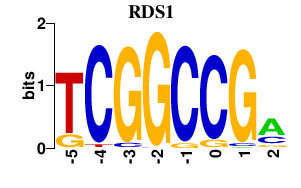
Results for RDS1
Z-value: 0.73
Transcription factors associated with RDS1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RDS1
|
S000000703 | Putative zinc cluster transcription factor |
Activity-expression correlation:
Activity profile of RDS1 motif
Sorted Z-values of RDS1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YCL025C | 3.54 |
AGP1
|
Low-affinity amino acid permease with broad substrate range, involved in uptake of asparagine, glutamine, and other amino acids; expression is regulated by the SPS plasma membrane amino acid sensor system (Ssy1p-Ptr3p-Ssy5p) |
|
| YKL217W | 2.90 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YML089C | 2.36 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression induced by calcium shortage |
||
| YJL089W | 2.35 |
SIP4
|
C6 zinc cluster transcriptional activator that binds to the carbon source-responsive element (CSRE) of gluconeogenic genes; involved in the positive regulation of gluconeogenesis; regulated by Snf1p protein kinase; localized to the nucleus |
|
| YBR230C | 2.16 |
OM14
|
Integral mitochondrial outer membrane protein; abundance is decreased in cells grown in glucose relative to other carbon sources; appears to contain 3 alpha-helical transmembrane segments; ORF encodes a 97-basepair intron |
|
| YDL210W | 2.12 |
UGA4
|
Permease that serves as a gamma-aminobutyrate (GABA) transport protein involved in the utilization of GABA as a nitrogen source; catalyzes the transport of putrescine and delta-aminolevulinic acid (ALA); localized to the vacuolar membrane |
|
| YDR536W | 2.11 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YJL133C-A | 1.99 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YOL131W | 1.89 |
Putative protein of unknown function |
||
| YGR236C | 1.75 |
SPG1
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YML042W | 1.55 |
CAT2
|
Carnitine acetyl-CoA transferase present in both mitochondria and peroxisomes, transfers activated acetyl groups to carnitine to form acetylcarnitine which can be shuttled across membranes |
|
| YAL054C | 1.53 |
ACS1
|
Acetyl-coA synthetase isoform which, along with Acs2p, is the nuclear source of acetyl-coA for histone acetlyation; expressed during growth on nonfermentable carbon sources and under aerobic conditions |
|
| YML090W | 1.52 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YML089C; exhibits growth defect on a non-fermentable (respiratory) carbon source |
||
| YNL144C | 1.52 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YNL144C is not an essential gene |
||
| YPR065W | 1.48 |
ROX1
|
Heme-dependent repressor of hypoxic genes; contains an HMG domain that is responsible for DNA bending activity |
|
| YNL179C | 1.46 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YLR296W | 1.44 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR127W | 1.41 |
Putative protein of unknown function; expression is activated by transcription factor YRM1/YOR172W; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and the nucleus |
||
| YKL016C | 1.39 |
ATP7
|
Subunit d of the stator stalk of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YNL180C | 1.37 |
RHO5
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely involved in protein kinase C (Pkc1p)-dependent signal transduction pathway that controls cell integrity |
|
| YPR036W-A | 1.37 |
Protein of unknown function; transcription is regulated by Pdr1p |
||
| YBL015W | 1.34 |
ACH1
|
Acetyl-coA hydrolase, primarily localized to mitochondria; phosphorylated; required for acetate utilization and for diploid pseudohyphal growth |
|
| YNR002C | 1.30 |
ATO2
|
Putative transmembrane protein involved in export of ammonia, a starvation signal that promotes cell death in aging colonies; phosphorylated in mitochondria; member of the TC 9.B.33 YaaH family; homolog of Ady2p and Y. lipolytica Gpr1p |
|
| YLR295C | 1.27 |
ATP14
|
Subunit h of the F0 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YJR078W | 1.25 |
BNA2
|
Putative tryptophan 2,3-dioxygenase or indoleamine 2,3-dioxygenase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p and Aft2p |
|
| YML091C | 1.25 |
RPM2
|
Protein subunit of mitochondrial RNase P, has roles in nuclear transcription, cytoplasmic and mitochondrial RNA processing, and mitochondrial translation; distributed to mitochondria, cytoplasmic processing bodies, and the nucleus |
|
| YMR194C-B | 1.20 |
Putative protein of unknown function |
||
| YAL039C | 1.18 |
CYC3
|
Cytochrome c heme lyase (holocytochrome c synthase), attaches heme to apo-cytochrome c (Cyc1p or Cyc7p) in the mitochondrial intermembrane space; human ortholog may have a role in microphthalmia with linear skin defects (MLS) |
|
| YKL031W | 1.15 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species |
||
| YNL305C | 1.12 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YNL305C is not an essential gene |
||
| YLR327C | 1.09 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YPL017C | 1.06 |
IRC15
|
Putative S-adenosylmethionine-dependent methyltransferase of the seven beta-strand family, required for accurate meiotic chromosome segregation; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YMR272C | 1.03 |
SCS7
|
Sphingolipid alpha-hydroxylase, functions in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids, has both cytochrome b5-like and hydroxylase/desaturase domains, not essential for growth |
|
| YNL142W | 1.02 |
MEP2
|
Ammonium permease involved in regulation of pseudohyphal growth; belongs to a ubiquitous family of cytoplasmic membrane proteins that transport only ammonium (NH4+); expression is under the nitrogen catabolite repression regulation |
|
| YBR201C-A | 1.00 |
Putative protein of unknown function |
||
| YGL184C | 0.99 |
STR3
|
Cystathionine beta-lyase, converts cystathionine into homocysteine |
|
| YMR107W | 0.99 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YNL143C | 0.97 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR030W | 0.96 |
CSR2
|
Nuclear protein with a potential regulatory role in utilization of galactose and nonfermentable carbon sources; overproduction suppresses the lethality at high temperature of a chs5 spa2 double null mutation; potential Cdc28p substrate |
|
| YOR152C | 0.94 |
Putative protein of unknown function; has no similarity to any known protein; YOR152C is not an essential gene |
||
| YDR530C | 0.93 |
APA2
|
Diadenosine 5',5''-P1,P4-tetraphosphate phosphorylase II (AP4A phosphorylase), involved in catabolism of bis(5'-nucleosidyl) tetraphosphates; has similarity to Apa1p |
|
| YKL109W | 0.92 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YAR035W | 0.92 |
YAT1
|
Outer mitochondrial carnitine acetyltransferase, minor ethanol-inducible enzyme involved in transport of activated acyl groups from the cytoplasm into the mitochondrial matrix; phosphorylated |
|
| YJL088W | 0.90 |
ARG3
|
Ornithine carbamoyltransferase (carbamoylphosphate:L-ornithine carbamoyltransferase), catalyzes the sixth step in the biosynthesis of the arginine precursor ornithine |
|
| YDR379C-A | 0.89 |
Hypothetical protein identified by homology. See FEBS Letters [2000] 487:31-36. |
||
| YJL045W | 0.89 |
Minor succinate dehydrogenase isozyme; homologous to Sdh1p, the major isozyme reponsible for the oxidation of succinate and transfer of electrons to ubiquinone; induced during the diauxic shift in a Cat8p-dependent manner |
||
| YFL024C | 0.88 |
EPL1
|
Component of NuA4, which is an essential histone H4/H2A acetyltransferase complex; homologous to Drosophila Enhancer of Polycomb |
|
| YKR009C | 0.83 |
FOX2
|
Multifunctional enzyme of the peroxisomal fatty acid beta-oxidation pathway; has 3-hydroxyacyl-CoA dehydrogenase and enoyl-CoA hydratase activities |
|
| YKR049C | 0.81 |
FMP46
|
Putative redox protein containing a thioredoxin fold; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YHR217C | 0.79 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; located in the telomeric region TEL08R. |
||
| YDR059C | 0.79 |
UBC5
|
Ubiquitin-conjugating enzyme that mediates selective degradation of short-lived and abnormal proteins, central component of the cellular stress response; expression is heat inducible |
|
| YJR077C | 0.77 |
MIR1
|
Mitochondrial phosphate carrier, imports inorganic phosphate into mitochondria; functionally redundant with Pic2p but more abundant than Pic2p under normal conditions; phosphorylated |
|
| YPL018W | 0.77 |
CTF19
|
Outer kinetochore protein, required for accurate mitotic chromosome segregation; component of the kinetochore sub-complex COMA (Ctf19p, Okp1p, Mcm21p, Ame1p) that functions as a platform for kinetochore assembly |
|
| YFL021W | 0.77 |
GAT1
|
Transcriptional activator of genes involved in nitrogen catabolite repression; contains a GATA-1-type zinc finger DNA-binding motif; activity and localization regulated by nitrogen limitation and Ure2p |
|
| YNL055C | 0.76 |
POR1
|
Mitochondrial porin (voltage-dependent anion channel), outer membrane protein required for the maintenance of mitochondrial osmotic stability and mitochondrial membrane permeability; phosphorylated |
|
| YOR072W-A | 0.75 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YOR072W; originally identified by fungal homology and RT-PCR |
||
| YDR231C | 0.75 |
COX20
|
Mitochondrial inner membrane protein, required for proteolytic processing of Cox2p and its assembly into cytochrome c oxidase |
|
| YBR230W-A | 0.75 |
Putative protein of unknown function |
||
| YOR065W | 0.74 |
CYT1
|
Cytochrome c1, component of the mitochondrial respiratory chain; expression is regulated by the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex |
|
| YMR195W | 0.73 |
ICY1
|
Protein of unknown function, required for viability in rich media of cells lacking mitochondrial DNA; mutants have an invasive growth defect with elongated morphology; induced by amino acid starvation |
|
| YPL054W | 0.71 |
LEE1
|
Zinc-finger protein of unknown function |
|
| YOR072W | 0.71 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOR072W-A; diploid deletion strains are methotrexate, paraquat and wortmannin sensitive |
||
| YFL062W | 0.70 |
COS4
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YLR004C | 0.70 |
THI73
|
Putative plasma membrane permease proposed to be involved in carboxylic acid uptake and repressed by thiamine; substrate of Dbf2p/Mob1p kinase; transcription is altered if mitochondrial dysfunction occurs |
|
| YOR071C | 0.69 |
NRT1
|
High-affinity nicotinamide riboside transporter; also transports thiamine with low affinity; shares sequence similarity with Thi7p and Thi72p; proposed to be involved in 5-fluorocytosine sensitivity |
|
| YLR297W | 0.69 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YLR297W is not an essential gene; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YFL064C | 0.69 |
Putative protein of unknown function |
||
| YPR192W | 0.69 |
AQY1
|
Spore-specific water channel that mediates the transport of water across cell membranes, developmentally controlled; may play a role in spore maturation, probably by allowing water outflow, may be involved in freeze tolerance |
|
| YGR067C | 0.68 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YMR280C | 0.67 |
CAT8
|
Zinc cluster transcriptional activator necessary for derepression of a variety of genes under non-fermentative growth conditions, active after diauxic shift, binds carbon source responsive elements |
|
| YHL036W | 0.66 |
MUP3
|
Low affinity methionine permease, similar to Mup1p |
|
| YER053C | 0.66 |
PIC2
|
Mitochondrial phosphate carrier, imports inorganic phosphate into mitochondria; functionally redundant with Mir1p but less abundant than Mir1p under normal conditions; expression is induced at high temperature |
|
| YFL063W | 0.66 |
Dubious open reading frame, based on available experimental and comparative sequence data |
||
| YDR406W | 0.66 |
PDR15
|
Plasma membrane ATP binding cassette (ABC) transporter, multidrug transporter and general stress response factor implicated in cellular detoxification; regulated by Pdr1p, Pdr3p and Pdr8p; promoter contains a PDR responsive element |
|
| YPL201C | 0.65 |
YIG1
|
Protein that interacts with glycerol 3-phosphatase and plays a role in anaerobic glycerol production; localizes to the nucleus and cytosol |
|
| YJR045C | 0.65 |
SSC1
|
Mitochondrial matrix ATPase, subunit of the presequence translocase-associated protein import motor (PAM) and of SceI endonuclease; involved in protein folding and translocation into the matrix; phosphorylated; member of HSP70 family |
|
| YOR140W | 0.65 |
SFL1
|
Transcriptional repressor and activator; involved in repression of flocculation-related genes, and activation of stress responsive genes; negatively regulated by cAMP-dependent protein kinase A subunit Tpk2p |
|
| YDR374C | 0.65 |
Putative protein of unknown function |
||
| YNL336W | 0.62 |
COS1
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YPR007C | 0.62 |
REC8
|
Meiosis-specific component of sister chromatid cohesion complex; maintains cohesion between sister chromatids during meiosis I; maintains cohesion between centromeres of sister chromatids until meiosis II; homolog of S. pombe Rec8p |
|
| YBR050C | 0.61 |
REG2
|
Regulatory subunit of the Glc7p type-1 protein phosphatase; involved with Reg1p, Glc7p, and Snf1p in regulation of glucose-repressible genes, also involved in glucose-induced proteolysis of maltose permease |
|
| YHR009C | 0.61 |
Putative protein of unknown function; not an essential gene |
||
| YIL125W | 0.61 |
KGD1
|
Component of the mitochondrial alpha-ketoglutarate dehydrogenase complex, which catalyzes a key step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation of alpha-ketoglutarate to form succinyl-CoA |
|
| YER091C-A | 0.61 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGL258W-A | 0.61 |
Putative protein of unknown function |
||
| YOR211C | 0.61 |
MGM1
|
Mitochondrial GTPase related to dynamin, present in a complex containing Ugo1p and Fzo1p; required for normal morphology of cristae and for stability of Tim11p; homolog of human OPA1 involved in autosomal dominant optic atrophy |
|
| YPL061W | 0.60 |
ALD6
|
Cytosolic aldehyde dehydrogenase, activated by Mg2+ and utilizes NADP+ as the preferred coenzyme; required for conversion of acetaldehyde to acetate; constitutively expressed; locates to the mitochondrial outer surface upon oxidative stress |
|
| YPL135W | 0.60 |
ISU1
|
Conserved protein of the mitochondrial matrix, performs a scaffolding function during assembly of iron-sulfur clusters, interacts physically and functionally with yeast frataxin (Yfh1p); isu1 isu2 double mutant is inviable |
|
| YOR139C | 0.60 |
Hypothetical protein |
||
| YPR027C | 0.60 |
Putative protein of unknown function |
||
| YOR084W | 0.59 |
LPX1
|
Oleic acid-inducible, peroxisomal matrix localized lipase; transcriptionally activated by Yrm1p along with genes involved in multidrug resistance; peroxisomal import is dependent on the PTS1 receptor, Pex5p and on self-interaction |
|
| YBR051W | 0.59 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partiallly overlaps the REG2/YBR050C regulatory subunit of the Glc7p type-1 protein phosphatase |
||
| YHR139C | 0.59 |
SPS100
|
Protein required for spore wall maturation; expressed during sporulation; may be a component of the spore wall |
|
| YNL013C | 0.58 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF HEF3/YNL014W |
||
| YML132W | 0.58 |
COS3
|
Protein involved in salt resistance; interacts with sodium:hydrogen antiporter Nha1p; member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YKL032C | 0.56 |
IXR1
|
Protein that binds DNA containing intrastrand cross-links formed by cisplatin, contains two HMG (high mobility group box) domains, which confer the ability to bend cisplatin-modified DNA; mediates aerobic transcriptional repression of COX5b |
|
| YKL177W | 0.56 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene STE3 |
||
| YER162C | 0.56 |
RAD4
|
Protein that recognizes and binds damaged DNA (with Rad23p) during nucleotide excision repair; subunit of Nuclear Excision Repair Factor 2 (NEF2); homolog of human XPC protein |
|
| YOR338W | 0.55 |
Putative protein of unknown function; YOR338W transcription is regulated by Azf1p and its transcript is a specific target of the G protein effector Scp160p; identified as being required for sporulation in a high-throughput mutant screen |
||
| YBL078C | 0.55 |
ATG8
|
Conserved protein that is a component of autophagosomes and Cvt vesicles; undergoes C-terminal conjugation to phosphatidylethanolamine (PE), Atg8p-PE is anchored to membranes and may mediate membrane fusion during autophagosome formation |
|
| YDR247W | 0.55 |
VHS1
|
Cytoplasmic serine/threonine protein kinase; identified as a high-copy suppressor of the synthetic lethality of a sis2 sit4 double mutant, suggesting a role in G1/S phase progression; homolog of Sks1p |
|
| YDR259C | 0.54 |
YAP6
|
Putative basic leucine zipper (bZIP) transcription factor; overexpression increases sodium and lithium tolerance |
|
| YBR224W | 0.54 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene TDP1 |
||
| YGL208W | 0.54 |
SIP2
|
One of three beta subunits of the Snf1 serine/threonine protein kinase complex involved in the response to glucose starvation; null mutants exhibit accelerated aging; N-myristoylprotein localized to the cytoplasm and the plasma membrane |
|
| YDR232W | 0.53 |
HEM1
|
5-aminolevulinate synthase, catalyzes the first step in the heme biosynthetic pathway; an N-terminal signal sequence is required for localization to the mitochondrial matrix; expression is regulated by Hap2p-Hap3p |
|
| YER101C | 0.53 |
AST2
|
Protein that may have a role in targeting of plasma membrane [H+]ATPase (Pma1p) to the plasma membrane, as suggested by analysis of genetic interactions |
|
| YHL024W | 0.53 |
RIM4
|
Putative RNA-binding protein required for the expression of early and middle sporulation genes |
|
| YBL094C | 0.53 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps uncharacterized ORF YBL095W |
||
| YEL009C-A | 0.52 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR017W | 0.52 |
SPO20
|
Meiosis-specific subunit of the t-SNARE complex, required for prospore membrane formation during sporulation; similar to but not functionally redundant with Sec9p; SNAP-25 homolog |
|
| YOL025W | 0.52 |
LAG2
|
Protein involved in determination of longevity; LAG2 gene is preferentially expressed in young cells; overexpression extends the mean and maximum life span of cells |
|
| YOR100C | 0.51 |
CRC1
|
Mitochondrial inner membrane carnitine transporter, required for carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria during fatty acid beta-oxidation |
|
| YNL036W | 0.51 |
NCE103
|
Carbonic anhydrase; poorly transcribed under aerobic conditions and at an undetectable level under anaerobic conditions; involved in non-classical protein export pathway |
|
| YDR010C | 0.51 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR138C | 0.50 |
RUP1
|
Protein involved in regulation of Rsp5p, which is an essential HECT ubiquitin ligase; required for binding of Rsp5p to Ubp2p; contains an UBA domain |
|
| YNL160W | 0.50 |
YGP1
|
Cell wall-related secretory glycoprotein; induced by nutrient deprivation-associated growth arrest and upon entry into stationary phase; may be involved in adaptation prior to stationary phase entry; has similarity to Sps100p |
|
| YLL019C | 0.50 |
KNS1
|
Nonessential putative protein kinase of unknown cellular role; member of the LAMMER family of protein kinases, which are serine/threonine kinases also capable of phosphorylating tyrosine residues |
|
| YPL200W | 0.50 |
CSM4
|
Protein required for accurate chromosome segregation during meiosis |
|
| YBR105C | 0.50 |
VID24
|
Peripheral membrane protein located at Vid (vacuole import and degradation) vesicles; regulates fructose-1,6-bisphosphatase (FBPase) targeting to the vacuole; involved in proteasome-dependent catabolite degradation of FBPase |
|
| YLR193C | 0.49 |
UPS1
|
Mitochondrial intermembrane space protein that regulates alternative processing and sorting of Mgm1p and other proteins; required for normal mitochondrial morphology; ortholog of human PRELI |
|
| YJR048W | 0.49 |
CYC1
|
Cytochrome c, isoform 1; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YOR220W | 0.48 |
RCN2
|
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and is induced in response to the DNA-damaging agent MMS |
|
| YFR034W-A | 0.48 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YFR035C; identified by gene-trapping, microarray expression analysis, and genome-wide homology searching |
||
| YAR019W-A | 0.48 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YKL093W | 0.47 |
MBR1
|
Protein involved in mitochondrial functions and stress response; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants |
|
| YDL199C | 0.47 |
Putative transporter, member of the sugar porter family |
||
| YDR269C | 0.47 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YEL010W | 0.47 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPR064W | 0.47 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YIL059C | 0.47 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YIL060W |
||
| YKL178C | 0.47 |
STE3
|
Receptor for a factor receptor, transcribed in alpha cells and required for mating by alpha cells, couples to MAP kinase cascade to mediate pheromone response; ligand bound receptors are endocytosed and recycled to the plasma membrane; GPCR |
|
| YIL033C | 0.46 |
BCY1
|
Regulatory subunit of the cyclic AMP-dependent protein kinase (PKA), a component of a signaling pathway that controls a variety of cellular processes, including metabolism, cell cycle, stress response, stationary phase, and sporulation |
|
| YKL163W | 0.46 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YBR250W | 0.46 |
SPO23
|
Protein of unknown function; associates with meiosis-specific protein Spo1p |
|
| YHR218W | 0.46 |
Helicase-like protein encoded within the telomeric Y' element |
||
| YDL174C | 0.46 |
DLD1
|
D-lactate dehydrogenase, oxidizes D-lactate to pyruvate, transcription is heme-dependent, repressed by glucose, and derepressed in ethanol or lactate; located in the mitochondrial inner membrane |
|
| YIR027C | 0.45 |
DAL1
|
Allantoinase, converts allantoin to allantoate in the first step of allantoin degradation; expression sensitive to nitrogen catabolite repression |
|
| YLR394W | 0.45 |
CST9
|
SUMO E3 ligase; required for synaptonemal complex formation; localizes to synapsis initiation sites on meiotic chromosomes; potential Cdc28p substrate |
|
| YBR117C | 0.45 |
TKL2
|
Transketolase, similar to Tkl1p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YER015W | 0.45 |
FAA2
|
Long chain fatty acyl-CoA synthetase; accepts a wider range of acyl chain lengths than Faa1p, preferring C9:0-C13:0; involved in the activation of endogenous pools of fatty acids |
|
| YOR316C-A | 0.45 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YBR223C | 0.45 |
TDP1
|
Tyrosyl-DNA Phosphodiesterase I, hydrolyzes 3'-phosphotyrosyl bonds to generate 3'-phosphate DNA and tyrosine, involved in the repair of DNA lesions created by topoisomerase I |
|
| YEL072W | 0.44 |
RMD6
|
Protein required for sporulation |
|
| YBR183W | 0.43 |
YPC1
|
Alkaline ceramidase that also has reverse (CoA-independent) ceramide synthase activity, catalyzes both breakdown and synthesis of phytoceramide; overexpression confers fumonisin B1 resistance |
|
| YDL181W | 0.42 |
INH1
|
Protein that inhibits ATP hydrolysis by the F1F0-ATP synthase; inhibitory function is enhanced by stabilizing proteins Stf1p and Stf2p; has similarity to Stf1p; has a calmodulin-binding motif and binds calmodulin in vitro |
|
| YKL067W | 0.42 |
YNK1
|
Nucleoside diphosphate kinase, catalyzes the transfer of gamma phosphates from nucleoside triphosphates, usually ATP, to nucleoside diphosphates by a mechanism that involves formation of an autophosphorylated enzyme intermediate |
|
| YDR359C | 0.42 |
EAF1
|
Component of the NuA4 histone acetyltransferase complex; required for initiation of pre-meiotic DNA replication, probably due to its requirement for significant expression of IME1 |
|
| YML133C | 0.42 |
Putative protein of unknown function with similarity to helicases; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YML133C contains an intron |
||
| YBR225W | 0.42 |
Putative protein of unknown function; non-essential gene identified in a screen for mutants affected in mannosylphophorylation of cell wall components |
||
| YGL163C | 0.41 |
RAD54
|
DNA-dependent ATPase, stimulates strand exchange by modifying the topology of double-stranded DNA; involved in the recombinational repair of double-strand breaks in DNA during vegetative growth and meiosis; member of the SWI/SNF family |
|
| YJL163C | 0.41 |
Putative protein of unknown function |
||
| YOR345C | 0.41 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene REV1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YDR461C-A | 0.41 |
Putative protein of unknown function |
||
| YOR346W | 0.41 |
REV1
|
Deoxycytidyl transferase, forms a complex with the subunits of DNA polymerase zeta, Rev3p and Rev7p; involved in repair of abasic sites in damaged DNA |
|
| YFR053C | 0.41 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YPL089C | 0.40 |
RLM1
|
MADS-box transcription factor, component of the protein kinase C-mediated MAP kinase pathway involved in the maintenance of cell integrity; phosphorylated and activated by the MAP-kinase Slt2p |
|
| YMR056C | 0.40 |
AAC1
|
Mitochondrial inner membrane ADP/ATP translocator, exchanges cytosolic ADP for mitochondrially synthesized ATP; phosphorylated; Aac1p is a minor isoform while Pet9p is the major ADP/ATP translocator |
|
| YNL125C | 0.40 |
ESBP6
|
Protein with similarity to monocarboxylate permeases, appears not to be involved in transport of monocarboxylates such as lactate, pyruvate or acetate across the plasma membrane |
|
| YJL152W | 0.40 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR270W | 0.39 |
CCC2
|
Cu(+2)-transporting P-type ATPase, required for export of copper from the cytosol into an extracytosolic compartment; has similarity to human proteins involved in Menkes and Wilsons diseases |
|
| YAR019C | 0.39 |
CDC15
|
Protein kinase of the Mitotic Exit Network that is localized to the spindle pole bodies at late anaphase; promotes mitotic exit by directly switching on the kinase activity of Dbf2p |
|
| YBR280C | 0.39 |
SAF1
|
F-Box protein involved in proteasome-dependent degradation of Aah1p during entry of cells into quiescence; interacts with Skp1 |
|
| YKL133C | 0.39 |
Putative protein of unknown function |
||
| YHR198C | 0.39 |
AIM18
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YAR023C | 0.39 |
Putative integral membrane protein, member of DUP240 gene family |
||
| YBR035C | 0.39 |
PDX3
|
Pyridoxine (pyridoxamine) phosphate oxidase, has homologs in E. coli and Myxococcus xanthus; transcription is under the general control of nitrogen metabolism |
|
| YLL029W | 0.38 |
FRA1
|
Protein involved in negative regulation of transcription of iron regulon; forms an iron independent complex with Fra2p, Grx3p, and Grx4p; cytosolic; mutant fails to repress transcription of iron regulon and is defective in spore formation |
|
| YMR256C | 0.38 |
COX7
|
Subunit VII of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain |
|
| YNL339C | 0.38 |
YRF1-6
|
Helicase encoded by the Y' element of subtelomeric regions, highly expressed in the mutants lacking the telomerase component TLC1; potentially phosphorylated by Cdc28p |
|
| YGR243W | 0.38 |
FMP43
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YHR199C | 0.38 |
AIM46
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YLR047C | 0.37 |
FRE8
|
Protein with sequence similarity to iron/copper reductases, involved in iron homeostasis; deletion mutant has iron deficiency/accumulation growth defects; expression increased in the absence of copper-responsive transcription factor Mac1p |
|
| YDR360W | 0.37 |
OPI7
|
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified gene VID21/YDR359C. |
|
| YFL052W | 0.37 |
Putative zinc cluster protein that contains a DNA binding domain; null mutant sensitive to calcofluor white, low osmolarity and heat, suggesting a role for YFL052Wp in cell wall integrity |
||
| YLR184W | 0.37 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDL222C | 0.36 |
FMP45
|
Integral membrane protein localized to mitochondria (untagged protein) and eisosomes, immobile patches at the cortex associated with endocytosis; sporulation and sphingolipid content are altered in mutants; has homologs SUR7 and YNL194C |
|
| YPL230W | 0.36 |
USV1
|
Putative transcription factor containing a C2H2 zinc finger; mutation affects transcriptional regulation of genes involved in protein folding, ATP binding, and cell wall biosynthesis |
|
| YDR092W | 0.36 |
UBC13
|
Ubiquitin-conjugating enzyme involved in the error-free DNA postreplication repair pathway; interacts with Mms2p to assemble ubiquitin chains at the Ub Lys-63 residue; DNA damage triggers redistribution from the cytoplasm to the nucleus |
|
| YLL055W | 0.36 |
YCT1
|
High-affinity cysteine-specific transporter with similarity to the Dal5p family of transporters; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; YCT1 is not an essential gene |
|
| YOR064C | 0.36 |
YNG1
|
Subunit of the NuA3 histone acetyltransferase complex that acetylates histone H3; contains PHD finger domain that interacts with methylated histone H3, has similarity to the human tumor suppressor ING1 |
|
| YKL171W | 0.36 |
Putative protein of unknown function; predicted protein kinase; implicated in proteasome function; epitope-tagged protein localizes to the cytoplasm |
||
| YHR082C | 0.36 |
KSP1
|
Nonessential putative serine/threonine protein kinase of unknown cellular role; overproduction causes allele-specific suppression of the prp20-10 mutation |
|
| YEL020C | 0.35 |
Hypothetical protein with low sequence identity to Pdc1p |
||
| YPL262W | 0.35 |
FUM1
|
Fumarase, converts fumaric acid to L-malic acid in the TCA cycle; cytosolic and mitochondrial localization determined by the N-terminal mitochondrial targeting sequence and protein conformation; phosphorylated in mitochondria |
|
| YBR019C | 0.34 |
GAL10
|
UDP-glucose-4-epimerase, catalyzes the interconversion of UDP-galactose and UDP-D-glucose in galactose metabolism; also catalyzes the conversion of alpha-D-glucose or alpha-D-galactose to their beta-anomers |
|
| YGL180W | 0.34 |
ATG1
|
Protein serine/threonine kinase required for vesicle formation in autophagy and the cytoplasm-to-vacuole targeting (Cvt) pathway; structurally required for pre-autophagosome formation; during autophagy forms a complex with Atg13p and Atg17p |
|
| YPR124W | 0.34 |
CTR1
|
High-affinity copper transporter of the plasma membrane, mediates nearly all copper uptake under low copper conditions; transcriptionally induced at low copper levels and degraded at high copper levels |
|
| YOR010C | 0.34 |
TIR2
|
Putative cell wall mannoprotein of the Srp1p/Tip1p family of serine-alanine-rich proteins; transcription is induced by cold shock and anaerobiosis |
|
| YBR016W | 0.34 |
Plasma membrane protein of unknown function; has similarity to hydrophilins, which are hydrophilic, glycine-rich proteins involved in the adaptive response to hyperosmotic conditions |
||
| YDL196W | 0.34 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; open reading frame is located in promoter region of essential gene SEC31 |
||
| YIL086C | 0.33 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YIL087C | 0.33 |
LRC2
|
Putative protein of unknown function; protein is detected in purified mitochondria in high-throughput studies; null mutant displays decreased mitochondrial genome loss (petite formation) and severe growth defect in minimal glycerol media |
|
| YNL196C | 0.33 |
SLZ1
|
Sporulation-specific protein with a leucine zipper motif |
|
| YFL036W | 0.33 |
RPO41
|
Mitochondrial RNA polymerase; single subunit enzyme similar to those of T3 and T7 bacteriophages; requires a specificity subunit encoded by MTF1 for promoter recognition |
|
| YJR115W | 0.33 |
Putative protein of unknown function |
||
| YGR194C | 0.33 |
XKS1
|
Xylulokinase, converts D-xylulose and ATP to xylulose 5-phosphate and ADP; rate limiting step in fermentation of xylulose; required for xylose fermentation by recombinant S. cerevisiae strains |
|
| YBL099W | 0.33 |
ATP1
|
Alpha subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; phosphorylated |
|
| YLL046C | 0.32 |
RNP1
|
Ribonucleoprotein that contains two RNA recognition motifs (RRM) |
|
| YIL058W | 0.32 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPL147W | 0.32 |
PXA1
|
Subunit of a heterodimeric peroxisomal ATP-binding cassette transporter complex (Pxa1p-Pxa2p), required for import of long-chain fatty acids into peroxisomes; similarity to human adrenoleukodystrophy transporter and ALD-related proteins |
Network of associatons between targets according to the STRING database.
First level regulatory network of RDS1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0061414 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 0.8 | 2.5 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.8 | 3.3 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.5 | 2.0 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.5 | 1.5 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.4 | 1.2 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.4 | 1.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.4 | 2.7 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.3 | 1.6 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.3 | 0.9 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.3 | 1.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 1.1 | GO:0032079 | positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.3 | 0.8 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.3 | 1.3 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.2 | 1.0 | GO:0019346 | transsulfuration(GO:0019346) |
| 0.2 | 0.7 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.2 | 0.6 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.2 | 0.8 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.2 | 1.0 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.2 | 0.6 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.2 | 0.4 | GO:0000101 | sulfur amino acid transport(GO:0000101) neutral amino acid transport(GO:0015804) |
| 0.2 | 0.7 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.2 | 0.7 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.5 | GO:0051469 | vesicle fusion with vacuole(GO:0051469) |
| 0.2 | 0.3 | GO:0006171 | cAMP biosynthetic process(GO:0006171) cyclic nucleotide biosynthetic process(GO:0009190) cyclic purine nucleotide metabolic process(GO:0052652) |
| 0.2 | 1.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 0.2 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.2 | 1.2 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.1 | 0.7 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.1 | 0.6 | GO:0045980 | negative regulation of nucleotide metabolic process(GO:0045980) |
| 0.1 | 0.4 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.1 | 1.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 2.7 | GO:0019740 | nitrogen utilization(GO:0019740) |
| 0.1 | 0.9 | GO:0009164 | nucleoside catabolic process(GO:0009164) glycosyl compound catabolic process(GO:1901658) |
| 0.1 | 0.4 | GO:0046039 | GTP biosynthetic process(GO:0006183) GTP metabolic process(GO:0046039) |
| 0.1 | 1.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.7 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.4 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 1.8 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.4 | GO:0010039 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.1 | 0.9 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 0.5 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.1 | 1.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.5 | GO:0043200 | response to amino acid(GO:0043200) |
| 0.1 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.4 | GO:0061416 | regulation of transcription from RNA polymerase II promoter in response to salt stress(GO:0061416) |
| 0.1 | 0.4 | GO:0010994 | free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.7 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.1 | 0.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.1 | 0.5 | GO:0051303 | meiotic telomere clustering(GO:0045141) establishment of chromosome localization(GO:0051303) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.1 | 0.2 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) regulation of membrane lipid metabolic process(GO:1905038) |
| 0.1 | 3.4 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.1 | 0.3 | GO:0070583 | spore membrane bending pathway(GO:0070583) |
| 0.1 | 0.6 | GO:0000255 | allantoin metabolic process(GO:0000255) allantoin catabolic process(GO:0000256) |
| 0.1 | 0.2 | GO:0072353 | cellular age-dependent response to reactive oxygen species(GO:0072353) |
| 0.1 | 0.6 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.1 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.4 | GO:0032105 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.1 | 0.6 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 1.5 | GO:0006101 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.1 | 0.3 | GO:2000911 | positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) positive regulation of sterol transport(GO:0032373) positive regulation of transmembrane transport(GO:0034764) regulation of sterol import by regulation of transcription from RNA polymerase II promoter(GO:0035968) positive regulation of sterol import by positive regulation of transcription from RNA polymerase II promoter(GO:0035969) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.1 | 0.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.4 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.1 | 0.2 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.1 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) positive regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061395) cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 0.3 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.1 | 0.8 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 | 0.4 | GO:0046184 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) aldehyde biosynthetic process(GO:0046184) |
| 0.1 | 0.1 | GO:0071475 | cellular response to salt stress(GO:0071472) cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 1.2 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 0.4 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) meiotic recombination checkpoint(GO:0051598) |
| 0.1 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.3 | GO:0070096 | mitochondrial outer membrane translocase complex assembly(GO:0070096) |
| 0.1 | 0.2 | GO:0015755 | fructose transport(GO:0015755) |
| 0.1 | 0.3 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.1 | GO:0051222 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 0.0 | 0.5 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.8 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.0 | 0.1 | GO:0061412 | cellular response to amino acid starvation(GO:0034198) positive regulation of transcription from RNA polymerase II promoter in response to amino acid starvation(GO:0061412) |
| 0.0 | 0.2 | GO:0006771 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.0 | 0.2 | GO:0015851 | nucleobase transport(GO:0015851) |
| 0.0 | 0.5 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.2 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 1.2 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) |
| 0.0 | 0.0 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) post-translational protein modification(GO:0043687) |
| 0.0 | 0.3 | GO:0006638 | neutral lipid metabolic process(GO:0006638) acylglycerol metabolic process(GO:0006639) triglyceride metabolic process(GO:0006641) |
| 0.0 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.3 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.4 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.0 | 0.3 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 0.0 | 0.1 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) |
| 0.0 | 0.1 | GO:0051101 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.0 | 0.4 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.0 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 1.2 | GO:0070726 | cell wall assembly(GO:0070726) |
| 0.0 | 0.2 | GO:0000755 | cytogamy(GO:0000755) |
| 0.0 | 0.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:0019543 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 0.0 | 0.1 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.0 | 0.1 | GO:0031062 | positive regulation of histone methylation(GO:0031062) regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 | 0.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.0 | 0.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0009065 | glutamine family amino acid catabolic process(GO:0009065) |
| 0.0 | 0.1 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.0 | 0.1 | GO:0019320 | hexose catabolic process(GO:0019320) |
| 0.0 | 0.1 | GO:0035459 | cargo loading into vesicle(GO:0035459) cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.0 | 0.1 | GO:0000114 | obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0006567 | glycine biosynthetic process(GO:0006545) threonine catabolic process(GO:0006567) |
| 0.0 | 0.2 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.0 | 0.1 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.2 | GO:1902532 | negative regulation of intracellular signal transduction(GO:1902532) |
| 0.0 | 0.1 | GO:0000272 | polysaccharide catabolic process(GO:0000272) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0045910 | negative regulation of DNA recombination(GO:0045910) |
| 0.0 | 0.0 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.2 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.0 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 1.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.0 | 0.2 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.0 | 0.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.0 | GO:0097035 | lipid translocation(GO:0034204) regulation of membrane lipid distribution(GO:0097035) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0007323 | peptide pheromone maturation(GO:0007323) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.0 | GO:0032048 | cardiolipin metabolic process(GO:0032048) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:0015758 | glucose transport(GO:0015758) |
| 0.0 | 0.3 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.2 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.1 | GO:0009099 | valine metabolic process(GO:0006573) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.0 | GO:2001025 | cellular response to drug(GO:0035690) regulation of response to drug(GO:2001023) positive regulation of response to drug(GO:2001025) regulation of cellular response to drug(GO:2001038) positive regulation of cellular response to drug(GO:2001040) |
| 0.0 | 0.8 | GO:0007005 | mitochondrion organization(GO:0007005) |
| 0.0 | 0.1 | GO:0001558 | regulation of cell growth(GO:0001558) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0045265 | mitochondrial proton-transporting ATP synthase, stator stalk(GO:0000274) proton-transporting ATP synthase, stator stalk(GO:0045265) |
| 0.3 | 1.6 | GO:0045263 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.3 | 0.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 1.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.2 | 0.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 0.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 0.8 | GO:0000817 | COMA complex(GO:0000817) |
| 0.2 | 0.6 | GO:0000111 | nucleotide-excision repair factor 2 complex(GO:0000111) |
| 0.1 | 0.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 2.4 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.7 | GO:0072380 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 0.1 | 1.2 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.1 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.8 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.8 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.1 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 3.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.3 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 3.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.2 | GO:0033309 | SBF transcription complex(GO:0033309) |
| 0.1 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.0 | 0.1 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.6 | GO:0005619 | ascospore wall(GO:0005619) |
| 0.0 | 0.1 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.0 | GO:0042763 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.0 | 4.8 | GO:0000329 | fungal-type vacuole membrane(GO:0000329) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.0 | 0.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 2.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.0 | GO:0005825 | half bridge of spindle pole body(GO:0005825) |
| 0.0 | 0.3 | GO:0044455 | mitochondrial membrane part(GO:0044455) |
| 0.0 | 0.3 | GO:0033698 | Rpd3L complex(GO:0033698) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 10.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0070274 | RES complex(GO:0070274) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.0 | GO:0070211 | Snt2C complex(GO:0070211) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.8 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle(GO:0030139) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.1 | GO:0016514 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.0 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.8 | 2.5 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.5 | 1.6 | GO:0016208 | AMP binding(GO:0016208) |
| 0.5 | 2.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.4 | 1.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.3 | 3.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 0.8 | GO:0015288 | anion channel activity(GO:0005253) voltage-gated anion channel activity(GO:0008308) porin activity(GO:0015288) wide pore channel activity(GO:0022829) |
| 0.2 | 0.8 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 0.5 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) |
| 0.2 | 1.6 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.2 | 0.5 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.2 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 0.6 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.2 | 1.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 0.6 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 2.5 | GO:0046906 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.1 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.5 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.2 | GO:0015116 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.1 | 1.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.3 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.1 | 5.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.4 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.1 | 0.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.3 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.6 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.4 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.1 | 0.2 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.1 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 0.7 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 1.0 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.1 | 0.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 0.2 | GO:0008897 | holo-[acyl-carrier-protein] synthase activity(GO:0008897) |
| 0.1 | 1.5 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.5 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.5 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.1 | 0.7 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.1 | GO:0019903 | phosphatase binding(GO:0019902) protein phosphatase binding(GO:0019903) |
| 0.1 | 0.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.2 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.2 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.1 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 1.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.4 | GO:0016723 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.4 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 0.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.9 | GO:0016298 | lipase activity(GO:0016298) |
| 0.0 | 0.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.0 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.0 | 0.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.1 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.0 | 0.2 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.0 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 0.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 25.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 0.6 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.1 | 0.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 0.4 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 25.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |