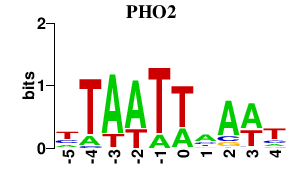
Results for PHO2
Z-value: 0.51
Transcription factors associated with PHO2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PHO2
|
S000002264 | Homeobox transcription factor |
Activity-expression correlation:
Activity profile of PHO2 motif
Sorted Z-values of PHO2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YOL086C | 1.46 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YGR108W | 1.33 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YOR101W | 1.18 |
RAS1
|
GTPase involved in G-protein signaling in the adenylate cyclase activating pathway, plays a role in cell proliferation; localized to the plasma membrane; homolog of mammalian RAS proto-oncogenes |
|
| YER055C | 1.18 |
HIS1
|
ATP phosphoribosyltransferase, a hexameric enzyme, catalyzes the first step in histidine biosynthesis; mutations cause histidine auxotrophy and sensitivity to Cu, Co, and Ni salts; transcription is regulated by general amino acid control |
|
| YPR119W | 1.00 |
CLB2
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YGL008C | 0.99 |
PMA1
|
Plasma membrane H+-ATPase, pumps protons out of the cell; major regulator of cytoplasmic pH and plasma membrane potential; part of the P2 subgroup of cation-transporting ATPases |
|
| YNR001W-A | 0.98 |
Dubious open reading frame unlikely to encode a functional protein; identified by homology |
||
| YFR056C | 0.98 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YCR009C | 0.95 |
RVS161
|
Amphiphysin-like lipid raft protein; subunit of a complex (Rvs161p-Rvs167p) that regulates polarization of the actin cytoskeleton, endocytosis, cell polarity, cell fusion and viability following starvation or osmotic stress |
|
| YFR055W | 0.95 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YDR399W | 0.94 |
HPT1
|
Dimeric hypoxanthine-guanine phosphoribosyltransferase, catalyzes the formation of both inosine monophosphate and guanosine monophosphate; mutations in the human homolog HPRT1 can cause Lesch-Nyhan syndrome and Kelley-Seegmiller syndrome |
|
| YDR050C | 0.90 |
TPI1
|
Triose phosphate isomerase, abundant glycolytic enzyme; mRNA half-life is regulated by iron availability; transcription is controlled by activators Reb1p, Gcr1p, and Rap1p through binding sites in the 5' non-coding region |
|
| YOR182C | 0.85 |
RPS30B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps30Ap and has similarity to rat S30 ribosomal protein |
|
| YDR417C | 0.82 |
Hypothetical protein |
||
| YNL189W | 0.80 |
SRP1
|
Karyopherin alpha homolog, forms a dimer with karyopherin beta Kap95p to mediate import of nuclear proteins, binds the nuclear localization signal of the substrate during import; may also play a role in regulation of protein degradation |
|
| YDR033W | 0.77 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YOL085C | 0.68 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOL085W-A |
||
| YDR064W | 0.67 |
RPS13
|
Protein component of the small (40S) ribosomal subunit; has similarity to E. coli S15 and rat S13 ribosomal proteins |
|
| YDR257C | 0.64 |
SET7
|
Nuclear protein that contains a SET-domain, which have been shown to mediate methyltransferase activity in other proteins |
|
| YLR339C | 0.63 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the essential gene RPP0 |
||
| YHR180W-A | 0.62 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps dubious ORF YHR180C-B and long terminal repeat YHRCsigma3 |
||
| YER117W | 0.62 |
RPL23B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl23Ap and has similarity to E. coli L14 and rat L23 ribosomal proteins |
|
| YMR281W | 0.61 |
GPI12
|
ER membrane protein involved in the second step of glycosylphosphatidylinositol (GPI) anchor assembly, the de-N-acetylation of the N-acetylglucosaminylphosphatidylinositol intermediate; functional homolog of human PIG-Lp |
|
| YGR118W | 0.59 |
RPS23A
|
Ribosomal protein 28 (rp28) of the small (40S) ribosomal subunit, required for translational accuracy; nearly identical to Rps23Bp and similar to E. coli S12 and rat S23 ribosomal proteins; deletion of both RPS23A and RPS23B is lethal |
|
| YLR301W | 0.54 |
Protein of unknown function that interacts with Sec72p |
||
| YNL024C | 0.54 |
Putative protein of unknown function with seven beta-strand methyltransferase motif; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YNL024C is not an essential gene |
||
| YLR340W | 0.54 |
RPP0
|
Conserved ribosomal protein P0 similar to rat P0, human P0, and E. coli L10e; shown to be phosphorylated on serine 302 |
|
| YPL007C | 0.51 |
TFC8
|
One of six subunits of RNA polymerase III transcription initiation factor complex (TFIIIC); part of TFIIIC TauB domain that binds BoxB promoter sites of tRNA and other genes; linker between TauB and TauA domains; human homolog is TFIIIC-90 |
|
| YHR181W | 0.49 |
SVP26
|
Integral membrane protein of the early Golgi apparatus and endoplasmic reticulum, involved in COP II vesicle transport; may also function to promote retention of proteins in the early Golgi compartment |
|
| YGR139W | 0.49 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR138C | 0.48 |
TPO2
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; transcription of TPO2 is regulated by Haa1p; member of the major facilitator superfamily |
|
| YER026C | 0.48 |
CHO1
|
Phosphatidylserine synthase, functions in phospholipid biosynthesis; catalyzes the reaction CDP-diaclyglycerol + L-serine = CMP + L-1-phosphatidylserine, transcriptionally repressed by myo-inositol and choline |
|
| YKL081W | 0.47 |
TEF4
|
Translation elongation factor EF-1 gamma |
|
| YLR341W | 0.46 |
SPO77
|
Meiosis-specific protein of unknown function, required for spore wall formation during sporulation; dispensable for both nuclear divisions during meiosis |
|
| YCR013C | 0.43 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; transcription of both YCR013C and the overlapping essential gene PGK1 is reduced in a gcr1 null mutant |
||
| YBR158W | 0.42 |
AMN1
|
Protein required for daughter cell separation, multiple mitotic checkpoints, and chromosome stability; contains 12 degenerate leucine-rich repeat motifs; expression is induced by the Mitotic Exit Network (MEN) |
|
| YGL123C-A | 0.41 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene RPS2/YGL123W |
||
| YOR047C | 0.40 |
STD1
|
Protein involved in control of glucose-regulated gene expression; interacts with protein kinase Snf1p, glucose sensors Snf3p and Rgt2p, and TATA-binding protein Spt15p; acts as a regulator of the transcription factor Rgt1p |
|
| YGL123W | 0.40 |
RPS2
|
Protein component of the small (40S) subunit, essential for control of translational accuracy; has similarity to E. coli S5 and rat S2 ribosomal proteins |
|
| YHR094C | 0.39 |
HXT1
|
Low-affinity glucose transporter of the major facilitator superfamily, expression is induced by Hxk2p in the presence of glucose and repressed by Rgt1p when glucose is limiting |
|
| YML075C | 0.39 |
HMG1
|
One of two isozymes of HMG-CoA reductase that catalyzes the conversion of HMG-CoA to mevalonate, which is a rate-limiting step in sterol biosynthesis; localizes to the nuclear envelope; overproduction induces the formation of karmellae |
|
| YDL228C | 0.38 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; almost completely overlaps the verified gene SSB1 |
||
| YOR232W | 0.38 |
MGE1
|
Protein of the mitochondrial matrix involved in protein import into mitochondria; acts as a cochaperone and a nucleotide release factor for Ssc1p; homolog of E. coli GrpE |
|
| YPL163C | 0.38 |
SVS1
|
Cell wall and vacuolar protein, required for wild-type resistance to vanadate |
|
| YHR174W | 0.38 |
ENO2
|
Enolase II, a phosphopyruvate hydratase that catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate during glycolysis and the reverse reaction during gluconeogenesis; expression is induced in response to glucose |
|
| YKL209C | 0.37 |
STE6
|
Plasma membrane ATP-binding cassette (ABC) transporter required for the export of a-factor, catalyzes ATP hydrolysis coupled to a-factor transport; contains 12 transmembrane domains and two ATP binding domains; expressed only in MATa cells |
|
| YML008C | 0.37 |
ERG6
|
Delta(24)-sterol C-methyltransferase, converts zymosterol to fecosterol in the ergosterol biosynthetic pathway by methylating position C-24; localized to both lipid particles and mitochondrial outer membrane |
|
| YDR450W | 0.36 |
RPS18A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps18Bp and has similarity to E. coli S13 and rat S18 ribosomal proteins |
|
| YBR085W | 0.36 |
AAC3
|
Mitochondrial inner membrane ADP/ATP translocator, exchanges cytosolic ADP for mitochondrially synthesized ATP; expressed under anaerobic conditions; similar to Pet9p and Aac1p; has roles in maintenance of viability and in respiration |
|
| YDR278C | 0.35 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YIL049W | 0.35 |
DFG10
|
Protein of unknown function, involved in filamentous growth |
|
| YFR001W | 0.35 |
LOC1
|
Nuclear protein involved in asymmetric localization of ASH1 mRNA; binds double-stranded RNA in vitro; constituent of 66S pre-ribosomal particles |
|
| YNL084C | 0.35 |
END3
|
EH domain-containing protein involved in endocytosis, actin cytoskeletal organization and cell wall morphogenesis; forms a complex with Sla1p and Pan1p |
|
| YKL153W | 0.35 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; transcription of both YLK153W and the overlapping essential gene GPM1 is reduced in the gcr1 null mutant |
||
| YIR021W | 0.34 |
MRS1
|
Protein required for the splicing of two mitochondrial group I introns (BI3 in COB and AI5beta in COX1); forms a splicing complex, containing four subunits of Mrs1p and two subunits of the BI3-encoded maturase, that binds to the BI3 RNA |
|
| YLR449W | 0.34 |
FPR4
|
Peptidyl-prolyl cis-trans isomerase (PPIase) (proline isomerase) localized to the nucleus; catalyzes isomerization of proline residues in histones H3 and H4, which affects lysine methylation of those histones |
|
| YKR041W | 0.34 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YDL134C | 0.33 |
PPH21
|
Catalytic subunit of protein phosphatase 2A, functionally redundant with Pph22p; methylated at C terminus; forms alternate complexes with several regulatory subunits; involved in signal transduction and regulation of mitosis |
|
| YLR261C | 0.33 |
VPS63
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 98% of ORF overlaps the verified gene YPT6; deletion causes a vacuolar protein sorting defect |
|
| YKR040C | 0.33 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YKR041W |
||
| YOR108W | 0.33 |
LEU9
|
Alpha-isopropylmalate synthase II (2-isopropylmalate synthase), catalyzes the first step in the leucine biosynthesis pathway; the minor isozyme, responsible for the residual alpha-IPMS activity detected in a leu4 null mutant |
|
| YCR004C | 0.33 |
YCP4
|
Protein of unknown function, has sequence and structural similarity to flavodoxins; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YGL077C | 0.33 |
HNM1
|
Choline/ethanolamine transporter; involved in the uptake of nitrogen mustard and the uptake of glycine betaine during hypersaline stress; co-regulated with phospholipid biosynthetic genes and negatively regulated by choline and myo-inositol |
|
| YNL090W | 0.33 |
RHO2
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, involved in the establishment of cell polarity and in microtubule assembly |
|
| YKL154W | 0.33 |
SRP102
|
Signal recognition particle (SRP) receptor beta subunit; involved in SRP-dependent protein targeting; anchors Srp101p to the ER membrane |
|
| YGR036C | 0.32 |
CAX4
|
Dolichyl pyrophosphate (Dol-P-P) phosphatase with a luminally oriented active site in the ER, cleaves the anhydride linkage in Dol-P-P, required for Dol-P-P-linked oligosaccharide intermediate synthesis and protein N-glycosylation |
|
| YLR103C | 0.32 |
CDC45
|
DNA replication initiation factor; recruited to MCM pre-RC complexes at replication origins; promotes release of MCM from Mcm10p, recruits elongation machinery; mutants in human homolog may cause velocardiofacial and DiGeorge syndromes |
|
| YKR094C | 0.31 |
RPL40B
|
Fusion protein, identical to Rpl40Ap, that is cleaved to yield ubiquitin and a ribosomal protein of the large (60S) ribosomal subunit with similarity to rat L40; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes |
|
| YNR016C | 0.31 |
ACC1
|
Acetyl-CoA carboxylase, biotin containing enzyme that catalyzes the carboxylation of acetyl-CoA to form malonyl-CoA; required for de novo biosynthesis of long-chain fatty acids |
|
| YDR279W | 0.31 |
RNH202
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YDR341C | 0.31 |
Arginyl-tRNA synthetase, proposed to be cytoplasmic but the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YJR054W | 0.31 |
Vacuolar protein of unknown function; potential Cdc28p substrate |
||
| YKL096W-A | 0.31 |
CWP2
|
Covalently linked cell wall mannoprotein, major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored |
|
| YBR084C-A | 0.31 |
RPL19A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl19Bp and has similarity to rat L19 ribosomal protein; rpl19a and rpl19b single null mutations result in slow growth, while the double null mutation is lethal |
|
| YEL040W | 0.30 |
UTR2
|
Cell wall protein that functions in the transfer of chitin to beta(1-6)glucan; putative chitin transglycosidase; glycosylphosphatidylinositol (GPI)-anchored protein localized to the bud neck; has a role in cell wall maintenance |
|
| YDR098C | 0.30 |
GRX3
|
Hydroperoxide and superoxide-radical responsive glutathione-dependent oxidoreductase; monothiol glutaredoxin subfamily member along with Grx4p and Grx5p; protects cells from oxidative damage |
|
| YFR044C | 0.29 |
DUG1
|
Probable di- and tri-peptidase; forms a complex with Dug2p and Dug3p to degrade glutathione (GSH) and other peptides containing a gamma-glu-X bond in an alternative pathway to GSH degradation by gamma-glutamyl transpeptidase (Ecm38p) |
|
| YLR328W | 0.29 |
NMA1
|
Nicotinic acid mononucleotide adenylyltransferase, involved in pathways of NAD biosynthesis, including the de novo, NAD(+) salvage, and nicotinamide riboside salvage pathways |
|
| YOR340C | 0.29 |
RPA43
|
RNA polymerase I subunit A43 |
|
| YGR107W | 0.29 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YIL116W | 0.29 |
HIS5
|
Histidinol-phosphate aminotransferase, catalyzes the seventh step in histidine biosynthesis; responsive to general control of amino acid biosynthesis; mutations cause histidine auxotrophy and sensitivity to Cu, Co, and Ni salts |
|
| YDR025W | 0.28 |
RPS11A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps11Bp and has similarity to E. coli S17 and rat S11 ribosomal proteins |
|
| YOR184W | 0.28 |
SER1
|
3-phosphoserine aminotransferase, catalyzes the formation of phosphoserine from 3-phosphohydroxypyruvate, required for serine and glycine biosynthesis; regulated by the general control of amino acid biosynthesis mediated by Gcn4p |
|
| YPL108W | 0.28 |
Cytoplasmic protein of unknown function; non-essential gene that is induced in a GDH1 deleted strain with altered redox metabolism; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YGR106C | 0.28 |
VOA1
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuolar memebrane |
|
| YML063W | 0.28 |
RPS1B
|
Ribosomal protein 10 (rp10) of the small (40S) subunit; nearly identical to Rps1Ap and has similarity to rat S3a ribosomal protein |
|
| YDR023W | 0.28 |
SES1
|
Cytosolic seryl-tRNA synthetase, class II aminoacyl-tRNA synthetase that aminoacylates tRNA(Ser), displays tRNA-dependent amino acid recognition which enhances discrimination of the serine substrate, interacts with peroxin Pex21p |
|
| YKL057C | 0.27 |
NUP120
|
Subunit of the Nup84p subcomplex of the nuclear pore complex (NPC), required for even distribution of NPCs around the nuclear envelope, involved in establishment of a normal nucleocytoplasmic concentration gradient of the GTPase Gsp1p |
|
| YCR099C | 0.27 |
Putative protein of unknown function |
||
| YGR090W | 0.27 |
UTP22
|
Possible U3 snoRNP protein involved in maturation of pre-18S rRNA, based on computational analysis of large-scale protein-protein interaction data |
|
| YMR123W | 0.27 |
PKR1
|
V-ATPase assembly factor, functions with other V-ATPase assembly factors in the ER to efficiently assemble the V-ATPase membrane sector (V0); overproduction confers resistance to Pichia farinosa killer toxin |
|
| YAL010C | 0.27 |
MDM10
|
Subunit of both the Mdm10-Mdm12-Mmm1 complex and the mitochondrial sorting and assembly machinery (SAM complex); functions in both the general and Tom40p-specific pathways for import and assembly of outer membrane beta-barrel proteins |
|
| YGR027C | 0.27 |
RPS25A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps25Bp and has similarity to rat S25 ribosomal protein |
|
| YPL197C | 0.26 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the ribosomal gene RPB7B |
||
| YGL145W | 0.26 |
TIP20
|
Peripheral membrane protein required for fusion of COPI vesicles with the ER, prohibits back-fusion of COPII vesicles with the ER, may act as a sensor for vesicles at the ER membrane; interacts with Sec20p |
|
| YOR287C | 0.26 |
Putative protein of unknown function; may play a role in the ribosome and rRNA biosynthesis based on expression profiles and mutant phenotype |
||
| YBL068W-A | 0.26 |
Dubious open reading frame unlikely to encode a protein; identified by fungal homology and RT-PCR |
||
| YJR123W | 0.26 |
RPS5
|
Protein component of the small (40S) ribosomal subunit, the least basic of the non-acidic ribosomal proteins; phosphorylated in vivo; essential for viability; has similarity to E. coli S7 and rat S5 ribosomal proteins |
|
| YKR038C | 0.26 |
KAE1
|
Putative glycoprotease proposed to be in transcription as a component of the EKC protein complex with Bud32p, Cgi121p, Pcc1p, and Gon7p; also identified as a component of the KEOPS protein complex |
|
| YNL300W | 0.26 |
Glycosylphosphatidylinositol-dependent cell wall protein, expression is periodic and decreases in respone to ergosterol perturbation or upon entry into stationary phase; depletion increases resistance to lactic acid |
||
| YKR061W | 0.26 |
KTR2
|
Mannosyltransferase involved in N-linked protein glycosylation; member of the KRE2/MNT1 mannosyltransferase family |
|
| YLR351C | 0.26 |
NIT3
|
Nit protein, one of two proteins in S. cerevisiae with similarity to the Nit domain of NitFhit from fly and worm and to the mouse and human Nit protein which interacts with the Fhit tumor suppressor; nitrilase superfamily member |
|
| YDR284C | 0.25 |
DPP1
|
Diacylglycerol pyrophosphate (DGPP) phosphatase, zinc-regulated vacuolar membrane-associated lipid phosphatase, dephosphorylates DGPP to phosphatidate (PA) and Pi, then PA to diacylglycerol; involved in lipid signaling and cell metabolism |
|
| YPL225W | 0.25 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YMR011W | 0.25 |
HXT2
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YPL143W | 0.24 |
RPL33A
|
N-terminally acetylated ribosomal protein L37 of the large (60S) ribosomal subunit, nearly identical to Rpl33Bp and has similarity to rat L35a; rpl33a null mutant exhibits slow growth while rpl33a rpl33b double null mutant is inviable |
|
| YLR441C | 0.24 |
RPS1A
|
Ribosomal protein 10 (rp10) of the small (40S) subunit; nearly identical to Rps1Bp and has similarity to rat S3a ribosomal protein |
|
| YOR309C | 0.24 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene NOP58 |
||
| YPL090C | 0.24 |
RPS6A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps6Bp and has similarity to rat S6 ribosomal protein |
|
| YDR210W | 0.24 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery |
||
| YJL002C | 0.24 |
OST1
|
Alpha subunit of the oligosaccharyltransferase complex of the ER lumen, which catalyzes asparagine-linked glycosylation of newly synthesized proteins |
|
| YDR267C | 0.24 |
CIA1
|
Essential protein involved in assembly of cytosolic and nuclear iron-sulfur proteins |
|
| YBL098W | 0.24 |
BNA4
|
Kynurenine 3-mono oxygenase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p; putative therapeutic target for Huntington disease |
|
| YHR143W | 0.24 |
DSE2
|
Daughter cell-specific secreted protein with similarity to glucanases, degrades cell wall from the daughter side causing daughter to separate from mother; expression is repressed by cAMP |
|
| YNL320W | 0.24 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YJR145C | 0.24 |
RPS4A
|
Protein component of the small (40S) ribosomal subunit; mutation affects 20S pre-rRNA processing; identical to Rps4Bp and has similarity to rat S4 ribosomal protein |
|
| YJR094W-A | 0.24 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YNL303W | 0.24 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YNL301C | 0.24 |
RPL18B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl18Ap and has similarity to rat L18 ribosomal protein |
|
| YFR054C | 0.23 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR117C | 0.23 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YNL079C | 0.23 |
TPM1
|
Major isoform of tropomyosin; binds to and stabilizes actin cables and filaments, which direct polarized cell growth and the distribution of several organelles; acetylated by the NatB complex and acetylated form binds actin most efficiently |
|
| YBR032W | 0.23 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YIL131C | 0.23 |
FKH1
|
Forkhead family transcription factor with a minor role in the expression of G2/M phase genes; negatively regulates transcriptional elongation; positive role in chromatin silencing at HML and HMR; regulates donor preference during switching |
|
| YCR098C | 0.23 |
GIT1
|
Plasma membrane permease, mediates uptake of glycerophosphoinositol and glycerophosphocholine as sources of the nutrients inositol and phosphate; expression and transport rate are regulated by phosphate and inositol availability |
|
| YLR154C | 0.23 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YKL014C | 0.22 |
URB1
|
Nucleolar protein required for the normal accumulation of 25S and 5.8S rRNAs, associated with the 27SA2 pre-ribosomal particle; proposed to be involved in the biogenesis of the 60S ribosomal subunit |
|
| YGR156W | 0.22 |
PTI1
|
Pta1p Interacting protein |
|
| YGL158W | 0.22 |
RCK1
|
Protein kinase involved in the response to oxidative stress; identified as suppressor of S. pombe cell cycle checkpoint mutations |
|
| YGR140W | 0.22 |
CBF2
|
Essential kinetochore protein, component of the CBF3 multisubunit complex that binds to the CDEIII region of the centromere; Cbf2p also binds to the CDEII region possibly forming a different multimeric complex, ubiquitinated in vivo |
|
| YIL124W | 0.22 |
AYR1
|
NADPH-dependent 1-acyl dihydroxyacetone phosphate reductase found in lipid particles, ER, and mitochondrial outer membrane; involved in phosphatidic acid biosynthesis; required for spore germination; capable of metabolizing steroid hormones |
|
| YIL018W | 0.22 |
RPL2B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl2Ap and has similarity to E. coli L2 and rat L8 ribosomal proteins; expression is upregulated at low temperatures |
|
| YIL052C | 0.22 |
RPL34B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl34Ap and has similarity to rat L34 ribosomal protein |
|
| YNL078W | 0.21 |
NIS1
|
Protein localized in the bud neck at G2/M phase; physically interacts with septins; possibly involved in a mitotic signaling network |
|
| YML073C | 0.21 |
RPL6A
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, has similarity to Rpl6Bp and to rat L6 ribosomal protein; binds to 5.8S rRNA |
|
| YNL222W | 0.21 |
SSU72
|
Transcription/RNA-processing factor that associates with TFIIB and cleavage/polyadenylation factor Pta1p; exhibits phosphatase activity on serine-5 of the RNA polymerase II C-terminal domain; affects start site selection in vivo |
|
| YEL027W | 0.21 |
CUP5
|
Proteolipid subunit of the vacuolar H(+)-ATPase V0 sector (subunit c; dicyclohexylcarbodiimide binding subunit); required for vacuolar acidification and important for copper and iron metal ion homeostasis |
|
| YGR148C | 0.21 |
RPL24B
|
Ribosomal protein L30 of the large (60S) ribosomal subunit, nearly identical to Rpl24Ap and has similarity to rat L24 ribosomal protein; not essential for translation but may be required for normal translation rate |
|
| YOR356W | 0.21 |
Mitochondrial protein with similarity to flavoprotein-type oxidoreductases; found in a large supramolecular complex with other mitochondrial dehydrogenases |
||
| YJL190C | 0.21 |
RPS22A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps22Bp and has similarity to E. coli S8 and rat S15a ribosomal proteins |
|
| YIR020C-B | 0.21 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified ORF MRS1 |
||
| YML036W | 0.21 |
CGI121
|
Protein involved in telomere uncapping and elongation as component of the KEOPS protein complex with Bud32p, Kae1p, Pcc1p, and Gon7p; also shown to be a component of the EKC protein complex; homolog of human CGI-121 |
|
| YAL007C | 0.21 |
ERP2
|
Protein that forms a heterotrimeric complex with Erp1p, Emp24p, and Erv25p; member, along with Emp24p and Erv25p, of the p24 family involved in ER to Golgi transport and localized to COPII-coated vesicles |
|
| YNR075W | 0.21 |
COS10
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YLR190W | 0.21 |
MMR1
|
Phosphorylated protein of the mitochondrial outer membrane, localizes only to mitochondria of the bud; interacts with Myo2p to mediate mitochondrial distribution to buds; mRNA is targeted to the bud via the transport system involving She2p |
|
| YDR041W | 0.20 |
RSM10
|
Mitochondrial ribosomal protein of the small subunit, has similarity to E. coli S10 ribosomal protein; essential for viability, unlike most other mitoribosomal proteins |
|
| YKR013W | 0.20 |
PRY2
|
Protein of unknown function, has similarity to Pry1p and Pry3p and to the plant PR-1 class of pathogen related proteins |
|
| YCR012W | 0.20 |
PGK1
|
3-phosphoglycerate kinase, catalyzes transfer of high-energy phosphoryl groups from the acyl phosphate of 1,3-bisphosphoglycerate to ADP to produce ATP; key enzyme in glycolysis and gluconeogenesis |
|
| YML115C | 0.20 |
VAN1
|
Component of the mannan polymerase I, which contains Van1p and Mnn9p and is involved in the first steps of mannan synthesis; mutants are vanadate-resistant |
|
| YBL004W | 0.20 |
UTP20
|
Component of the small-subunit (SSU) processome, which is involved in the biogenesis of the 18S rRNA |
|
| YPL241C | 0.20 |
CIN2
|
GTPase-activating protein (GAP) for Cin4p; tubulin folding factor C involved in beta-tubulin (Tub2p) folding; mutants display increased chromosome loss and benomyl sensitivity; deletion complemented by human GAP, retinitis pigmentosa 2 |
|
| YMR108W | 0.20 |
ILV2
|
Acetolactate synthase, catalyses the first common step in isoleucine and valine biosynthesis and is the target of several classes of inhibitors, localizes to the mitochondria; expression of the gene is under general amino acid control |
|
| YLR344W | 0.20 |
RPL26A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl26Bp and has similarity to E. coli L24 and rat L26 ribosomal proteins; binds to 5.8S rRNA |
|
| YNL178W | 0.20 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YGL135W | 0.20 |
RPL1B
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, nearly identical to Rpl1Ap and has similarity to E. coli L1 and rat L10a ribosomal proteins; rpl1a rpl1b double null mutation is lethal |
|
| YMR082C | 0.20 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR361C | 0.20 |
BCP1
|
Essential protein involved in nuclear export of Mss4p, which is a lipid kinase that generates phosphatidylinositol 4,5-biphosphate and plays a role in actin cytoskeleton organization and vesicular transport |
|
| YPL273W | 0.19 |
SAM4
|
S-adenosylmethionine-homocysteine methyltransferase, functions along with Mht1p in the conversion of S-adenosylmethionine (AdoMet) to methionine to control the methionine/AdoMet ratio |
|
| YDL143W | 0.19 |
CCT4
|
Subunit of the cytosolic chaperonin Cct ring complex, related to Tcp1p, required for the assembly of actin and tubulins in vivo |
|
| YDL111C | 0.19 |
RRP42
|
Protein involved in rRNA processing; component of the exosome 3->5 exonuclease complex with Rrp4p, Rrp41p, Rrp43p and Dis3p |
|
| YOR133W | 0.19 |
EFT1
|
Elongation factor 2 (EF-2), also encoded by EFT2; catalyzes ribosomal translocation during protein synthesis; contains diphthamide, the unique posttranslationally modified histidine residue specifically ADP-ribosylated by diphtheria toxin |
|
| YEL056W | 0.19 |
HAT2
|
Subunit of the Hat1p-Hat2p histone acetyltransferase complex; required for high affinity binding of the complex to free histone H4, thereby enhancing Hat1p activity; similar to human RbAp46 and 48; has a role in telomeric silencing |
|
| YJR001W | 0.19 |
AVT1
|
Vacuolar transporter, imports large neutral amino acids into the vacuole; member of a family of seven S. cerevisiae genes (AVT1-7) related to vesicular GABA-glycine transporters |
|
| YDR183C-A | 0.19 |
Dubious open reading frame unlikely to encode a functional protein; identified by fungal homology and RT-PCR |
||
| YOR063W | 0.19 |
RPL3
|
Protein component of the large (60S) ribosomal subunit, has similarity to E. coli L3 and rat L3 ribosomal proteins; involved in the replication and maintenance of killer double stranded RNA virus |
|
| YGL229C | 0.19 |
SAP4
|
Protein required for function of the Sit4p protein phosphatase, member of a family of similar proteins that form complexes with Sit4p, including Sap155p, Sap185p, and Sap190p |
|
| YKR012C | 0.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene PRY2 |
||
| YOR315W | 0.19 |
SFG1
|
Nuclear protein, putative transcription factor required for growth of superficial pseudohyphae (which do not invade the agar substrate) but not for invasive pseudohyphal growth; may act together with Phd1p; potential Cdc28p substrate |
|
| YJL158C | 0.18 |
CIS3
|
Mannose-containing glycoprotein constituent of the cell wall; member of the PIR (proteins with internal repeats) family |
|
| YDR507C | 0.18 |
GIN4
|
Protein kinase involved in bud growth and assembly of the septin ring, proposed to have kinase-dependent and kinase-independent activities; undergoes autophosphorylation; similar to Kcc4p and Hsl1p |
|
| YHR173C | 0.18 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YKR026C | 0.18 |
GCN3
|
Alpha subunit of the translation initiation factor eIF2B, the guanine-nucleotide exchange factor for eIF2; activity subsequently regulated by phosphorylated eIF2; first identified as a positive regulator of GCN4 expression |
|
| YHR021C | 0.18 |
RPS27B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps27Ap and has similarity to rat S27 ribosomal protein |
|
| YJR010C-A | 0.18 |
SPC1
|
Subunit of the signal peptidase complex (SPC), which cleaves the signal sequence from proteins targeted to the endoplasmic reticulum (ER), homolog of the SPC12 subunit of mammalian signal peptidase complex |
|
| YMR307W | 0.18 |
GAS1
|
Beta-1,3-glucanosyltransferase, required for cell wall assembly; localizes to the cell surface via a glycosylphosphatidylinositol (GPI) anchor |
|
| YNL112W | 0.18 |
DBP2
|
Essential ATP-dependent RNA helicase of the DEAD-box protein family, involved in nonsense-mediated mRNA decay and rRNA processing |
|
| YMR143W | 0.18 |
RPS16A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps16Bp and has similarity to E. coli S9 and rat S16 ribosomal proteins |
|
| YPL235W | 0.18 |
RVB2
|
Essential protein involved in transcription regulation; component of chromatin remodeling complexes; required for assembly and function of the INO80 complex; member of the RUVB-like protein family |
|
| YGL028C | 0.17 |
SCW11
|
Cell wall protein with similarity to glucanases; may play a role in conjugation during mating based on its regulation by Ste12p |
|
| YIL008W | 0.17 |
URM1
|
Ubiquitin-like protein with weak sequence similarity to ubiquitin; depends on the E1-like activating enzyme Uba4p; molecular function of the Urm1p pathway is unknown, but it is required for normal growth, particularly at high temperature |
|
| YER044C | 0.17 |
ERG28
|
Endoplasmic reticulum membrane protein, may facilitate protein-protein interactions between the Erg26p dehydrogenase and the Erg27p 3-ketoreductase and/or tether these enzymes to the ER, also interacts with Erg6p |
|
| YLR448W | 0.17 |
RPL6B
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl6Ap and to rat L6 ribosomal protein; binds to 5.8S rRNA |
|
| YHR140W | 0.17 |
Putative integral membrane protein of unknown function |
||
| YML019W | 0.17 |
OST6
|
Subunit of the oligosaccharyltransferase complex of the ER lumen, which catalyzes asparagine-linked glycosylation of newly synthesized proteins; similar to and partially functionally redundant with Ost3p |
|
| YDR418W | 0.17 |
RPL12B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl12Ap; rpl12a rpl12b double mutant exhibits slow growth and slow translation; has similarity to E. coli L11 and rat L12 ribosomal proteins |
|
| YPL137C | 0.17 |
GIP3
|
Glc7-interacting protein whose overexpression relocalizes Glc7p from the nucleus and prevents chromosome segregation; may interact with ribosomes, based on co-purification experiments |
|
| YKR079C | 0.17 |
TRZ1
|
tRNase Z, involved in RNA processing, has two putative nucleotide triphosphate-binding motifs (P-loop) and a conserved histidine motif, homolog of the human candidate prostate cancer susceptibility gene ELAC2 |
|
| YOR226C | 0.17 |
ISU2
|
Conserved protein of the mitochondrial matrix, required for synthesis of mitochondrial and cytosolic iron-sulfur proteins, performs a scaffolding function in mitochondria during Fe/S cluster assembly; isu1 isu2 double mutant is inviable |
|
| YPL243W | 0.16 |
SRP68
|
Core component of the signal recognition particle (SRP) ribonucleoprotein (RNP) complex that functions in targeting nascent secretory proteins to the endoplasmic reticulum (ER) membrane |
|
| YMR142C | 0.16 |
RPL13B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl13Ap; not essential for viability; has similarity to rat L13 ribosomal protein |
|
| YOR183W | 0.16 |
FYV12
|
Protein of unknown function, required for survival upon exposure to K1 killer toxin |
|
| YBR067C | 0.16 |
TIP1
|
Major cell wall mannoprotein with possible lipase activity; transcription is induced by heat- and cold-shock; member of the Srp1p/Tip1p family of serine-alanine-rich proteins |
|
| YGR164W | 0.16 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR385W | 0.16 |
EFT2
|
Elongation factor 2 (EF-2), also encoded by EFT1; catalyzes ribosomal translocation during protein synthesis; contains diphthamide, the unique posttranslationally modified histidine residue specifically ADP-ribosylated by diphtheria toxin |
|
| YEL029C | 0.16 |
BUD16
|
Putative pyridoxal kinase, key enzyme in vitamin B6 metabolism; involved in maintaining levels of pyridoxal 5'-phosphate, the active form of vitamin B6; required for genome integrity; homolog of E. coli PdxK; involved in bud-site selection |
|
| YLR292C | 0.16 |
SEC72
|
Non-essential subunit of Sec63 complex (Sec63p, Sec62p, Sec66p and Sec72p); with Sec61 complex, Kar2p/BiP and Lhs1p forms a channel competent for SRP-dependent and post-translational SRP-independent protein targeting and import into the ER |
|
| YPL141C | 0.16 |
Putative protein kinase; similar to Kin4p; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YPL141C is not an essential gene |
||
| YLR370C | 0.16 |
ARC18
|
Subunit of the ARP2/3 complex, which is required for the motility and integrity of cortical actin patches |
|
| YMR198W | 0.16 |
CIK1
|
Kinesin-associated protein required for both karyogamy and mitotic spindle organization, interacts stably and specifically with Kar3p and may function to target this kinesin to a specific cellular role; has similarity to Vik1p |
|
| YOL011W | 0.16 |
PLB3
|
Phospholipase B (lysophospholipase) involved in phospholipid metabolism; hydrolyzes phosphatidylinositol and phosphatidylserine and displays transacylase activity in vitro |
Network of associatons between targets according to the STRING database.
First level regulatory network of PHO2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.3 | 2.4 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 0.3 | 1.0 | GO:0060988 | lipid tube assembly(GO:0060988) protein-lipid complex assembly(GO:0065005) protein-lipid complex subunit organization(GO:0071825) |
| 0.3 | 0.8 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.2 | 1.5 | GO:0019660 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.2 | 0.9 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 0.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.2 | 0.6 | GO:0000296 | spermine transport(GO:0000296) |
| 0.1 | 0.4 | GO:0070637 | nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) |
| 0.1 | 1.3 | GO:0052803 | histidine biosynthetic process(GO:0000105) histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.1 | 0.5 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.1 | 0.4 | GO:0019408 | polyprenol metabolic process(GO:0016093) polyprenol biosynthetic process(GO:0016094) dolichol metabolic process(GO:0019348) dolichol biosynthetic process(GO:0019408) |
| 0.1 | 0.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.4 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.4 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.4 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.1 | 0.3 | GO:0031057 | negative regulation of histone modification(GO:0031057) |
| 0.1 | 0.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.4 | GO:0001041 | transcription from RNA polymerase III type 2 promoter(GO:0001009) transcription from a RNA polymerase III hybrid type promoter(GO:0001041) |
| 0.1 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 0.2 | GO:0031555 | transcriptional attenuation(GO:0031555) transcription antitermination(GO:0031564) |
| 0.1 | 1.3 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.1 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.3 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 9.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 1.6 | GO:0006885 | regulation of pH(GO:0006885) |
| 0.1 | 0.3 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 1.6 | GO:0006757 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.1 | 0.4 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.3 | GO:0035392 | maintenance of chromatin silencing(GO:0006344) maintenance of chromatin silencing at telomere(GO:0035392) |
| 0.1 | 0.2 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.1 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.2 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.0 | 0.2 | GO:0072668 | obsolete tubulin complex biogenesis(GO:0072668) |
| 0.0 | 0.1 | GO:0071168 | establishment of protein localization to chromosome(GO:0070199) protein localization to chromatin(GO:0071168) establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.5 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0009202 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.2 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.4 | GO:0000372 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:0006798 | polyphosphate catabolic process(GO:0006798) phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0051320 | mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.0 | 0.1 | GO:0010942 | response to singlet oxygen(GO:0000304) positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) UDP-glucose transport(GO:0015786) |
| 0.0 | 0.7 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.1 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.3 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.2 | GO:0034755 | intracellular copper ion transport(GO:0015680) iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.4 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.0 | 0.4 | GO:0000921 | septin ring assembly(GO:0000921) |
| 0.0 | 0.6 | GO:0010383 | cell wall polysaccharide metabolic process(GO:0010383) |
| 0.0 | 0.1 | GO:0001111 | promoter clearance during DNA-templated transcription(GO:0001109) promoter clearance from RNA polymerase II promoter(GO:0001111) |
| 0.0 | 0.1 | GO:0031292 | gene conversion at mating-type locus, DNA double-strand break processing(GO:0031292) |
| 0.0 | 0.3 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.1 | GO:0007188 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.8 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) phytosteroid biosynthetic process(GO:0016129) cellular alcohol biosynthetic process(GO:0044108) cellular lipid biosynthetic process(GO:0097384) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.5 | GO:0001100 | negative regulation of exit from mitosis(GO:0001100) |
| 0.0 | 0.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 1.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.1 | GO:2000222 | positive regulation of cell growth(GO:0030307) positive regulation of pseudohyphal growth by positive regulation of transcription from RNA polymerase II promoter(GO:1900461) positive regulation of pseudohyphal growth(GO:2000222) |
| 0.0 | 0.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.2 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 0.4 | GO:0015833 | peptide transport(GO:0015833) |
| 0.0 | 0.7 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.7 | GO:0009247 | GPI anchor biosynthetic process(GO:0006506) glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.2 | GO:0030846 | termination of RNA polymerase II transcription, poly(A)-coupled(GO:0030846) |
| 0.0 | 0.1 | GO:0009615 | immune effector process(GO:0002252) immune system process(GO:0002376) response to virus(GO:0009615) response to external biotic stimulus(GO:0043207) defense response to virus(GO:0051607) response to other organism(GO:0051707) defense response to other organism(GO:0098542) |
| 0.0 | 0.0 | GO:0042710 | biofilm formation(GO:0042710) |
| 0.0 | 0.0 | GO:0071478 | cellular response to radiation(GO:0071478) cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.1 | GO:1903726 | negative regulation of phospholipid biosynthetic process(GO:0071072) negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.0 | 0.2 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of mitotic spindle localization(GO:0040001) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.2 | GO:0043487 | regulation of RNA stability(GO:0043487) regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0006116 | amino acid catabolic process to alcohol via Ehrlich pathway(GO:0000947) NADH oxidation(GO:0006116) |
| 0.0 | 0.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 0.1 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.3 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0060188 | regulation of protein desumoylation(GO:0060188) |
| 0.0 | 0.1 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.6 | GO:1902593 | protein import into nucleus(GO:0006606) nuclear import(GO:0051170) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.0 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.2 | GO:0000147 | actin cortical patch assembly(GO:0000147) actin cortical patch organization(GO:0044396) |
| 0.0 | 0.2 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.3 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 | 0.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0007535 | donor selection(GO:0007535) |
| 0.0 | 0.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.0 | 0.2 | GO:0071042 | nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.1 | GO:0070481 | nuclear-transcribed mRNA catabolic process, non-stop decay(GO:0070481) |
| 0.0 | 0.1 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.1 | GO:0001402 | signal transduction involved in filamentous growth(GO:0001402) |
| 0.0 | 0.0 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.2 | GO:0009304 | tRNA transcription(GO:0009304) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.0 | GO:0051469 | vesicle fusion with vacuole(GO:0051469) |
| 0.0 | 0.1 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0018208 | protein peptidyl-prolyl isomerization(GO:0000413) peptidyl-proline modification(GO:0018208) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 2.5 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 6.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.3 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.9 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 5.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.2 | GO:0031518 | CBF3 complex(GO:0031518) |
| 0.1 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.3 | GO:0032545 | CURI complex(GO:0032545) |
| 0.0 | 0.2 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum(GO:0005791) rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0043529 | GET complex(GO:0043529) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.3 | GO:0000144 | cellular bud neck septin ring(GO:0000144) |
| 0.0 | 0.2 | GO:0000136 | alpha-1,6-mannosyltransferase complex(GO:0000136) |
| 0.0 | 0.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0070860 | RNA polymerase I core factor complex(GO:0070860) |
| 0.0 | 0.2 | GO:0034044 | exomer complex(GO:0034044) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0000817 | COMA complex(GO:0000817) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0000818 | MIS12/MIND type complex(GO:0000444) nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.4 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex, spindle pole body(GO:0000928) gamma-tubulin complex(GO:0000930) gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.0 | 0.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 1.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.0 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.0 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.0 | GO:0097344 | Rix1 complex(GO:0097344) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.0 | GO:0034448 | EGO complex(GO:0034448) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.3 | 1.6 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 1.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 0.6 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.1 | 0.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 2.2 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.1 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.4 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 1.0 | GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism(GO:0015662) |
| 0.1 | 0.4 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.1 | 2.0 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 0.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0016885 | ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.1 | 0.4 | GO:0001004 | RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) transcription factor activity, RNA polymerase III promoter sequence-specific binding, TFIIIB recruiting(GO:0001004) transcription factor activity, RNA polymerase III type 1 promoter sequence-specific binding, TFIIIB recruiting(GO:0001005) transcription factor activity, RNA polymerase III type 2 promoter sequence-specific binding, TFIIIB recruiting(GO:0001008) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.1 | 0.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.3 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 0.1 | 0.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.3 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 0.1 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.2 | GO:0015188 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.1 | 0.6 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.1 | 0.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.3 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.1 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 2.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 7.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0042973 | glucan endo-1,3-beta-D-glucosidase activity(GO:0042973) |
| 0.0 | 0.2 | GO:0043141 | 5'-3' DNA helicase activity(GO:0043139) ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0001187 | transcription factor activity, RNA polymerase I CORE element sequence-specific binding(GO:0001169) transcription factor activity, RNA polymerase I CORE element binding transcription factor recruiting(GO:0001187) |
| 0.0 | 0.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0042124 | glucanosyltransferase activity(GO:0042123) 1,3-beta-glucanosyltransferase activity(GO:0042124) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.6 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.0 | 0.5 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.4 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.3 | GO:0032934 | steroid binding(GO:0005496) sterol binding(GO:0032934) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0001139 | transcription factor activity, core RNA polymerase II recruiting(GO:0001139) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
| 0.0 | 0.1 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) |
| 0.0 | 0.3 | GO:0015605 | organophosphate ester transmembrane transporter activity(GO:0015605) |
| 0.0 | 0.0 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0016769 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.0 | 0.3 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 0.1 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.3 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.4 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.6 | GO:0008092 | cytoskeletal protein binding(GO:0008092) |
| 0.0 | 0.0 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 1.5 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.1 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.5 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME GPCR DOWNSTREAM SIGNALING | Genes involved in GPCR downstream signaling |
| 0.0 | 0.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |