA-type snoRNPs, which are involved in 18S rRNA processing |
|
| YER151C | 0.68 |
UBP3
|
Ubiquitin-specific protease that interacts with Bre5p to co-regulate anterograde and retrograde transport between endoplasmic reticulum and Golgi compartments; inhibitor of gene silencing; cleaves ubiquitin fusions but not polyubiquitin |
|
| YKL119C | 0.68 |
VPH2
|
Integral membrane protein required for vacuolar H+-ATPase (V-ATPase) function, although not an actual component of the V-ATPase complex; functions in the assembly of the V-ATPase; localized to the endoplasmic reticulum (ER) |
|
| YOR074C | 0.68 |
CDC21
|
Thymidylate synthase, required for de novo biosynthesis of pyrimidine deoxyribonucleotides; expression is induced at G1/S |
|
| YBR034C | 0.68 |
HMT1
|
Nuclear SAM-dependent mono- and asymmetric arginine dimethylating methyltransferase that modifies hnRNPs, including Npl3p and Hrp1p, thus facilitating nuclear export of these proteins; required for viability of npl3 mutants |
|
| YOR276W | 0.68 |
CAF20
|
Phosphoprotein of the mRNA cap-binding complex involved in translational control, repressor of cap-dependent translation initiation, competes with eIF4G for binding to eIF4E |
|
| YNL079C | 0.67 |
TPM1
|
Major isoform of tropomyosin; binds to and stabilizes actin cables and filaments, which direct polarized cell growth and the distribution of several organelles; acetylated by the NatB complex and acetylated form binds actin most efficiently |
|
| YHR203C | 0.67 |
RPS4B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps4Ap and has similarity to rat S4 ribosomal protein |
|
| YMR015C | 0.67 |
ERG5
|
C-22 sterol desaturase, a cytochrome P450 enzyme that catalyzes the formation of the C-22(23) double bond in the sterol side chain in ergosterol biosynthesis; may be a target of azole antifungal drugs |
|
| YHR146W | 0.67 |
CRP1
|
Protein that binds to cruciform DNA structures |
|
| YDL226C | 0.67 |
GCS1
|
ADP-ribosylation factor GTPase activating protein (ARF GAP), involved in ER-Golgi transport; shares functional similarity with Glo3p |
|
| YDR086C | 0.67 |
SSS1
|
Subunit of the Sec61p translocation complex (Sec61p-Sss1p-Sbh1p) that forms a channel for passage of secretory proteins through the endoplasmic reticulum membrane, and of the Ssh1p complex (Ssh1p-Sbh2p-Sss1p); interacts with Ost4p and Wbp1p |
|
| YBR257W | 0.66 |
POP4
|
Subunit of both RNase MRP, which cleaves pre-rRNA, and nuclear RNase P, which cleaves tRNA precursors to generate mature 5' ends; binds to the RPR1 RNA subunit in Rnase P |
|
| YJL076W | 0.66 |
NET1
|
Core subunit of the RENT complex, which is a complex involved in nucleolar silencing and telophase exit; stimulates transcription by RNA polymerase I and regulates nucleolar structure |
|
| YOR063W | 0.66 |
RPL3
|
Protein component of the large (60S) ribosomal subunit, has similarity to E. coli L3 and rat L3 ribosomal proteins; involved in the replication and maintenance of killer double stranded RNA virus |
|
| YIL117C | 0.65 |
PRM5
|
Pheromone-regulated protein, predicted to have 1 transmembrane segment; induced during cell integrity signaling |
|
| YMR091C | 0.65 |
NPL6
|
Component of the RSC chromatin remodeling complex; interacts with Rsc3p, Rsc30p, Ldb7p, and Htl1p to form a module important for a broad range of RSC functions; involved in nuclear protein import and maintenance of proper telomere length |
|
| YGR079W | 0.65 |
Putative protein of unknown function; YGR079W is not an essential gene |
||
| YBR171W | 0.65 |
SEC66
|
Non-essential subunit of Sec63 complex (Sec63p, Sec62p, Sec66p and Sec72p); with Sec61 complex, Kar2p/BiP and Lhs1p forms a channel competent for SRP-dependent and post-translational SRP-independent protein targeting and import into the ER |
|
| YPR142C | 0.65 |
Hypothetical protein; open reading frame overlaps 5' end of essential RRP15 gene required for ribosomal RNA processing |
||
| YCR051W | 0.65 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; contains ankyrin (Ank) repeats; YCR051W is not an essential gene |
||
| YOR198C | 0.65 |
BFR1
|
Component of mRNP complexes associated with polyribosomes; implicated in secretion and nuclear segregation; multicopy suppressor of BFA (Brefeldin A) sensitivity |
|
| YFR056C | 0.65 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YPR157W | 0.65 |
Putative protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YGR285C | 0.65 |
ZUO1
|
Cytosolic ribosome-associated chaperone that acts, together with Ssz1p and the Ssb proteins, as a chaperone for nascent polypeptide chains; contains a DnaJ domain and functions as a J-protein partner for Ssb1p and Ssb2p |
|
| YDR412W | 0.64 |
RRP17
|
Component of the pre-60S pre-ribosomal particle; required for cell viability |
|
| YJR092W | 0.64 |
BUD4
|
Protein involved in bud-site selection and required for axial budding pattern; localizes with septins to bud neck in mitosis and may constitute an axial landmark for next round of budding; potential Cdc28p substrate |
|
| YDR101C | 0.64 |
ARX1
|
Shuttling pre-60S factor; involved in the biogenesis of ribosomal large subunit biogenesis; interacts directly with Alb1; responsible for Tif6 recycling defects in absence of Rei1; associated with the ribosomal export complex |
|
| YGL120C | 0.64 |
PRP43
|
RNA helicase in the DEAH-box family, functions in both RNA polymerase I and polymerase II transcript metabolism, involved in release of the lariat-intron from the spliceosome |
|
| YKR035W-A | 0.64 |
DID2
|
Class E protein of the vacuolar protein-sorting (Vps) pathway; binds Vps4p and directs it to dissociate ESCRT-III complexes; forms a functional and physical complex with Ist1p; human ortholog may be altered in breast tumors |
|
| YJR003C | 0.64 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YOL038W | 0.63 |
PRE6
|
Alpha 4 subunit of the 20S proteasome; may replace alpha 3 subunit (Pre9p) under stress conditions to create a more active proteasomal isoform; GFP-fusion protein relocates from cytosol to the mitochondrial surface upon oxidative stress |
|
| YGL177W | 0.63 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YML001W | 0.63 |
YPT7
|
GTPase; GTP-binding protein of the rab family; required for homotypic fusion event in vacuole inheritance, for endosome-endosome fusion, similar to mammalian Rab7 |
|
| YNL216W | 0.63 |
RAP1
|
DNA-binding protein involved in either activation or repression of transcription, depending on binding site context; also binds telomere sequences and plays a role in telomeric position effect (silencing) and telomere structure |
|
| YLR206W | 0.63 |
ENT2
|
Epsin-like protein required for endocytosis and actin patch assembly and functionally redundant with Ent1p; contains clathrin-binding motif at C-terminus |
|
| YDR207C | 0.63 |
UME6
|
Key transcriptional regulator of early meiotic genes, binds URS1 upstream regulatory sequence, couples metabolic responses to nutritional cues with initiation and progression of meiosis, forms complex with Ime1p, and also with Sin3p-Rpd3p |
|
| YGR060W | 0.63 |
ERG25
|
C-4 methyl sterol oxidase, catalyzes the first of three steps required to remove two C-4 methyl groups from an intermediate in ergosterol biosynthesis; mutants accumulate the sterol intermediate 4,4-dimethylzymosterol |
|
| YGR052W | 0.63 |
FMP48
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YOR319W | 0.62 |
HSH49
|
U2-snRNP associated splicing factor with similarity to the mammalian splicing factor SAP49; proposed to function as a U2-snRNP assembly factor along with Hsh155p and binding partner Cus1p; contains two RNA recognition motifs (RRM) |
|
| YFR037C | 0.62 |
RSC8
|
Component of the RSC chromatin remodeling complex; essential for viability and mitotic growth; homolog of SWI/SNF subunit Swi3p, but unlike Swi3p, does not activate transcription of reporters |
|
| YMR106C | 0.62 |
YKU80
|
Subunit of the telomeric Ku complex (Yku70p-Yku80p), involved in telomere length maintenance, structure and telomere position effect; relocates to sites of double-strand cleavage to promote nonhomologous end joining during DSB repair |
|
| YGL148W | 0.62 |
ARO2
|
Bifunctional chorismate synthase and flavin reductase, catalyzes the conversion of 5-enolpyruvylshikimate 3-phosphate (EPSP) to form chorismate, which is a precursor to aromatic amino acids |
|
| YBR143C | 0.62 |
SUP45
|
Polypeptide release factor involved in translation termination; mutant form acts as a recessive omnipotent suppressor |
|
| YDR450W | 0.62 |
RPS18A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps18Bp and has similarity to E. coli S13 and rat S18 ribosomal proteins |
|
| YGR051C | 0.62 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YGR051C is not an essential gene |
||
| YPL237W | 0.61 |
SUI3
|
Beta subunit of the translation initiation factor eIF2, involved in the identification of the start codon; proposed to be involved in mRNA binding |
|
| YOL031C | 0.61 |
SIL1
|
Nucleotide exchange factor for the endoplasmic reticulum (ER) lumenal Hsp70 chaperone Kar2p, required for protein translocation into the ER; homolog of Yarrowia lipolytica SLS1; GrpE-like protein |
|
| YPR051W | 0.61 |
MAK3
|
Catalytic subunit of N-terminal acetyltransferase of the NatC type; required for replication of dsRNA virus |
|
| YBR130C | 0.60 |
SHE3
|
Protein that acts as an adaptor between Myo4p and the She2p-mRNA complex; part of the mRNA localization machinery that restricts accumulation of certain proteins to the bud; also required for cortical ER inheritance |
|
| YJL105W | 0.60 |
SET4
|
Protein of unknown function, contains a SET domain |
|
| YML082W | 0.60 |
Putative protein predicted to have carbon-sulfur lyase activity; green fluorescent protein (GFP)-fusion protein localizes to the nucleus and the cytoplasm; YNML082W is not an essential gene |
||
| YLR033W | 0.60 |
RSC58
|
Component of the RSC chromatin remodeling complex; RSC functions in transcriptional regulation and elongation, chromosome stability, and establishing sister chromatid cohesion; involved in telomere maintenance58KDa subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of REB1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 0.6 | 5.4 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.6 | 1.7 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.6 | 2.8 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin structure organization(GO:0031032) actomyosin contractile ring organization(GO:0044837) |
| 0.5 | 2.0 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.5 | 1.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.5 | 1.4 | GO:0060548 | negative regulation of apoptotic process(GO:0043066) negative regulation of programmed cell death(GO:0043069) negative regulation of cell death(GO:0060548) |
| 0.5 | 1.9 | GO:0048313 | Golgi inheritance(GO:0048313) |
| 0.5 | 2.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.5 | 3.2 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.5 | 2.7 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) |
| 0.4 | 3.1 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 0.4 | 0.4 | GO:0001178 | regulation of transcriptional start site selection at RNA polymerase II promoter(GO:0001178) |
| 0.4 | 1.3 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.4 | 2.1 | GO:0070649 | parallel actin filament bundle assembly(GO:0030046) formin-nucleated actin cable assembly(GO:0070649) |
| 0.4 | 1.7 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.4 | 2.9 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.4 | 2.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.4 | 1.2 | GO:0009177 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.4 | 1.1 | GO:0070637 | nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) |
| 0.4 | 1.1 | GO:0031384 | regulation of initiation of mating projection growth(GO:0031384) |
| 0.3 | 1.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 0.3 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.3 | 1.0 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.3 | 1.0 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.3 | 1.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.3 | 1.2 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.3 | 1.5 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.3 | 1.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 1.2 | GO:0071216 | cellular response to biotic stimulus(GO:0071216) response to cell cycle checkpoint signaling(GO:0072396) response to mitotic cell cycle checkpoint signaling(GO:0072414) response to spindle checkpoint signaling(GO:0072417) response to mitotic spindle checkpoint signaling(GO:0072476) response to mitotic cell cycle spindle assembly checkpoint signaling(GO:0072479) response to spindle assembly checkpoint signaling(GO:0072485) negative regulation of protein import into nucleus during spindle assembly checkpoint(GO:1901925) |
| 0.3 | 0.9 | GO:0016093 | polyprenol metabolic process(GO:0016093) polyprenol biosynthetic process(GO:0016094) dolichol metabolic process(GO:0019348) dolichol biosynthetic process(GO:0019408) |
| 0.3 | 0.6 | GO:0061422 | positive regulation of transcription from RNA polymerase II promoter in response to alkaline pH(GO:0061422) |
| 0.3 | 0.5 | GO:0071044 | histone mRNA metabolic process(GO:0008334) histone mRNA catabolic process(GO:0071044) |
| 0.3 | 1.1 | GO:0046898 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 0.3 | 0.8 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) snRNA modification(GO:0040031) |
| 0.2 | 0.7 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 1.0 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) |
| 0.2 | 0.2 | GO:2000879 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 0.2 | 4.3 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.2 | 0.7 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.2 | 7.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 0.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 3.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 0.4 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) |
| 0.2 | 0.7 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.2 | 0.2 | GO:0032186 | cellular bud neck septin ring organization(GO:0032186) |
| 0.2 | 3.0 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 1.3 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.2 | 0.8 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.2 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 0.4 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.2 | 0.2 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.2 | 5.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 1.2 | GO:0043242 | negative regulation of protein complex disassembly(GO:0043242) |
| 0.2 | 1.0 | GO:0032845 | negative regulation of telomere maintenance(GO:0032205) negative regulation of homeostatic process(GO:0032845) |
| 0.2 | 0.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.2 | 0.6 | GO:0001015 | snoRNA transcription from an RNA polymerase II promoter(GO:0001015) |
| 0.2 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 4.6 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 2.4 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.2 | 0.7 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.2 | 2.9 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 1.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 0.7 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 | 0.9 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.2 | 3.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 0.9 | GO:0070481 | nuclear-transcribed mRNA catabolic process, non-stop decay(GO:0070481) |
| 0.2 | 0.9 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.2 | 0.3 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.2 | 3.0 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.2 | 0.6 | GO:0097271 | protein localization to bud neck(GO:0097271) |
| 0.2 | 2.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.2 | 1.1 | GO:0000743 | nuclear migration involved in conjugation with cellular fusion(GO:0000743) |
| 0.2 | 0.3 | GO:0042992 | negative regulation of transcription factor import into nucleus(GO:0042992) |
| 0.2 | 0.8 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.2 | 0.6 | GO:0007535 | donor selection(GO:0007535) |
| 0.2 | 0.6 | GO:0009423 | chorismate biosynthetic process(GO:0009423) |
| 0.2 | 0.5 | GO:0019419 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.2 | 0.6 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.2 | 0.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 0.9 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.7 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.1 | 4.6 | GO:0098876 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.1 | 1.0 | GO:0042991 | transcription factor import into nucleus(GO:0042991) |
| 0.1 | 1.6 | GO:0045860 | positive regulation of protein kinase activity(GO:0045860) |
| 0.1 | 1.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 1.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.6 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 1.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.4 | GO:0034517 | ribophagy(GO:0034517) |
| 0.1 | 0.4 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.4 | GO:0032204 | regulation of telomere maintenance(GO:0032204) regulation of telomere maintenance via telomerase(GO:0032210) regulation of telomere maintenance via telomere lengthening(GO:1904356) regulation of DNA biosynthetic process(GO:2000278) |
| 0.1 | 0.5 | GO:0015883 | FAD transport(GO:0015883) |
| 0.1 | 0.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.8 | GO:0043489 | RNA stabilization(GO:0043489) mRNA stabilization(GO:0048255) |
| 0.1 | 0.4 | GO:2000001 | regulation of cell cycle checkpoint(GO:1901976) negative regulation of cell cycle checkpoint(GO:1901977) regulation of DNA damage checkpoint(GO:2000001) negative regulation of DNA damage checkpoint(GO:2000002) negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.1 | 0.3 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.1 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.8 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.1 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.3 | GO:0043270 | positive regulation of ion transport(GO:0043270) |
| 0.1 | 0.7 | GO:0048309 | endoplasmic reticulum inheritance(GO:0048309) |
| 0.1 | 13.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 2.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.2 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.1 | 1.2 | GO:0000947 | amino acid catabolic process to alcohol via Ehrlich pathway(GO:0000947) |
| 0.1 | 0.4 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 3.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.6 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 1.2 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.1 | 1.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.8 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 16.6 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.1 | 0.7 | GO:0031134 | sister chromatid biorientation(GO:0031134) |
| 0.1 | 1.5 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) nucleus localization(GO:0051647) |
| 0.1 | 1.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.0 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.8 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 2.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.9 | GO:0032042 | mitochondrial DNA metabolic process(GO:0032042) |
| 0.1 | 0.3 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 0.1 | 0.5 | GO:0009071 | serine family amino acid catabolic process(GO:0009071) |
| 0.1 | 1.1 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 0.3 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 2.3 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.1 | 1.1 | GO:0090662 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 0.4 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 0.3 | GO:0034661 | rRNA catabolic process(GO:0016075) ncRNA catabolic process(GO:0034661) |
| 0.1 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.4 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.1 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.1 | 0.4 | GO:0000296 | spermine transport(GO:0000296) |
| 0.1 | 0.8 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 0.3 | GO:0009106 | lipoate metabolic process(GO:0009106) lipoate biosynthetic process(GO:0009107) protein lipoylation(GO:0009249) |
| 0.1 | 0.5 | GO:0031081 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.1 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.5 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 0.4 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 0.2 | GO:0061412 | positive regulation of transcription from RNA polymerase II promoter in response to amino acid starvation(GO:0061412) |
| 0.1 | 0.8 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.1 | 1.4 | GO:0007020 | microtubule nucleation(GO:0007020) microtubule polymerization(GO:0046785) |
| 0.1 | 0.7 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 0.6 | GO:0045014 | negative regulation of transcription from RNA polymerase II promoter by glucose(GO:0000433) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 1.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.2 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.1 | 0.2 | GO:0019673 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 0.3 | GO:0060277 | obsolete negative regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0060277) |
| 0.1 | 0.1 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 0.4 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.1 | 0.2 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.3 | GO:0019218 | regulation of steroid metabolic process(GO:0019218) regulation of ergosterol biosynthetic process(GO:0032443) regulation of steroid biosynthetic process(GO:0050810) |
| 0.1 | 0.1 | GO:1903726 | negative regulation of phospholipid biosynthetic process(GO:0071072) negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.1 | 0.5 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.6 | GO:0007155 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.1 | 2.5 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 0.2 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.1 | 0.2 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.3 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 0.4 | GO:1903338 | regulation of cell wall organization or biogenesis(GO:1903338) |
| 0.1 | 0.2 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 0.2 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.1 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.8 | GO:0044396 | actin cortical patch assembly(GO:0000147) actin cortical patch organization(GO:0044396) |
| 0.1 | 0.9 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.5 | GO:0016126 | steroid biosynthetic process(GO:0006694) sterol biosynthetic process(GO:0016126) |
| 0.1 | 1.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.2 | GO:0006032 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.1 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.3 | GO:0010833 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) telomere maintenance via telomere lengthening(GO:0010833) |
| 0.0 | 0.3 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0031382 | mating projection assembly(GO:0031382) |
| 0.0 | 1.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0009262 | deoxyribonucleotide metabolic process(GO:0009262) deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.3 | GO:0070096 | mitochondrial outer membrane translocase complex assembly(GO:0070096) |
| 0.0 | 0.3 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0048211 | Golgi vesicle budding(GO:0048194) Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.3 | GO:0031570 | DNA integrity checkpoint(GO:0031570) |
| 0.0 | 0.1 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.3 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.0 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.0 | GO:1902749 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.1 | GO:0030308 | negative regulation of cell growth(GO:0030308) negative regulation of pseudohyphal growth(GO:2000221) |
| 0.0 | 1.1 | GO:0032200 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) anatomical structure homeostasis(GO:0060249) |
| 0.0 | 1.7 | GO:0007346 | regulation of mitotic cell cycle(GO:0007346) |
| 0.0 | 0.3 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 1.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.0 | GO:0033567 | defense response(GO:0006952) DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 | 0.1 | GO:0046039 | GTP biosynthetic process(GO:0006183) GTP metabolic process(GO:0046039) |
| 0.0 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.4 | GO:0016125 | steroid metabolic process(GO:0008202) sterol metabolic process(GO:0016125) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.0 | 0.1 | GO:0071825 | lipid tube assembly(GO:0060988) protein-lipid complex assembly(GO:0065005) protein-lipid complex subunit organization(GO:0071825) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine metabolic process(GO:0006047) UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.4 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.1 | GO:0016073 | snRNA metabolic process(GO:0016073) |
| 0.0 | 0.4 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.1 | GO:0000730 | meiotic DNA recombinase assembly(GO:0000707) DNA recombinase assembly(GO:0000730) |
| 0.0 | 0.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 0.2 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0006038 | cell wall chitin biosynthetic process(GO:0006038) fungal-type cell wall chitin biosynthetic process(GO:0034221) fungal-type cell wall polysaccharide biosynthetic process(GO:0051278) cell wall polysaccharide biosynthetic process(GO:0070592) fungal-type cell wall polysaccharide metabolic process(GO:0071966) |
| 0.0 | 0.2 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.0 | 0.2 | GO:0010526 | negative regulation of transposition, RNA-mediated(GO:0010526) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0035376 | sterol import(GO:0035376) |
| 0.0 | 0.6 | GO:1902408 | cellular bud site selection(GO:0000282) cytokinesis, site selection(GO:0007105) mitotic cytokinesis, site selection(GO:1902408) |
| 0.0 | 0.1 | GO:0043007 | maintenance of rDNA(GO:0043007) |
| 0.0 | 0.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0023058 | adaptation of signaling pathway by response to pheromone involved in conjugation with cellular fusion(GO:0000754) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.1 | GO:0090294 | nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) nitrogen catabolite activation of transcription(GO:0090294) |
| 0.0 | 0.2 | GO:0015918 | sterol transport(GO:0015918) |
| 0.0 | 0.1 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.0 | 0.1 | GO:0051666 | actin cortical patch localization(GO:0051666) |
| 0.0 | 0.0 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.0 | GO:0000196 | MAPK cascade involved in cell wall organization or biogenesis(GO:0000196) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) regulation of cellular amine catabolic process(GO:0033241) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.1 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.1 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.4 | GO:0051783 | regulation of nuclear division(GO:0051783) |
| 0.0 | 0.4 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.0 | 0.5 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.0 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.0 | GO:0043419 | urea catabolic process(GO:0043419) |
| 0.0 | 0.2 | GO:0006541 | glutamine metabolic process(GO:0006541) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.7 | 2.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.5 | 1.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.5 | 1.5 | GO:0071261 | Ssh1 translocon complex(GO:0071261) |
| 0.5 | 2.4 | GO:0030689 | Noc complex(GO:0030689) |
| 0.5 | 1.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.4 | 1.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.4 | 2.0 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.4 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.4 | 3.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.4 | 1.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.4 | 1.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.4 | 1.4 | GO:0070860 | RNA polymerase I core factor complex(GO:0070860) |
| 0.3 | 1.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.3 | 1.0 | GO:0005827 | polar microtubule(GO:0005827) tubulin complex(GO:0045298) |
| 0.3 | 1.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 1.0 | GO:0030874 | nucleolar chromatin(GO:0030874) |
| 0.3 | 1.8 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.3 | 4.2 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.3 | 0.9 | GO:0051286 | cell tip(GO:0051286) |
| 0.3 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.3 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 0.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 1.1 | GO:0005822 | inner plaque of spindle pole body(GO:0005822) |
| 0.3 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 3.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 1.7 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.2 | 1.4 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.2 | 1.6 | GO:0033180 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 0.4 | GO:0000229 | cytoplasmic chromosome(GO:0000229) mitochondrial chromosome(GO:0000262) |
| 0.2 | 1.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 2.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 0.4 | GO:0032545 | CURI complex(GO:0032545) |
| 0.2 | 1.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 11.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 0.9 | GO:0034044 | exomer complex(GO:0034044) |
| 0.2 | 0.5 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.2 | 0.9 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 3.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 0.7 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 0.9 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.2 | 7.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 0.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.2 | 1.0 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 15.4 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.2 | 0.7 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) Sec62/Sec63 complex(GO:0031207) |
| 0.2 | 0.6 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.2 | 0.9 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.2 | 0.6 | GO:0005724 | nuclear heterochromatin(GO:0005720) nuclear telomeric heterochromatin(GO:0005724) telomeric heterochromatin(GO:0031933) |
| 0.2 | 0.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 0.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 0.3 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.2 | 1.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.9 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.4 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 2.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 1.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.5 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.1 | 0.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 2.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 4.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.5 | GO:0030869 | RENT complex(GO:0030869) |
| 0.1 | 0.5 | GO:0032221 | Rpd3S complex(GO:0032221) |
| 0.1 | 0.4 | GO:0033254 | vacuolar transporter chaperone complex(GO:0033254) |
| 0.1 | 0.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 0.8 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 10.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 0.6 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.5 | GO:0000136 | alpha-1,6-mannosyltransferase complex(GO:0000136) |
| 0.1 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.6 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 0.3 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.9 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 1.6 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.4 | GO:0032126 | eisosome(GO:0032126) |
| 0.1 | 0.6 | GO:0090544 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.1 | 0.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 1.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.1 | GO:0071819 | DUBm complex(GO:0071819) |
| 0.1 | 0.5 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) |
| 0.1 | 0.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.9 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) vacuolar proton-transporting V-type ATPase complex(GO:0016471) proton-transporting V-type ATPase complex(GO:0033176) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.2 | GO:0044462 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 0.1 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0030907 | MBF transcription complex(GO:0030907) |
| 0.0 | 6.0 | GO:0005935 | cellular bud neck(GO:0005935) |
| 0.0 | 0.3 | GO:0070772 | PAS complex(GO:0070772) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0031518 | CBF3 complex(GO:0031518) |
| 0.0 | 1.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.2 | GO:0035361 | Cul8-RING ubiquitin ligase complex(GO:0035361) |
| 0.0 | 0.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.0 | 0.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.0 | 0.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.2 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.5 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.2 | GO:0033698 | Rpd3L complex(GO:0033698) |
| 0.0 | 0.5 | GO:0005933 | cellular bud(GO:0005933) |
| 0.0 | 0.6 | GO:0061645 | actin cortical patch(GO:0030479) endocytic patch(GO:0061645) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.1 | GO:0000818 | MIS12/MIND type complex(GO:0000444) nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.3 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.0 | 0.0 | GO:0000124 | SAGA complex(GO:0000124) SAGA-type complex(GO:0070461) |
| 0.0 | 0.7 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.0 | 0.6 | GO:0000785 | chromatin(GO:0000785) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.6 | 2.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.5 | 2.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.5 | 2.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.5 | 1.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.5 | 1.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.5 | 1.4 | GO:0001169 | transcription factor activity, RNA polymerase I CORE element sequence-specific binding(GO:0001169) transcription factor activity, RNA polymerase I CORE element binding transcription factor recruiting(GO:0001187) |
| 0.5 | 5.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.4 | 3.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.4 | 1.8 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.3 | 2.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.3 | 0.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 2.3 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.3 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 2.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.3 | 0.8 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.3 | 1.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 0.8 | GO:0097617 | RNA strand annealing activity(GO:0033592) annealing activity(GO:0097617) |
| 0.3 | 3.0 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.2 | 0.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.2 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.2 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.2 | 0.7 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 0.9 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 1.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 1.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.2 | 6.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.2 | 1.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 4.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.2 | 0.8 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 0.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 2.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 1.2 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.2 | 2.0 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.2 | 1.0 | GO:0008186 | RNA-dependent ATPase activity(GO:0008186) |
| 0.2 | 0.8 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 0.8 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.2 | 5.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.2 | 0.6 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.2 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 0.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 0.9 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.2 | 2.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 3.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.4 | GO:0001179 | RNA polymerase I transcription factor binding(GO:0001179) transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.1 | 2.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 1.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 8.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.5 | GO:0043141 | 5'-3' DNA helicase activity(GO:0043139) ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.5 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.1 | 1.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.8 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 1.0 | GO:0015081 | sodium ion transmembrane transporter activity(GO:0015081) |
| 0.1 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.3 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 0.4 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 0.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.5 | GO:0000772 | mating pheromone activity(GO:0000772) receptor binding(GO:0005102) pheromone activity(GO:0005186) |
| 0.1 | 1.2 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 4.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.6 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 3.0 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 6.0 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.6 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.3 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 1.2 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 1.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.4 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.1 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 2.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) thiol oxidase activity(GO:0016972) |
| 0.1 | 0.4 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 15.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.5 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 0.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 1.6 | GO:0019001 | GTP binding(GO:0005525) guanyl nucleotide binding(GO:0019001) guanyl ribonucleotide binding(GO:0032561) |
| 0.1 | 0.9 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 1.0 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.1 | 0.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.3 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 0.6 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.1 | 0.9 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 0.5 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
| 0.1 | 0.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 1.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 0.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.5 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.1 | 0.5 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.1 | 0.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.7 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.1 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 3.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 0.5 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0032934 | steroid binding(GO:0005496) sterol binding(GO:0032934) |
| 0.0 | 0.9 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.2 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.0 | GO:0043178 | alcohol binding(GO:0043178) |
| 0.0 | 0.4 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 5.6 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.4 | GO:0019205 | nucleobase-containing compound kinase activity(GO:0019205) |
| 0.0 | 0.1 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.0 | 0.7 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.5 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.2 | GO:0016791 | phosphatase activity(GO:0016791) |
| 0.0 | 0.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) |
| 0.0 | 0.0 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.5 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 2.1 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.3 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 0.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.1 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.3 | 1.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 0.9 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 0.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 0.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.2 | 0.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.5 | 1.5 | REACTOME SIGNALLING TO ERKS | Genes involved in Signalling to ERKs |
| 0.4 | 1.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.4 | 1.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.4 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.3 | 2.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 0.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 1.6 | REACTOME DEADENYLATION DEPENDENT MRNA DECAY | Genes involved in Deadenylation-dependent mRNA decay |
| 0.1 | 0.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.4 | REACTOME TRANSCRIPTION | Genes involved in Transcription |
| 0.1 | 0.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.2 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.2 | REACTOME METABOLISM OF RNA | Genes involved in Metabolism of RNA |
| 0.1 | 0.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME MITOTIC G1 G1 S PHASES | Genes involved in Mitotic G1-G1/S phases |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |