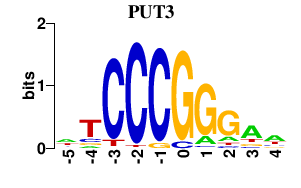
Results for PUT3
Z-value: 0.69
Transcription factors associated with PUT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PUT3
|
S000001498 | Transcriptional activator of proline utilization genes, constitutivel |
Activity-expression correlation:
Activity profile of PUT3 motif
Sorted Z-values of PUT3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YAR035W | 3.93 |
YAT1
|
Outer mitochondrial carnitine acetyltransferase, minor ethanol-inducible enzyme involved in transport of activated acyl groups from the cytoplasm into the mitochondrial matrix; phosphorylated |
|
| YMR017W | 3.79 |
SPO20
|
Meiosis-specific subunit of the t-SNARE complex, required for prospore membrane formation during sporulation; similar to but not functionally redundant with Sec9p; SNAP-25 homolog |
|
| YPL026C | 3.28 |
SKS1
|
Putative serine/threonine protein kinase; involved in the adaptation to low concentrations of glucose independent of the SNF3 regulated pathway |
|
| YIL057C | 2.94 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YAR053W | 2.92 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR377C | 2.85 |
FBP1
|
Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway, required for glucose metabolism |
|
| YJR048W | 2.56 |
CYC1
|
Cytochrome c, isoform 1; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YJR095W | 2.39 |
SFC1
|
Mitochondrial succinate-fumarate transporter, transports succinate into and fumarate out of the mitochondrion; required for ethanol and acetate utilization |
|
| YPL024W | 2.34 |
RMI1
|
Involved in response to DNA damage; null mutants have increased rates of recombination and delayed S phase; interacts physically and genetically with Sgs1p (RecQ family member) and Top3p (topoisomerase III) |
|
| YJL045W | 1.95 |
Minor succinate dehydrogenase isozyme; homologous to Sdh1p, the major isozyme reponsible for the oxidation of succinate and transfer of electrons to ubiquinone; induced during the diauxic shift in a Cat8p-dependent manner |
||
| YNL117W | 1.93 |
MLS1
|
Malate synthase, enzyme of the glyoxylate cycle, involved in utilization of non-fermentable carbon sources; expression is subject to carbon catabolite repression; localizes in peroxisomes during growth in oleic acid medium |
|
| YOR348C | 1.81 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YKL163W | 1.80 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YKL109W | 1.80 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YHR212W-A | 1.66 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YDR275W | 1.65 |
BSC2
|
Protein of unknown function, ORF exhibits genomic organization compatible with a translational readthrough-dependent mode of expression |
|
| YAR060C | 1.65 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR096C | 1.64 |
HXT5
|
Hexose transporter with moderate affinity for glucose, induced in the presence of non-fermentable carbon sources, induced by a decrease in growth rate, contains an extended N-terminal domain relative to other HXTs |
|
| YBL064C | 1.63 |
PRX1
|
Mitochondrial peroxiredoxin (1-Cys Prx) with thioredoxin peroxidase activity, has a role in reduction of hydroperoxides; induced during respiratory growth and under conditions of oxidative stress; phosphorylated |
|
| YCR007C | 1.55 |
Putative integral membrane protein, member of DUP240 gene family; YCR007C is not an essential gene |
||
| YDR536W | 1.48 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YHR212C | 1.34 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR040W | 1.34 |
YET2
|
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; homolog of human BAP31 protein |
|
| YFR053C | 1.31 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YLR174W | 1.30 |
IDP2
|
Cytosolic NADP-specific isocitrate dehydrogenase, catalyzes oxidation of isocitrate to alpha-ketoglutarate; levels are elevated during growth on non-fermentable carbon sources and reduced during growth on glucose |
|
| YBL015W | 1.29 |
ACH1
|
Acetyl-coA hydrolase, primarily localized to mitochondria; phosphorylated; required for acetate utilization and for diploid pseudohyphal growth |
|
| YPR007C | 1.27 |
REC8
|
Meiosis-specific component of sister chromatid cohesion complex; maintains cohesion between sister chromatids during meiosis I; maintains cohesion between centromeres of sister chromatids until meiosis II; homolog of S. pombe Rec8p |
|
| YPL061W | 1.24 |
ALD6
|
Cytosolic aldehyde dehydrogenase, activated by Mg2+ and utilizes NADP+ as the preferred coenzyme; required for conversion of acetaldehyde to acetate; constitutively expressed; locates to the mitochondrial outer surface upon oxidative stress |
|
| YDL138W | 1.23 |
RGT2
|
Plasma membrane glucose receptor, highly similar to Snf3p; both Rgt2p and Snf3p serve as transmembrane glucose sensors generating an intracellular signal that induces expression of glucose transporter (HXT) genes |
|
| YGR243W | 1.22 |
FMP43
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YNR073C | 1.22 |
Putative mannitol dehydrogenase |
||
| YBR230C | 1.20 |
OM14
|
Integral mitochondrial outer membrane protein; abundance is decreased in cells grown in glucose relative to other carbon sources; appears to contain 3 alpha-helical transmembrane segments; ORF encodes a 97-basepair intron |
|
| YGL183C | 1.18 |
MND1
|
Protein required for recombination and meiotic nuclear division; forms a complex with Hop2p, which is involved in chromosome pairing and repair of meiotic double-strand breaks |
|
| YDR313C | 1.16 |
PIB1
|
RING-type ubiquitin ligase of the endosomal and vacuolar membranes, binds phosphatidylinositol(3)-phosphate; contains a FYVE finger domain |
|
| YOR152C | 1.13 |
Putative protein of unknown function; has no similarity to any known protein; YOR152C is not an essential gene |
||
| YER067C-A | 1.10 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YER067W |
||
| YLR296W | 1.09 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR169W-A | 1.08 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps two other dubious ORFs: YPR170C and YPR170W-B |
||
| YDR216W | 1.08 |
ADR1
|
Carbon source-responsive zinc-finger transcription factor, required for transcription of the glucose-repressed gene ADH2, of peroxisomal protein genes, and of genes required for ethanol, glycerol, and fatty acid utilization |
|
| YMR081C | 1.06 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YOR120W | 1.04 |
GCY1
|
Putative NADP(+) coupled glycerol dehydrogenase, proposed to be involved in an alternative pathway for glycerol catabolism; member of the aldo-keto reductase (AKR) family |
|
| YNL305C | 1.01 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YNL305C is not an essential gene |
||
| YIL024C | 1.00 |
Putative protein of unknown function; non-essential gene; expression directly regulated by the metabolic and meiotic transcriptional regulator Ume6p |
||
| YGR032W | 0.97 |
GSC2
|
Catalytic subunit of 1,3-beta-glucan synthase, involved in formation of the inner layer of the spore wall; activity positively regulated by Rho1p and negatively by Smk1p; has similarity to an alternate catalytic subunit, Fks1p (Gsc1p) |
|
| YGL033W | 0.95 |
HOP2
|
Meiosis-specific protein that localizes to chromosomes, preventing synapsis between nonhomologous chromosomes and ensuring synapsis between homologs; complexes with Mnd1p to promote homolog pairing and meiotic double-strand break repair |
|
| YER150W | 0.95 |
SPI1
|
GPI-anchored cell wall protein involved in weak acid resistance; basal expression requires Msn2p/Msn4p; expression is induced under conditions of stress and during the diauxic shift; similar to Sed1p |
|
| YGL191W | 0.93 |
COX13
|
Subunit VIa of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; not essential for cytochrome c oxidase activity but may modulate activity in response to ATP |
|
| YPL156C | 0.93 |
PRM4
|
Pheromone-regulated protein, predicted to have 1 transmembrane segment; transcriptionally regulated by Ste12p during mating and by Cat8p during the diauxic shift |
|
| YHR126C | 0.92 |
Putative protein of unknown function; transcription dependent upon Azf1p |
||
| YGL193C | 0.91 |
Haploid-specific gene repressed by a1-alpha2, turned off in sir3 null strains, absence enhances the sensitivity of rad52-327 cells to campothecin almost 100-fold |
||
| YPL230W | 0.90 |
USV1
|
Putative transcription factor containing a C2H2 zinc finger; mutation affects transcriptional regulation of genes involved in protein folding, ATP binding, and cell wall biosynthesis |
|
| YEL070W | 0.90 |
DSF1
|
Deletion suppressor of mpt5 mutation |
|
| YNL180C | 0.89 |
RHO5
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely involved in protein kinase C (Pkc1p)-dependent signal transduction pathway that controls cell integrity |
|
| YDR077W | 0.89 |
SED1
|
Major stress-induced structural GPI-cell wall glycoprotein in stationary-phase cells, associates with translating ribosomes, possible role in mitochondrial genome maintenance; ORF contains two distinct variable minisatellites |
|
| YDL139C | 0.89 |
SCM3
|
Nonhistone component of centromeric chromatin that binds stoichiometrically to CenH3-H4 histones, required for kinetochore assembly; contains nuclear export signal (NES); required for G2/M progression and localization of Cse4p |
|
| YBR144C | 0.89 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR144C is not an essential gene |
||
| YMR090W | 0.88 |
Putative protein of unknown function with similarity to DTDP-glucose 4,6-dehydratases; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YMR090W is not an essential gene |
||
| YDL244W | 0.87 |
THI13
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YDR178W | 0.87 |
SDH4
|
Membrane anchor subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
|
| YER015W | 0.86 |
FAA2
|
Long chain fatty acyl-CoA synthetase; accepts a wider range of acyl chain lengths than Faa1p, preferring C9:0-C13:0; involved in the activation of endogenous pools of fatty acids |
|
| YNL144C | 0.86 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YNL144C is not an essential gene |
||
| YFR029W | 0.86 |
PTR3
|
Component of the SPS plasma membrane amino acid sensor system (Ssy1p-Ptr3p-Ssy5p), which senses external amino acid concentration and transmits intracellular signals that result in regulation of expression of amino acid permease genes |
|
| YEL060C | 0.86 |
PRB1
|
Vacuolar proteinase B (yscB), a serine protease of the subtilisin family; involved in protein degradation in the vacuole and required for full protein degradation during sporulation |
|
| YLR312C | 0.85 |
QNQ1
|
Protein of unknown function; null mutant quiescent and non-quiescent cells exhibit reduced reproductive capacity |
|
| YPR036W-A | 0.84 |
Protein of unknown function; transcription is regulated by Pdr1p |
||
| YNL179C | 0.83 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YAL054C | 0.83 |
ACS1
|
Acetyl-coA synthetase isoform which, along with Acs2p, is the nuclear source of acetyl-coA for histone acetlyation; expressed during growth on nonfermentable carbon sources and under aerobic conditions |
|
| YOR121C | 0.82 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; open reading frame overlaps the verified gene GCY1/YOR120W |
||
| YHR139C | 0.82 |
SPS100
|
Protein required for spore wall maturation; expressed during sporulation; may be a component of the spore wall |
|
| YHR211W | 0.81 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YDR277C | 0.80 |
MTH1
|
Negative regulator of the glucose-sensing signal transduction pathway, required for repression of transcription by Rgt1p; interacts with Rgt1p and the Snf3p and Rgt2p glucose sensors; phosphorylated by Yck1p, triggering Mth1p degradation |
|
| YGL192W | 0.80 |
IME4
|
Probable mRNA N6-adenosine methyltransferase that is required for IME1 transcript accumulation and for sporulation; expression is induced in starved MATa/MAT alpha diploid cells |
|
| YDR259C | 0.80 |
YAP6
|
Putative basic leucine zipper (bZIP) transcription factor; overexpression increases sodium and lithium tolerance |
|
| YDL020C | 0.79 |
RPN4
|
Transcription factor that stimulates expression of proteasome genes; Rpn4p levels are in turn regulated by the 26S proteasome in a negative feedback control mechanism; RPN4 is transcriptionally regulated by various stress responses |
|
| YCR005C | 0.78 |
CIT2
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate, peroxisomal isozyme involved in glyoxylate cycle; expression is controlled by Rtg1p and Rtg2p transcription factors |
|
| YGR286C | 0.77 |
BIO2
|
Biotin synthase, catalyzes the conversion of dethiobiotin to biotin, which is the last step of the biotin biosynthesis pathway; complements E. coli bioB mutant |
|
| YPR127W | 0.76 |
Putative protein of unknown function; expression is activated by transcription factor YRM1/YOR172W; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and the nucleus |
||
| YKL171W | 0.76 |
Putative protein of unknown function; predicted protein kinase; implicated in proteasome function; epitope-tagged protein localizes to the cytoplasm |
||
| YLR055C | 0.76 |
SPT8
|
Subunit of the SAGA transcriptional regulatory complex but not present in SAGA-like complex SLIK/SALSA, required for SAGA-mediated inhibition at some promoters |
|
| YMR016C | 0.75 |
SOK2
|
Nuclear protein that plays a regulatory role in the cyclic AMP (cAMP)-dependent protein kinase (PKA) signal transduction pathway; negatively regulates pseudohyphal differentiation; homologous to several transcription factors |
|
| YDR441C | 0.75 |
APT2
|
Apparent pseudogene, not transcribed or translated under normal conditions; encodes a protein with similarity to adenine phosphoribosyltransferase, but artificially expressed protein exhibits no enzymatic activity |
|
| YLR295C | 0.74 |
ATP14
|
Subunit h of the F0 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YBR230W-A | 0.74 |
Putative protein of unknown function |
||
| YCL001W-B | 0.74 |
Putative protein of unknown function; identified by homology |
||
| YML042W | 0.71 |
CAT2
|
Carnitine acetyl-CoA transferase present in both mitochondria and peroxisomes, transfers activated acetyl groups to carnitine to form acetylcarnitine which can be shuttled across membranes |
|
| YBL103C | 0.70 |
RTG3
|
Basic helix-loop-helix-leucine zipper (bHLH/Zip) transcription factor that forms a complex with another bHLH/Zip protein, Rtg1p, to activate the retrograde (RTG) and TOR pathways |
|
| YMR002W | 0.70 |
MIC17
|
Mitochondrial intermembrane space cysteine motif protein; MIC17 is not an essential gene |
|
| YNL190W | 0.70 |
Cell wall protein of unknown function; proposed role as a hydrophilin induced by osmotic stress; contains a putative GPI-attachment site |
||
| YNL142W | 0.69 |
MEP2
|
Ammonium permease involved in regulation of pseudohyphal growth; belongs to a ubiquitous family of cytoplasmic membrane proteins that transport only ammonium (NH4+); expression is under the nitrogen catabolite repression regulation |
|
| YPL062W | 0.68 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YPL062W is not an essential gene; homozygous diploid mutant shows a decrease in glycogen accumulation |
||
| YGL056C | 0.68 |
SDS23
|
One of two S. cerevisiae homologs (Sds23p and Sds24p) of the Schizosaccharomyces pombe Sds23 protein, which genetic studies have implicated in APC/cyclosome regulation |
|
| YKL093W | 0.68 |
MBR1
|
Protein involved in mitochondrial functions and stress response; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants |
|
| YDR096W | 0.68 |
GIS1
|
JmjC domain-containing histone demethylase; transcription factor involved in the expression of genes during nutrient limitation; also involved in the negative regulation of DPP1 and PHR1 |
|
| YLR260W | 0.67 |
LCB5
|
Minor sphingoid long-chain base kinase, paralog of Lcb4p responsible for few percent of the total activity, possibly involved in synthesis of long-chain base phosphates, which function as signaling molecules |
|
| YDR102C | 0.67 |
Dubious open reading frame; homozygous diploid deletion strain exhibits high budding index |
||
| YPL089C | 0.66 |
RLM1
|
MADS-box transcription factor, component of the protein kinase C-mediated MAP kinase pathway involved in the maintenance of cell integrity; phosphorylated and activated by the MAP-kinase Slt2p |
|
| YPR065W | 0.65 |
ROX1
|
Heme-dependent repressor of hypoxic genes; contains an HMG domain that is responsible for DNA bending activity |
|
| YPL025C | 0.65 |
Hypothetical protein |
||
| YER103W | 0.65 |
SSA4
|
Heat shock protein that is highly induced upon stress; plays a role in SRP-dependent cotranslational protein-membrane targeting and translocation; member of the HSP70 family; cytoplasmic protein that concentrates in nuclei upon starvation |
|
| YKR034W | 0.65 |
DAL80
|
Negative regulator of genes in multiple nitrogen degradation pathways; expression is regulated by nitrogen levels and by Gln3p; member of the GATA-binding family, forms homodimers and heterodimers with Deh1p |
|
| YDL223C | 0.65 |
HBT1
|
Substrate of the Hub1p ubiquitin-like protein that localizes to the shmoo tip (mating projection); mutants are defective for mating projection formation, thereby implicating Hbt1p in polarized cell morphogenesis |
|
| YMR103C | 0.65 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR343C | 0.64 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YDR406W | 0.64 |
PDR15
|
Plasma membrane ATP binding cassette (ABC) transporter, multidrug transporter and general stress response factor implicated in cellular detoxification; regulated by Pdr1p, Pdr3p and Pdr8p; promoter contains a PDR responsive element |
|
| YBR280C | 0.63 |
SAF1
|
F-Box protein involved in proteasome-dependent degradation of Aah1p during entry of cells into quiescence; interacts with Skp1 |
|
| YDR022C | 0.63 |
CIS1
|
Autophagy-specific protein required for autophagosome formation; may form a complex with Atg17p and Atg29p that localizes other proteins to the pre-autophagosomal structure; high-copy suppressor of CIK1 deletion |
|
| YGR039W | 0.63 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the Autonomously Replicating Sequence ARS722 |
||
| YKL102C | 0.61 |
Dubious open reading frame unlikely to encode a functional protein; deletion confers sensitivity to citric acid; predicted protein would include a thiol-disulfide oxidoreductase active site |
||
| YML081C-A | 0.61 |
ATP18
|
Subunit of the mitochondrial F1F0 ATP synthase, which is a large enzyme complex required for ATP synthesis; termed subunit I or subunit j; does not correspond to known ATP synthase subunits in other organisms |
|
| YIL125W | 0.61 |
KGD1
|
Component of the mitochondrial alpha-ketoglutarate dehydrogenase complex, which catalyzes a key step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation of alpha-ketoglutarate to form succinyl-CoA |
|
| YGL194C | 0.60 |
HOS2
|
Histone deacetylase required for gene activation via specific deacetylation of lysines in H3 and H4 histone tails; subunit of the Set3 complex, a meiotic-specific repressor of sporulation specific genes that contains deacetylase activity |
|
| YDR452W | 0.59 |
PPN1
|
Vacuolar endopolyphosphatase with a role in phosphate metabolism; functions as a homodimer |
|
| YNL143C | 0.59 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR161C | 0.59 |
RTS3
|
Putative component of the protein phosphatase type 2A complex |
|
| YDR298C | 0.58 |
ATP5
|
Subunit 5 of the stator stalk of mitochondrial F1F0 ATP synthase, which is an evolutionarily conserved enzyme complex required for ATP synthesis; homologous to bovine subunit OSCP (oligomycin sensitivity-conferring protein); phosphorylated |
|
| YOL084W | 0.58 |
PHM7
|
Protein of unknown function, expression is regulated by phosphate levels; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery and vacuole |
|
| YFR012W | 0.57 |
Putative protein of unknown function |
||
| YPR026W | 0.57 |
ATH1
|
Acid trehalase required for utilization of extracellular trehalose |
|
| YGL035C | 0.57 |
MIG1
|
Transcription factor involved in glucose repression; sequence specific DNA binding protein containing two Cys2His2 zinc finger motifs; regulated by the SNF1 kinase and the GLC7 phosphatase |
|
| YKR039W | 0.56 |
GAP1
|
General amino acid permease; localization to the plasma membrane is regulated by nitrogen source |
|
| YLR124W | 0.56 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR056W | 0.56 |
RSC1
|
Component of the RSC chromatin remodeling complex; required for expression of mid-late sporulation-specific genes; contains two essential bromodomains, a bromo-adjacent homology (BAH) domain, and an AT hook |
|
| YBR051W | 0.56 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partiallly overlaps the REG2/YBR050C regulatory subunit of the Glc7p type-1 protein phosphatase |
||
| YOR306C | 0.55 |
MCH5
|
Plasma membrane riboflavin transporter; facilitates the uptake of vitamin B2; required for FAD-dependent processes; sequence similarity to mammalian monocarboxylate permeases, however mutants are not deficient in monocarboxylate transport |
|
| YMR141W-A | 0.55 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene RPL13B/YMR142C |
||
| YBR180W | 0.55 |
DTR1
|
Putative dityrosine transporter, required for spore wall synthesis; expressed during sporulation; member of the major facilitator superfamily (DHA1 family) of multidrug resistance transporters |
|
| YDR542W | 0.54 |
PAU10
|
Hypothetical protein |
|
| YPR013C | 0.54 |
Putative zinc finger protein; YPR013C is not an essential gene |
||
| YOR139C | 0.53 |
Hypothetical protein |
||
| YBR297W | 0.53 |
MAL33
|
MAL-activator protein, part of complex locus MAL3; nonfunctional in genomic reference strain S288C |
|
| YJL116C | 0.52 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YDR442W | 0.52 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR123C | 0.52 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR122C; contains characteristic aminoacyl-tRNA motif |
||
| YOL138C | 0.52 |
RTC1
|
Putative protein of unknown function; may interact with ribosomes, based on co-purification experiments; null mutation suppresses cdc13-1 temperature sensitivity |
|
| YOL051W | 0.52 |
GAL11
|
Subunit of the RNA polymerase II mediator complex; associates with core polymerase subunits to form the RNA polymerase II holoenzyme; affects transcription by acting as target of activators and repressors |
|
| YDR223W | 0.51 |
CRF1
|
Transcriptional corepressor involved in the regulation of ribosomal protein gene transcription via the TOR signaling pathway and protein kinase A, phosphorylated by activated Yak1p which promotes accumulation of Crf1p in the nucleus |
|
| YJR094C | 0.51 |
IME1
|
Master regulator of meiosis that is active only during meiotic events, activates transcription of early meiotic genes through interaction with Ume6p, degraded by the 26S proteasome following phosphorylation by Ime2p |
|
| YLR122C | 0.50 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR123C |
||
| YBR050C | 0.50 |
REG2
|
Regulatory subunit of the Glc7p type-1 protein phosphatase; involved with Reg1p, Glc7p, and Snf1p in regulation of glucose-repressible genes, also involved in glucose-induced proteolysis of maltose permease |
|
| YOR140W | 0.50 |
SFL1
|
Transcriptional repressor and activator; involved in repression of flocculation-related genes, and activation of stress responsive genes; negatively regulated by cAMP-dependent protein kinase A subunit Tpk2p |
|
| YEL012W | 0.50 |
UBC8
|
Ubiquitin-conjugating enzyme that negatively regulates gluconeogenesis by mediating the glucose-induced ubiquitination of fructose-1,6-bisphosphatase (FBPase); cytoplasmic enzyme that catalyzes the ubiquitination of histones in vitro |
|
| YOR178C | 0.49 |
GAC1
|
Regulatory subunit for Glc7p type-1 protein phosphatase (PP1), tethers Glc7p to Gsy2p glycogen synthase, binds Hsf1p heat shock transcription factor, required for induction of some HSF-regulated genes under heat shock |
|
| YPL222C-A | 0.49 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YEL049W | 0.49 |
PAU2
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YKR100C | 0.48 |
SKG1
|
Transmembrane protein with a role in cell wall polymer composition; localizes on the inner surface of the plasma membrane at the bud and in the daughter cell |
|
| YKL112W | 0.48 |
ABF1
|
DNA binding protein with possible chromatin-reorganizing activity involved in transcriptional activation, gene silencing, and DNA replication and repair |
|
| YOL085W-A | 0.48 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YOL085C |
||
| YBR179C | 0.48 |
FZO1
|
Mitochondrial integral membrane protein involved in mitochondrial fusion and maintenance of the mitochondrial genome; contains N-terminal GTPase domain |
|
| YDR242W | 0.47 |
AMD2
|
Putative amidase |
|
| YBR021W | 0.47 |
FUR4
|
Uracil permease, localized to the plasma membrane; expression is tightly regulated by uracil levels and environmental cues |
|
| YAL018C | 0.47 |
Putative protein of unknown function |
||
| YDR171W | 0.47 |
HSP42
|
Small heat shock protein (sHSP) with chaperone activity; forms barrel-shaped oligomers that suppress unfolded protein aggregation; involved in cytoskeleton reorganization after heat shock |
|
| YDR103W | 0.46 |
STE5
|
Pheromone-response scaffold protein; binds kinases Ste11p, Ste7p, and Fus3p to form a MAPK cascade complex that interacts with the plasma membrane, via a PH (pleckstrin homology) and PM/NLS domain, and with Ste4p-Ste18p, during signaling |
|
| YCR033W | 0.46 |
SNT1
|
Subunit of the Set3C deacetylase complex that interacts directly with the Set3C subunit, Sif2p; putative DNA-binding protein |
|
| YDL245C | 0.46 |
HXT15
|
Protein of unknown function with similarity to hexose transporter family members, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YDR036C | 0.46 |
EHD3
|
3-hydroxyisobutyryl-CoA hydrolase, member of a family of enoyl-CoA hydratase/isomerases; non-tagged protein is detected in highly purified mitochondria in high-throughput studies; phosphorylated; mutation affects fluid-phase endocytosis |
|
| YKR101W | 0.45 |
SIR1
|
Protein involved in repression of transcription at the silent mating-type loci HML and HMR; recruitment to silent chromatin requires interactions with Orc1p and with Sir4p, through a common Sir1p domain; binds to centromeric chromatin |
|
| YPL088W | 0.45 |
Putative aryl alcohol dehydrogenase; transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in multidrug resistance |
||
| YMR284W | 0.45 |
YKU70
|
Subunit of the telomeric Ku complex (Yku70p-Yku80p), involved in telomere length maintenance, structure and telomere position effect; relocates to sites of double-strand cleavage to promote nonhomologous end joining during DSB repair |
|
| YDL110C | 0.45 |
TMA17
|
Protein of unknown function that associates with ribosomes |
|
| YHR210C | 0.45 |
Putative protein of unknown function; non-essential gene; highly expressed under anaeorbic conditions; sequence similarity to aldose 1-epimerases such as GAL10 |
||
| YGL133W | 0.45 |
ITC1
|
Component of the ATP-dependent Isw2p-Itc1p chromatin remodeling complex, required for repression of a-specific genes, repression of early meiotic genes during mitotic growth, and repression of INO1 |
|
| YPL171C | 0.45 |
OYE3
|
Widely conserved NADPH oxidoreductase containing flavin mononucleotide (FMN), homologous to Oye2p with slight differences in ligand binding and catalytic properties; may be involved in sterol metabolism |
|
| YLR383W | 0.45 |
SMC6
|
Protein involved in structural maintenance of chromosomes; essential subunit of Mms21-Smc5-Smc6 complex; required for growth, DNA repair, interchromosomal and sister chromatid recombination; homologous to S. pombe rad18 |
|
| YKR073C | 0.44 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YER158C | 0.44 |
Protein of unknown function, has similarity to Afr1p; potentially phosphorylated by Cdc28p |
||
| YGL177W | 0.44 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR402W | 0.44 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR067C | 0.44 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YMR280C | 0.44 |
CAT8
|
Zinc cluster transcriptional activator necessary for derepression of a variety of genes under non-fermentative growth conditions, active after diauxic shift, binds carbon source responsive elements |
|
| YPR010C-A | 0.44 |
Putative protein of unknown function; conserved among Saccharomyces sensu stricto species |
||
| YHR145C | 0.43 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR371C | 0.43 |
GPB1
|
Multistep regulator of cAMP-PKA signaling; inhibits PKA downstream of Gpa2p and Cyr1p, thereby increasing cAMP dependency; inhibits Ras activity through direct interactions with Ira1p/2p; regulated by G-alpha protein Gpa2p; homolog of Gpb2p |
|
| YPL119C | 0.43 |
DBP1
|
Putative ATP-dependent RNA helicase of the DEAD-box protein family; mutants show reduced stability of the 40S ribosomal subunit scanning through 5' untranslated regions of mRNAs |
|
| YDL181W | 0.42 |
INH1
|
Protein that inhibits ATP hydrolysis by the F1F0-ATP synthase; inhibitory function is enhanced by stabilizing proteins Stf1p and Stf2p; has similarity to Stf1p; has a calmodulin-binding motif and binds calmodulin in vitro |
|
| YGR045C | 0.42 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR403W | 0.42 |
SFP1
|
Transcription factor that controls expression of many ribosome biogenesis genes in response to nutrients and stress, regulates G2/M transitions during mitotic cell cycle and DNA-damage response, involved in cell size modulation |
|
| YOR223W | 0.42 |
Putative protein of unknown function |
||
| YGL117W | 0.42 |
Putative protein of unknown function |
||
| YNL139C | 0.42 |
THO2
|
Subunit of the THO complex, which is required for efficient transcription elongation and involved in transcriptional elongation-associated recombination; required for LacZ RNA expression from certain plasmids |
|
| YLR304C | 0.42 |
ACO1
|
Aconitase, required for the tricarboxylic acid (TCA) cycle and also independently required for mitochondrial genome maintenance; phosphorylated; component of the mitochondrial nucleoid; mutation leads to glutamate auxotrophy |
|
| YLR326W | 0.41 |
Hypothetical protein |
||
| YLR390W-A | 0.41 |
CCW14
|
Covalently linked cell wall glycoprotein, present in the inner layer of the cell wall |
|
| YPL282C | 0.40 |
PAU22
|
Hypothetical protein |
|
| YNL282W | 0.40 |
POP3
|
Subunit of both RNase MRP, which cleaves pre-rRNA, and nuclear RNase P, which cleaves tRNA precursors to generate mature 5' ends |
|
| YJR089W | 0.40 |
BIR1
|
Essential chromosomal passenger protein involved in coordinating cell cycle events for proper chromosome segregation; C-terminal region binds Sli15p, and the middle region, upon phosphorylation, localizes Cbf2p to the spindle at anaphase |
|
| YPL262W | 0.40 |
FUM1
|
Fumarase, converts fumaric acid to L-malic acid in the TCA cycle; cytosolic and mitochondrial localization determined by the N-terminal mitochondrial targeting sequence and protein conformation; phosphorylated in mitochondria |
|
| YGR046W | 0.40 |
TAM41
|
Mitochondrial protein involved in protein import into the mitochondrial matrix; maintains the functional integrity of the TIM23 protein translocator complex; viability of null mutant is strain-dependent; mRNA is targeted to the bud |
|
| YOR235W | 0.40 |
IRC13
|
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YKR011C | 0.40 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus |
||
| YAL039C | 0.39 |
CYC3
|
Cytochrome c heme lyase (holocytochrome c synthase), attaches heme to apo-cytochrome c (Cyc1p or Cyc7p) in the mitochondrial intermembrane space; human ortholog may have a role in microphthalmia with linear skin defects (MLS) |
|
| YFL013W-A | 0.39 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps IES1/YFL013C; identified by gene-trapping, microarray expression analysis, and genome-wide homology searching |
||
| YKL217W | 0.39 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YOR394W | 0.39 |
PAU21
|
Hypothetical protein |
|
| YGR144W | 0.39 |
THI4
|
Thiazole synthase, catalyzes formation of the thiazole moiety of thiamin pyrophosphate; required for thiamine biosynthesis and for mitochondrial genome stability |
|
| YMR124W | 0.38 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery, cytoplasm, bud, and bud neck; interacts with Crm1p in two-hybrid assay; YMR124W is not an essential gene |
||
| YMR194C-B | 0.38 |
Putative protein of unknown function |
||
| YOL055C | 0.38 |
THI20
|
Multifunctional protein with both hydroxymethylpyrimidine kinase and thiaminase activities; involved in thiamine biosynthesis and also in thiamine degradation; member of a gene family with THI21 and THI22; functionally redundant with Thi21p |
|
| YDL010W | 0.38 |
GRX6
|
Cis-golgi localized monothiol glutaredoxin that binds an iron-sulfur cluster; more similar in activity to dithiol than other monothiol glutaredoxins; involved in the oxidative stress response; functional overlap with GRX6 |
|
| YML025C | 0.38 |
YML6
|
Mitochondrial ribosomal protein of the large subunit, has similarity to E. coli L4 ribosomal protein and human mitoribosomal MRP-L4 protein; essential for viability, unlike most other mitoribosomal proteins |
Network of associatons between targets according to the STRING database.
First level regulatory network of PUT3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.8 | 2.3 | GO:0034310 | ethanol catabolic process(GO:0006068) primary alcohol catabolic process(GO:0034310) |
| 0.7 | 2.2 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.7 | 8.0 | GO:0015758 | glucose transport(GO:0015758) |
| 0.6 | 2.3 | GO:0043388 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.5 | 4.8 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.4 | 1.3 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.4 | 2.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.4 | 1.2 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) |
| 0.4 | 0.4 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.4 | 1.1 | GO:0070304 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.3 | 1.0 | GO:0042843 | D-xylose catabolic process(GO:0042843) |
| 0.3 | 1.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.3 | 1.0 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.3 | 2.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 0.9 | GO:2000058 | regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.3 | 1.5 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.3 | 1.7 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.3 | 0.3 | GO:0036095 | positive regulation of invasive growth in response to glucose limitation by positive regulation of transcription from RNA polymerase II promoter(GO:0036095) positive regulation of invasive growth in response to glucose limitation(GO:2000219) |
| 0.3 | 3.8 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 0.3 | 0.8 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.2 | 1.0 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.2 | 0.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 1.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.2 | 0.6 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.2 | 0.9 | GO:0043200 | response to amino acid(GO:0043200) |
| 0.2 | 0.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 1.0 | GO:0061416 | regulation of transcription from RNA polymerase II promoter in response to salt stress(GO:0061416) |
| 0.2 | 1.9 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 0.6 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 0.2 | 2.8 | GO:0072593 | reactive oxygen species metabolic process(GO:0072593) |
| 0.2 | 0.7 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 0.2 | 1.3 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.2 | 2.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 0.9 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.2 | 0.5 | GO:0006343 | establishment of chromatin silencing(GO:0006343) |
| 0.1 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.6 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.1 | 0.6 | GO:0042173 | regulation of sporulation resulting in formation of a cellular spore(GO:0042173) |
| 0.1 | 1.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 2.2 | GO:0001101 | response to acid chemical(GO:0001101) |
| 0.1 | 0.7 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 0.4 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 0.4 | GO:0036257 | multivesicular body organization(GO:0036257) |
| 0.1 | 0.8 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.1 | 0.4 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.1 | 0.5 | GO:0042357 | thiamine diphosphate biosynthetic process(GO:0009229) thiamine diphosphate metabolic process(GO:0042357) |
| 0.1 | 0.2 | GO:0019629 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 0.1 | 0.7 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.2 | GO:0042773 | respiratory electron transport chain(GO:0022904) ATP synthesis coupled electron transport(GO:0042773) mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 0.7 | GO:0071322 | cellular response to carbohydrate stimulus(GO:0071322) |
| 0.1 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.5 | GO:0032048 | cardiolipin metabolic process(GO:0032048) |
| 0.1 | 0.7 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.4 | GO:1900436 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.1 | 0.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.1 | GO:1900544 | positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.1 | 0.1 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.1 | 0.6 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.6 | GO:0015851 | nucleobase transport(GO:0015851) |
| 0.1 | 1.2 | GO:0000767 | cell morphogenesis involved in conjugation with cellular fusion(GO:0000753) cell morphogenesis involved in conjugation(GO:0000767) |
| 0.1 | 1.0 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.1 | 4.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 1.3 | GO:0042724 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.1 | 0.5 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 0.9 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 0.4 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.3 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 0.6 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.1 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.4 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.1 | 0.2 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 0.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.5 | GO:0001079 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) |
| 0.1 | 0.2 | GO:2000284 | positive regulation of cellular amino acid metabolic process(GO:0045764) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 0.2 | GO:0010994 | free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 1.3 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 1.1 | GO:0007129 | synapsis(GO:0007129) |
| 0.1 | 0.3 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.3 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.2 | GO:0034395 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.1 | 0.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.2 | GO:0019346 | transsulfuration(GO:0019346) |
| 0.1 | 0.4 | GO:0031134 | sister chromatid biorientation(GO:0031134) |
| 0.1 | 0.3 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.1 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.3 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.1 | 0.3 | GO:0034487 | amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 0.1 | 0.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.1 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.1 | 0.2 | GO:0006827 | high-affinity iron ion transmembrane transport(GO:0006827) |
| 0.1 | 0.2 | GO:0009411 | response to UV(GO:0009411) response to light stimulus(GO:0009416) |
| 0.1 | 0.2 | GO:0051222 | positive regulation of protein transport(GO:0051222) positive regulation of protein localization to nucleus(GO:1900182) positive regulation of cellular protein localization(GO:1903829) positive regulation of establishment of protein localization(GO:1904951) |
| 0.1 | 0.3 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.1 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 1.0 | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress(GO:0036003) |
| 0.1 | 0.1 | GO:0052547 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 0.1 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.3 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.1 | 0.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 0.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.2 | GO:0000821 | regulation of glutamine family amino acid metabolic process(GO:0000820) regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.7 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.0 | 0.2 | GO:0098610 | agglutination involved in conjugation with cellular fusion(GO:0000752) agglutination involved in conjugation(GO:0000771) heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) cell-cell adhesion(GO:0098609) adhesion between unicellular organisms(GO:0098610) multi organism cell adhesion(GO:0098740) cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.0 | 0.0 | GO:0071406 | response to methylmercury(GO:0051597) cellular response to methylmercury(GO:0071406) |
| 0.0 | 0.1 | GO:0016241 | regulation of macroautophagy(GO:0016241) |
| 0.0 | 0.2 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0043954 | cellular component maintenance(GO:0043954) |
| 0.0 | 0.1 | GO:0060194 | antisense RNA transcription(GO:0009300) regulation of antisense RNA transcription(GO:0060194) negative regulation of antisense RNA transcription(GO:0060195) |
| 0.0 | 0.1 | GO:0071050 | snoRNA polyadenylation(GO:0071050) |
| 0.0 | 0.5 | GO:0030474 | spindle pole body duplication(GO:0030474) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.2 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.1 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.0 | 0.2 | GO:0009164 | nucleoside catabolic process(GO:0009164) glycosyl compound catabolic process(GO:1901658) |
| 0.0 | 0.7 | GO:0006915 | apoptotic process(GO:0006915) cell death(GO:0008219) programmed cell death(GO:0012501) |
| 0.0 | 0.3 | GO:0045996 | negative regulation of transcription by pheromones(GO:0045996) negative regulation of transcription from RNA polymerase II promoter by pheromones(GO:0046020) |
| 0.0 | 0.1 | GO:1903313 | positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.8 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:0072668 | obsolete tubulin complex biogenesis(GO:0072668) |
| 0.0 | 1.4 | GO:0010927 | cellular component assembly involved in morphogenesis(GO:0010927) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.3 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.4 | GO:0001934 | positive regulation of protein phosphorylation(GO:0001934) |
| 0.0 | 0.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0031292 | gene conversion at mating-type locus, DNA double-strand break processing(GO:0031292) negative regulation of reciprocal meiotic recombination(GO:0045128) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.2 | GO:0051447 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.1 | GO:0072414 | cellular response to biotic stimulus(GO:0071216) response to cell cycle checkpoint signaling(GO:0072396) response to mitotic cell cycle checkpoint signaling(GO:0072414) response to spindle checkpoint signaling(GO:0072417) response to mitotic spindle checkpoint signaling(GO:0072476) response to mitotic cell cycle spindle assembly checkpoint signaling(GO:0072479) response to spindle assembly checkpoint signaling(GO:0072485) negative regulation of protein import into nucleus during spindle assembly checkpoint(GO:1901925) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0048313 | Golgi inheritance(GO:0048313) |
| 0.0 | 0.1 | GO:0060237 | regulation of fungal-type cell wall organization(GO:0060237) |
| 0.0 | 0.1 | GO:0045144 | meiosis II(GO:0007135) meiotic sister chromatid segregation(GO:0045144) |
| 0.0 | 0.2 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 1.2 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 1.8 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.6 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.3 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.3 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.6 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.2 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 0.9 | GO:0007124 | pseudohyphal growth(GO:0007124) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.0 | 0.2 | GO:0070987 | error-prone translesion synthesis(GO:0042276) error-free translesion synthesis(GO:0070987) |
| 0.0 | 1.2 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.3 | GO:0008645 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.0 | 0.1 | GO:0072369 | positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) positive regulation of sterol transport(GO:0032373) positive regulation of transmembrane transport(GO:0034764) regulation of sterol import by regulation of transcription from RNA polymerase II promoter(GO:0035968) positive regulation of sterol import by positive regulation of transcription from RNA polymerase II promoter(GO:0035969) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.0 | 0.7 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.0 | 0.1 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0000372 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.0 | 0.0 | GO:0046033 | AMP biosynthetic process(GO:0006167) AMP metabolic process(GO:0046033) |
| 0.0 | 0.1 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.0 | 0.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) peptidyl-serine modification(GO:0018209) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) proton-transporting ATP synthase complex assembly(GO:0043461) |
| 0.0 | 0.1 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 0.1 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.3 | GO:1905039 | organic acid transmembrane transport(GO:1903825) carboxylic acid transmembrane transport(GO:1905039) |
| 0.0 | 0.1 | GO:0007532 | regulation of mating-type specific transcription, DNA-templated(GO:0007532) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.5 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 0.1 | GO:0034087 | establishment of sister chromatid cohesion(GO:0034085) establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.0 | 0.1 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0008361 | regulation of cell size(GO:0008361) |
| 0.0 | 0.1 | GO:0035542 | regulation of vesicle fusion(GO:0031338) regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.0 | GO:0051098 | regulation of binding(GO:0051098) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.5 | 2.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.4 | 2.8 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.2 | 1.0 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.2 | 2.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.2 | 1.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 0.6 | GO:0009353 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial oxoglutarate dehydrogenase complex(GO:0009353) dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 0.6 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.2 | 0.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 1.7 | GO:0031160 | ascospore wall(GO:0005619) spore wall(GO:0031160) |
| 0.2 | 4.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 1.3 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.2 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 0.7 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.2 | 0.3 | GO:0001400 | mating projection base(GO:0001400) |
| 0.2 | 4.6 | GO:0042763 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.1 | 1.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.4 | GO:0000446 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) nucleoplasmic THO complex(GO:0000446) |
| 0.1 | 0.3 | GO:0005756 | mitochondrial proton-transporting ATP synthase, central stalk(GO:0005756) proton-transporting ATP synthase, central stalk(GO:0045269) |
| 0.1 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.8 | GO:0031307 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.3 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.3 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.1 | 0.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.3 | GO:0030907 | MBF transcription complex(GO:0030907) |
| 0.1 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.2 | GO:0097002 | mitochondrial inner boundary membrane(GO:0097002) |
| 0.1 | 8.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 0.8 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 0.4 | GO:0005724 | nuclear heterochromatin(GO:0005720) nuclear telomeric heterochromatin(GO:0005724) telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0000111 | nucleotide-excision repair factor 2 complex(GO:0000111) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 4.1 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 1.2 | GO:0031970 | organelle envelope lumen(GO:0031970) |
| 0.0 | 0.3 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.0 | 0.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0032221 | Rpd3S complex(GO:0032221) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 2.4 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.2 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 0.0 | 0.0 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.2 | GO:0098797 | plasma membrane protein complex(GO:0098797) |
| 0.0 | 0.1 | GO:0005823 | central plaque of spindle pole body(GO:0005823) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0032176 | split septin rings(GO:0032176) cellular bud neck split septin rings(GO:0032177) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0005940 | septin ring(GO:0005940) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.2 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.0 | 0.2 | GO:0033698 | Rpd3L complex(GO:0033698) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0000133 | polarisome(GO:0000133) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.0 | 0.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0098589 | membrane region(GO:0098589) |
| 0.0 | 0.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.0 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.0 | GO:0098562 | side of membrane(GO:0098552) cytoplasmic side of membrane(GO:0098562) |
| 0.0 | 0.0 | GO:0072380 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.6 | 2.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.6 | 1.7 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.6 | 2.2 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.5 | 2.7 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.5 | 1.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.4 | 1.3 | GO:0005536 | glucose binding(GO:0005536) |
| 0.4 | 2.1 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.4 | 1.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.4 | 2.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.4 | 1.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.4 | 1.1 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.3 | 1.0 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.3 | 0.9 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.3 | 0.9 | GO:0048038 | quinone binding(GO:0048038) |
| 0.3 | 1.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.3 | 1.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 1.9 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 2.5 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.2 | 2.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.6 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.2 | 0.5 | GO:0008902 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.2 | 4.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.2 | 0.8 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.1 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.7 | GO:0015294 | solute:cation symporter activity(GO:0015294) |
| 0.1 | 0.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.5 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 1.8 | GO:0015578 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.1 | 0.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.6 | GO:0097372 | NAD-dependent histone deacetylase activity (H3-K18 specific)(GO:0097372) |
| 0.1 | 0.4 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.6 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 1.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 1.0 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.5 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.5 | GO:0015038 | peptide disulfide oxidoreductase activity(GO:0015037) glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 1.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 0.3 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 0.1 | GO:0042947 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.1 | 0.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 0.3 | GO:0015343 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.1 | 0.4 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 0.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 3.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 0.3 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.1 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.6 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.1 | 0.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.2 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 2.8 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.0 | 0.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.0 | 0.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0000772 | mating pheromone activity(GO:0000772) pheromone activity(GO:0005186) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 2.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0060089 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
| 0.0 | 1.6 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 3.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.8 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.7 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.2 | GO:0016723 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.2 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.0 | GO:0015188 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.0 | 0.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.0 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.0 | 0.3 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.0 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.3 | 0.9 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.3 | 2.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 0.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 0.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 15.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.4 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 2.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.2 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.3 | REACTOME CHROMOSOME MAINTENANCE | Genes involved in Chromosome Maintenance |
| 0.0 | 0.2 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME HEMOSTASIS | Genes involved in Hemostasis |
| 0.0 | 0.1 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.0 | REACTOME TRANSCRIPTION | Genes involved in Transcription |
| 0.0 | 15.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |