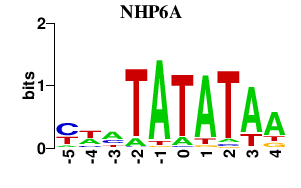
Results for NHP6A
Z-value: 0.41
Transcription factors associated with NHP6A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NHP6A
|
S000006256 | High-mobility group (HMG) protein, binds to and remodels nucleosomes |
Activity-expression correlation:
Activity profile of NHP6A motif
Sorted Z-values of NHP6A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YGL177W | 1.06 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YKR034W | 0.91 |
DAL80
|
Negative regulator of genes in multiple nitrogen degradation pathways; expression is regulated by nitrogen levels and by Gln3p; member of the GATA-binding family, forms homodimers and heterodimers with Deh1p |
|
| YPR169W-A | 0.84 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps two other dubious ORFs: YPR170C and YPR170W-B |
||
| YDR043C | 0.73 |
NRG1
|
Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters; mediates glucose repression and negatively regulates a variety of processes including filamentous growth and alkaline pH response |
|
| YGR161C | 0.70 |
RTS3
|
Putative component of the protein phosphatase type 2A complex |
|
| YLR307C-A | 0.69 |
Putative protein of unknown function |
||
| YJR094C | 0.63 |
IME1
|
Master regulator of meiosis that is active only during meiotic events, activates transcription of early meiotic genes through interaction with Ume6p, degraded by the 26S proteasome following phosphorylation by Ime2p |
|
| YGR039W | 0.58 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the Autonomously Replicating Sequence ARS722 |
||
| YJL149W | 0.58 |
DAS1
|
Putative SCF ubiquitin ligase F-box protein; interacts physically with both Cdc53p and Skp1 and genetically with CDC34; similar to putative F-box protein YDR131C; null mutant suppresses dst1delta sensitivity for 6-azauracil |
|
| YJL107C | 0.54 |
Putative protein of unknown function; expression is induced by activation of the HOG1 mitogen-activated signaling pathway and this induction is Hog1p/Pbs2p dependent; YJL107C and adjacent ORF, YJL108C are merged in related fungi |
||
| YAL062W | 0.54 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YMR175W | 0.54 |
SIP18
|
Protein of unknown function whose expression is induced by osmotic stress |
|
| YJL150W | 0.54 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDL023C | 0.52 |
Dubious open reading frame, unlikely to encode a protein; not conserved in other Saccharomyces species; overlaps the verified gene GPD1; deletion confers sensitivity to GSAO; deletion in cyr1 mutant results in loss of stress resistance |
||
| YDL022W | 0.51 |
GPD1
|
NAD-dependent glycerol-3-phosphate dehydrogenase, key enzyme of glycerol synthesis, essential for growth under osmotic stress; expression regulated by high-osmolarity glycerol response pathway; homolog of Gpd2p |
|
| YFL033C | 0.49 |
RIM15
|
Glucose-repressible protein kinase involved in signal transduction during cell proliferation in response to nutrients, specifically the establishment of stationary phase; identified as a regulator of IME2; substrate of Pho80p-Pho85p kinase |
|
| YGL178W | 0.48 |
MPT5
|
Member of the Puf family of RNA-binding proteins; binds to mRNAs encoding chromatin modifiers and spindle pole body components; involved in longevity, maintenance of cell wall integrity, and sensitivity to and recovery from pheromone arrest |
|
| YHR096C | 0.47 |
HXT5
|
Hexose transporter with moderate affinity for glucose, induced in the presence of non-fermentable carbon sources, induced by a decrease in growth rate, contains an extended N-terminal domain relative to other HXTs |
|
| YER158W-A | 0.46 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YKL065W-A | 0.45 |
Putative protein of unknown function |
||
| YOL034W | 0.45 |
SMC5
|
Structural maintenance of chromosomes (SMC) protein; essential subunit of the Mms21-Smc5-Smc6 complex; required for growth and DNA repair; S. pombe homolog forms a heterodimer with S. pombe Rad18p that is involved in DNA repair |
|
| YIL057C | 0.44 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YLR408C | 0.43 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the endosome; YLR408C is not an essential gene |
||
| YDL223C | 0.42 |
HBT1
|
Substrate of the Hub1p ubiquitin-like protein that localizes to the shmoo tip (mating projection); mutants are defective for mating projection formation, thereby implicating Hbt1p in polarized cell morphogenesis |
|
| YLR300W | 0.42 |
EXG1
|
Major exo-1,3-beta-glucanase of the cell wall, involved in cell wall beta-glucan assembly; exists as three differentially glycosylated isoenzymes |
|
| YJL200C | 0.42 |
ACO2
|
Putative mitochondrial aconitase isozyme; similarity to Aco1p, an aconitase required for the TCA cycle; expression induced during growth on glucose, by amino acid starvation via Gcn4p, and repressed on ethanol |
|
| YBR076C-A | 0.42 |
Dubious open reading frame unlikely to encode a protein; partially overlaps verified gene ECM8; identified by fungal homology and RT-PCR |
||
| YGR258C | 0.42 |
RAD2
|
Single-stranded DNA endonuclease, cleaves single-stranded DNA during nucleotide excision repair to excise damaged DNA; subunit of Nucleotide Excision Repair Factor 3 (NEF3); homolog of human XPG protein |
|
| YFL011W | 0.41 |
HXT10
|
Putative hexose transporter, expressed at low levels and expression is repressed by glucose |
|
| YLL046C | 0.41 |
RNP1
|
Ribonucleoprotein that contains two RNA recognition motifs (RRM) |
|
| YDR085C | 0.41 |
AFR1
|
Alpha-factor pheromone receptor regulator, negatively regulates pheromone receptor signaling; required for normal mating projection (shmoo) formation; required for Spa2p to recruit Mpk1p to shmoo tip during mating; interacts with Cdc12p |
|
| YER187W | 0.41 |
Putative protein of unknown function; induced in respiratory-deficient cells |
||
| YEL065W | 0.41 |
SIT1
|
Ferrioxamine B transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; transcription is induced during iron deprivation and diauxic shift; potentially phosphorylated by Cdc28p |
|
| YKL221W | 0.40 |
MCH2
|
Protein with similarity to mammalian monocarboxylate permeases, which are involved in transport of monocarboxylic acids across the plasma membrane; mutant is not deficient in monocarboxylate transport |
|
| YIL117C | 0.39 |
PRM5
|
Pheromone-regulated protein, predicted to have 1 transmembrane segment; induced during cell integrity signaling |
|
| YLR308W | 0.39 |
CDA2
|
Chitin deacetylase, together with Cda1p involved in the biosynthesis ascospore wall component, chitosan; required for proper rigidity of the ascospore wall |
|
| YKL105C | 0.38 |
Putative protein of unknown function |
||
| YJL194W | 0.38 |
CDC6
|
Essential ATP-binding protein required for DNA replication, component of the pre-replicative complex (pre-RC) which requires ORC to associate with chromatin and is in turn required for Mcm2-7p DNA association; homologous to S. pombe Cdc18p |
|
| YBR240C | 0.37 |
THI2
|
Zinc finger protein of the Zn(II)2Cys6 type, probable transcriptional activator of thiamine biosynthetic genes |
|
| YGR286C | 0.37 |
BIO2
|
Biotin synthase, catalyzes the conversion of dethiobiotin to biotin, which is the last step of the biotin biosynthesis pathway; complements E. coli bioB mutant |
|
| YMR193C-A | 0.37 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YML047W-A | 0.37 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOL159C | 0.37 |
Soluble protein of unknown function; deletion mutants are viable and have elevated levels of Ty1 retrotransposition and Ty1 cDNA |
||
| YDL138W | 0.36 |
RGT2
|
Plasma membrane glucose receptor, highly similar to Snf3p; both Rgt2p and Snf3p serve as transmembrane glucose sensors generating an intracellular signal that induces expression of glucose transporter (HXT) genes |
|
| YAL063C | 0.36 |
FLO9
|
Lectin-like protein with similarity to Flo1p, thought to be expressed and involved in flocculation |
|
| YOL123W | 0.36 |
HRP1
|
Subunit of cleavage factor I, a five-subunit complex required for the cleavage and polyadenylation of pre-mRNA 3' ends; RRM-containing heteronuclear RNA binding protein and hnRNPA/B family member that binds to poly (A) signal sequences |
|
| YKL188C | 0.36 |
PXA2
|
Subunit of a heterodimeric peroxisomal ATP-binding cassette transporter complex (Pxa1p-Pxa2p), required for import of long-chain fatty acids into peroxisomes; similarity to human adrenoleukodystrophy transporter and ALD-related proteins |
|
| YER084W | 0.36 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPL156C | 0.35 |
PRM4
|
Pheromone-regulated protein, predicted to have 1 transmembrane segment; transcriptionally regulated by Ste12p during mating and by Cat8p during the diauxic shift |
|
| YML016C | 0.35 |
PPZ1
|
Serine/threonine protein phosphatase Z, isoform of Ppz2p; involved in regulation of potassium transport, which affects osmotic stability, cell cycle progression, and halotolerance |
|
| YJL007C | 0.35 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHR126C | 0.34 |
Putative protein of unknown function; transcription dependent upon Azf1p |
||
| YGL089C | 0.34 |
MF(ALPHA)2
|
Mating pheromone alpha-factor, made by alpha cells; interacts with mating type a cells to induce cell cycle arrest and other responses leading to mating; also encoded by MF(ALPHA)1, which is more highly expressed than MF(ALPHA)2 |
|
| YKR033C | 0.34 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene DAL80 |
||
| YJL135W | 0.33 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified genes YJL134W/LCB3 |
||
| YCR007C | 0.33 |
Putative integral membrane protein, member of DUP240 gene family; YCR007C is not an essential gene |
||
| YKL050C | 0.33 |
Protein of unknown function; the YKL050W protein is a target of the SCFCdc4 ubiquitin ligase complex and YKL050W transcription is regulated by Azf1p |
||
| YGR087C | 0.33 |
PDC6
|
Minor isoform of pyruvate decarboxylase, decarboxylates pyruvate to acetaldehyde, involved in amino acid catabolism; transcription is glucose- and ethanol-dependent, and is strongly induced during sulfur limitation |
|
| YOR178C | 0.32 |
GAC1
|
Regulatory subunit for Glc7p type-1 protein phosphatase (PP1), tethers Glc7p to Gsy2p glycogen synthase, binds Hsf1p heat shock transcription factor, required for induction of some HSF-regulated genes under heat shock |
|
| YKR032W | 0.32 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR142W | 0.32 |
PUT1
|
Proline oxidase, nuclear-encoded mitochondrial protein involved in utilization of proline as sole nitrogen source; PUT1 transcription is induced by Put3p in the presence of proline and the absence of a preferred nitrogen source |
|
| YKL001C | 0.31 |
MET14
|
Adenylylsulfate kinase, required for sulfate assimilation and involved in methionine metabolism |
|
| YLR049C | 0.31 |
Putative protein of unknown function |
||
| YPR078C | 0.31 |
Putative protein of unknown function; possible role in DNA metabolism and/or in genome stability; expression is heat-inducible |
||
| YML047C | 0.31 |
PRM6
|
Pheromone-regulated protein, predicted to have 2 transmembrane segments; regulated by Ste12p during mating |
|
| YBL052C | 0.31 |
SAS3
|
Histone acetyltransferase catalytic subunit of NuA3 complex that acetylates histone H3, involved in transcriptional silencing; homolog of the mammalian MOZ proto-oncogene; sas3 gcn5 double mutation confers lethality |
|
| YLR358C | 0.30 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF RSC2/YLR357W |
||
| YJR005C-A | 0.30 |
Putative protein of unknown function, originally identified as a syntenic homolog of an Ashbya gossypii gene |
||
| YLR296W | 0.30 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR071W | 0.30 |
PCL5
|
Cyclin, interacts with Pho85p cyclin-dependent kinase (Cdk), induced by Gcn4p at level of transcription, specifically required for Gcn4p degradation, may be sensor of cellular protein biosynthetic capacity |
|
| YBR208C | 0.29 |
DUR1,2
|
Urea amidolyase, contains both urea carboxylase and allophanate hydrolase activities, degrades urea to CO2 and NH3; expression sensitive to nitrogen catabolite repression and induced by allophanate, an intermediate in allantoin degradation |
|
| YKL163W | 0.29 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YPL062W | 0.28 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YPL062W is not an essential gene; homozygous diploid mutant shows a decrease in glycogen accumulation |
||
| YBR094W | 0.28 |
PBY1
|
Putative tubulin tyrosine ligase associated with P-bodies |
|
| YBR056W-A | 0.28 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR056C-B |
||
| YLR136C | 0.28 |
TIS11
|
mRNA-binding protein expressed during iron starvation; binds to a sequence element in the 3'-untranslated regions of specific mRNAs to mediate their degradation; involved in iron homeostasis |
|
| YMR104C | 0.28 |
YPK2
|
Protein kinase with similarityto serine/threonine protein kinase Ypk1p; functionally redundant with YPK1 at the genetic level; participates in a signaling pathway required for optimal cell wall integrity; homolog of mammalian kinase SGK |
|
| YGR067C | 0.27 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YLR164W | 0.27 |
Mitochondrial inner membrane of unknown function; similar to Tim18p and Sdh4p; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
||
| YBR292C | 0.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR292C is not an essential gene |
||
| YJL106W | 0.27 |
IME2
|
Serine/threonine protein kinase involved in activation of meiosis, associates with Ime1p and mediates its stability, activates Ndt80p; IME2 expression is positively regulated by Ime1p |
|
| YIL099W | 0.27 |
SGA1
|
Intracellular sporulation-specific glucoamylase involved in glycogen degradation; induced during starvation of a/a diploids late in sporulation, but dispensable for sporulation |
|
| YCR001W | 0.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YCR001W is not an essential gene |
||
| YER167W | 0.27 |
BCK2
|
Protein rich in serine and threonine residues involved in protein kinase C signaling pathway, which controls cell integrity; overproduction suppresses pkc1 mutations |
|
| YDR258C | 0.26 |
HSP78
|
Oligomeric mitochondrial matrix chaperone that cooperates with Ssc1p in mitochondrial thermotolerance after heat shock; prevents the aggregation of misfolded matrix proteins; component of the mitochondrial proteolysis system |
|
| YGR146C | 0.26 |
Putative protein of unknown function; induced by iron homeostasis transcription factor Aft2p; multicopy suppressor of a temperature sensitive hsf1 mutant; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YIL119C | 0.26 |
RPI1
|
Putative transcriptional regulator; overexpression suppresses the heat shock sensitivity of wild-type RAS2 overexpression and also suppresses the cell lysis defect of an mpk1 mutation |
|
| YDL020C | 0.26 |
RPN4
|
Transcription factor that stimulates expression of proteasome genes; Rpn4p levels are in turn regulated by the 26S proteasome in a negative feedback control mechanism; RPN4 is transcriptionally regulated by various stress responses |
|
| YER158C | 0.26 |
Protein of unknown function, has similarity to Afr1p; potentially phosphorylated by Cdc28p |
||
| YNL007C | 0.26 |
SIS1
|
Type II HSP40 co-chaperone that interacts with the HSP70 protein Ssa1p; not functionally redundant with Ydj1p due to due to substrate specificity; shares similarity with bacterial DnaJ proteins |
|
| YBR157C | 0.26 |
ICS2
|
Protein of unknown function; null mutation does not confer any obvious defects in growth, spore germination, viability, or carbohydrate utilization |
|
| YNL093W | 0.26 |
YPT53
|
GTPase, similar to Ypt51p and Ypt52p and to mammalian rab5; required for vacuolar protein sorting and endocytosis |
|
| YPL026C | 0.26 |
SKS1
|
Putative serine/threonine protein kinase; involved in the adaptation to low concentrations of glucose independent of the SNF3 regulated pathway |
|
| YEL014C | 0.26 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL179C | 0.26 |
TOS3
|
Protein kinase, related to and functionally redundant with Elm1p and Sak1p for the phosphorylation and activation of Snf1p; functionally orthologous to LKB1, a mammalian kinase associated with Peutz-Jeghers cancer-susceptibility syndrome |
|
| YGR060W | 0.26 |
ERG25
|
C-4 methyl sterol oxidase, catalyzes the first of three steps required to remove two C-4 methyl groups from an intermediate in ergosterol biosynthesis; mutants accumulate the sterol intermediate 4,4-dimethylzymosterol |
|
| YDR528W | 0.25 |
HLR1
|
Protein involved in regulation of cell wall composition and integrity and response to osmotic stress; overproduction suppresses a lysis sensitive PKC mutation; similar to Lre1p, which functions antagonistically to protein kinase A |
|
| YLR260W | 0.25 |
LCB5
|
Minor sphingoid long-chain base kinase, paralog of Lcb4p responsible for few percent of the total activity, possibly involved in synthesis of long-chain base phosphates, which function as signaling molecules |
|
| YIL101C | 0.25 |
XBP1
|
Transcriptional repressor that binds to promoter sequences of the cyclin genes, CYS3, and SMF2; expression is induced by stress or starvation during mitosis, and late in meiosis; member of the Swi4p/Mbp1p family; potential Cdc28p substrate |
|
| YJR047C | 0.25 |
ANB1
|
Translation initiation factor eIF-5A, promotes formation of the first peptide bond; similar to and functionally redundant with Hyp2p; undergoes an essential hypusination modification; expressed under anaerobic conditions |
|
| YDL246C | 0.25 |
SOR2
|
Protein of unknown function, computational analysis of large-scale protein-protein interaction data suggests a possible role in fructose or mannose metabolism |
|
| YBR071W | 0.25 |
Putative protein of unknown function; (GFP)-fusion and epitope-tagged proteins localize to the cytoplasm; mRNA expression may be regulated by the cell cycle and/or cell wall stress |
||
| YHL024W | 0.25 |
RIM4
|
Putative RNA-binding protein required for the expression of early and middle sporulation genes |
|
| YPL110C | 0.25 |
GDE1
|
Glycerophosphocholine (GroPCho) phosphodiesterase; hydrolyzes GroPCho to choline and glycerolphosphate, for use as a phosphate source and as a precursor for phosphocholine synthesis; may interact with ribosomes |
|
| YGL227W | 0.25 |
VID30
|
Protein involved in proteasome-dependent catabolite degradation of fructose-1,6-bisphosphatase (FBPase); shifts the balance of nitrogen metabolism toward the production of glutamate; localized to the nucleus and the cytoplasm |
|
| YJL116C | 0.24 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YDL003W | 0.24 |
MCD1
|
Essential protein required for sister chromatid cohesion in mitosis and meiosis; subunit of the cohesin complex; expression is cell cycle regulated and peaks in S phase |
|
| YLR390W-A | 0.23 |
CCW14
|
Covalently linked cell wall glycoprotein, present in the inner layer of the cell wall |
|
| YLR431C | 0.23 |
ATG23
|
Peripheral membrane protein required for the cytoplasm-to-vacuole targeting (Cvt) pathway; cycles between the pre-autophagosome (PAS) and non-PAS locations; forms a complex with Atg9p and Atg27p |
|
| YHR095W | 0.23 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPL258C | 0.23 |
THI21
|
Hydroxymethylpyrimidine phosphate kinase, involved in the last steps in thiamine biosynthesis; member of a gene family with THI20 and THI22; functionally redundant with Thi20p |
|
| YMR174C | 0.23 |
PAI3
|
Cytoplasmic proteinase A (Pep4p) inhibitor, dependent on Pbs2p and Hog1p protein kinases for osmotic induction; intrinsically unstructured, N-terminal half becomes ordered in the active site of proteinase A upon contact |
|
| YGL033W | 0.23 |
HOP2
|
Meiosis-specific protein that localizes to chromosomes, preventing synapsis between nonhomologous chromosomes and ensuring synapsis between homologs; complexes with Mnd1p to promote homolog pairing and meiotic double-strand break repair |
|
| YCR040W | 0.23 |
MATALPHA1
|
Transcriptional co-activator involved in regulation of mating-type-specific gene expression; targets the transcription factor Mcm1p to the promoters of alpha-specific genes; one of two genes encoded by the MATalpha mating type cassette |
|
| YAL024C | 0.23 |
LTE1
|
Putative GDP/GTP exchange factor required for mitotic exit at low temperatures; acts as a guanine nucleotide exchange factor (GEF) for Tem1p, which is a key regulator of mitotic exit; physically associates with Ras2p-GTP |
|
| YHR006W | 0.23 |
STP2
|
Transcription factor, activated by proteolytic processing in response to signals from the SPS sensor system for external amino acids; activates transcription of amino acid permease genes |
|
| YCR099C | 0.23 |
Putative protein of unknown function |
||
| YKL086W | 0.22 |
SRX1
|
Sulfiredoxin, contributes to oxidative stress resistance by reducing cysteine-sulfinic acid groups in the peroxiredoxins Tsa1p and Ahp1p that are formed upon exposure to oxidants; conserved in higher eukaryotes |
|
| YKR102W | 0.22 |
FLO10
|
Lectin-like protein with similarity to Flo1p, thought to be involved in flocculation |
|
| YDL028C | 0.22 |
MPS1
|
Dual-specificity kinase required for spindle pole body (SPB) duplication and spindle checkpoint function; substrates include SPB proteins Spc42p, Spc110p, and Spc98p, mitotic exit network protein Mob1p, and checkpoint protein Mad1p |
|
| YMR015C | 0.22 |
ERG5
|
C-22 sterol desaturase, a cytochrome P450 enzyme that catalyzes the formation of the C-22(23) double bond in the sterol side chain in ergosterol biosynthesis; may be a target of azole antifungal drugs |
|
| YNL142W | 0.22 |
MEP2
|
Ammonium permease involved in regulation of pseudohyphal growth; belongs to a ubiquitous family of cytoplasmic membrane proteins that transport only ammonium (NH4+); expression is under the nitrogen catabolite repression regulation |
|
| YIL056W | 0.22 |
VHR1
|
Transcriptional activator, required for the vitamin H-responsive element (VHRE) mediated induction of VHT1 (Vitamin H transporter) and BIO5 (biotin biosynthesis intermediate transporter) in response to low biotin concentrations |
|
| YER109C | 0.22 |
FLO8
|
Transcription factor required for flocculation, diploid filamentous growth, and haploid invasive growth; genome reference strain S288C and most laboratory strains have a mutation in this gene |
|
| YPL054W | 0.22 |
LEE1
|
Zinc-finger protein of unknown function |
|
| YPR193C | 0.22 |
HPA2
|
Tetrameric histone acetyltransferase with similarity to Gcn5p, Hat1p, Elp3p, and Hpa3p; acetylates histones H3 and H4 in vitro and exhibits autoacetylation activity |
|
| YCL048W | 0.22 |
SPS22
|
Protein of unknown function, redundant with Sps2p for the organization of the beta-glucan layer of the spore wall |
|
| YOR355W | 0.22 |
GDS1
|
Protein of unknown function, required for growth on glycerol as a carbon source; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YCL069W | 0.21 |
VBA3
|
Permease of basic amino acids in the vacuolar membrane |
|
| YDR510W | 0.21 |
SMT3
|
Ubiquitin-like protein of the SUMO family, conjugated to lysine residues of target proteins; regulates chromatid cohesion, chromosome segregation, APC-mediated proteolysis, DNA replication and septin ring dynamics |
|
| YCR098C | 0.21 |
GIT1
|
Plasma membrane permease, mediates uptake of glycerophosphoinositol and glycerophosphocholine as sources of the nutrients inositol and phosphate; expression and transport rate are regulated by phosphate and inositol availability |
|
| YDR409W | 0.21 |
SIZ1
|
SUMO/Smt3 ligase that promotes the attachment of sumo (Smt3p; small ubiquitin-related modifier) to proteins; binds Ubc9p and may bind septins; specifically required for sumoylation of septins in vivo; localized to the septin ring |
|
| YLR307W | 0.21 |
CDA1
|
Chitin deacetylase, together with Cda2p involved in the biosynthesis ascospore wall component, chitosan; required for proper rigidity of the ascospore wall |
|
| YOR231W | 0.21 |
MKK1
|
Mitogen-activated kinase kinase involved in protein kinase C signaling pathway that controls cell integrity; upon activation by Bck1p phosphorylates downstream target, Slt2p; functionally redundant with Mkk2p |
|
| YJR159W | 0.21 |
SOR1
|
Sorbitol dehydrogenase; expression is induced in the presence of sorbitol |
|
| YKL123W | 0.21 |
Dubious open reading frame, unlikely to encode a protein; partially overlaps the verified gene SSH4 |
||
| YOR306C | 0.21 |
MCH5
|
Plasma membrane riboflavin transporter; facilitates the uptake of vitamin B2; required for FAD-dependent processes; sequence similarity to mammalian monocarboxylate permeases, however mutants are not deficient in monocarboxylate transport |
|
| YMR101C | 0.20 |
SRT1
|
Cis-prenyltransferase involved in synthesis of long-chain dolichols (19-22 isoprene units; as opposed to Rer2p which synthesizes shorter-chain dolichols); localizes to lipid bodies; transcription is induced during stationary phase |
|
| YGR226C | 0.20 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; overlaps significantly with a verified ORF, AMA1/YGR225W |
||
| YIL072W | 0.20 |
HOP1
|
Meiosis-specific DNA binding protein that displays Red1p dependent localization to the unsynapsed axial-lateral elements of the synaptonemal complex; required for homologous chromosome synapsis and chiasma formation |
|
| YMR306C-A | 0.20 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDL211C | 0.20 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YPL230W | 0.20 |
USV1
|
Putative transcription factor containing a C2H2 zinc finger; mutation affects transcriptional regulation of genes involved in protein folding, ATP binding, and cell wall biosynthesis |
|
| YIL094C | 0.20 |
LYS12
|
Homo-isocitrate dehydrogenase, an NAD-linked mitochondrial enzyme required for the fourth step in the biosynthesis of lysine, in which homo-isocitrate is oxidatively decarboxylated to alpha-ketoadipate |
|
| YOR113W | 0.20 |
AZF1
|
Zinc-finger transcription factor, involved in induction of CLN3 transcription in response to glucose; genetic and physical interactions indicate a possible role in mitochondrial transcription or genome maintenance |
|
| YDR192C | 0.20 |
NUP42
|
Subunit of the nuclear pore complex (NPC) that localizes exclusively to the cytoplasmic side; involved in RNA export, most likely at a terminal step; interacts with Gle1p |
|
| YKL180W | 0.20 |
RPL17A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl17Bp and has similarity to E. coli L22 and rat L17 ribosomal proteins; copurifies with the Dam1 complex (aka DASH complex) |
|
| YBR124W | 0.20 |
Putative protein of unknown function |
||
| YCL066W | 0.20 |
HMLALPHA1
|
Silenced copy of ALPHA1 at HML, encoding a transcriptional coactivator involved in the regulation of mating-type alpha-specific gene expression |
|
| YBL043W | 0.20 |
ECM13
|
Non-essential protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YDL139C | 0.20 |
SCM3
|
Nonhistone component of centromeric chromatin that binds stoichiometrically to CenH3-H4 histones, required for kinetochore assembly; contains nuclear export signal (NES); required for G2/M progression and localization of Cse4p |
|
| YFR056C | 0.19 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YNL171C | 0.19 |
Dubious open reading frame unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YFL020C | 0.19 |
PAU5
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YLR037C | 0.19 |
DAN2
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YBR040W | 0.19 |
FIG1
|
Integral membrane protein required for efficient mating; may participate in or regulate the low affinity Ca2+ influx system, which affects intracellular signaling and cell-cell fusion during mating |
|
| YNR062C | 0.19 |
Putative membrane protein of unknown function |
||
| YPL027W | 0.18 |
SMA1
|
Protein of unknown function involved in the assembly of the prospore membrane during sporulation |
|
| YFL058W | 0.18 |
THI5
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YKL017C | 0.18 |
HCS1
|
Hexameric DNA polymerase alpha-associated DNA helicase A involved in lagging strand DNA synthesis; contains single-stranded DNA stimulated ATPase and dATPase activities; replication protein A stimulates helicase and ATPase activities |
|
| YLR111W | 0.18 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YJL036W | 0.18 |
SNX4
|
Sorting nexin, involved in the retrieval of late-Golgi SNAREs from the post-Golgi endosome to the trans-Golgi network and in cytoplasm to vacuole transport; contains a PX domain; forms complexes with Snx41p and with Atg20p |
|
| YBL029C-A | 0.18 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery; has potential orthologs in Saccharomyces species and in Yarrowia lipolytica |
||
| YIL069C | 0.18 |
RPS24B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps24Ap and has similarity to rat S24 ribosomal protein |
|
| YMR118C | 0.18 |
Protein of unknown function with similarity to succinate dehydrogenase cytochrome b subunit; YMR118C is not an essential gene |
||
| YDL210W | 0.18 |
UGA4
|
Permease that serves as a gamma-aminobutyrate (GABA) transport protein involved in the utilization of GABA as a nitrogen source; catalyzes the transport of putrescine and delta-aminolevulinic acid (ALA); localized to the vacuolar membrane |
|
| YPR030W | 0.18 |
CSR2
|
Nuclear protein with a potential regulatory role in utilization of galactose and nonfermentable carbon sources; overproduction suppresses the lethality at high temperature of a chs5 spa2 double null mutation; potential Cdc28p substrate |
|
| YBR123C | 0.18 |
TFC1
|
One of six subunits of the RNA polymerase III transcription initiation factor complex (TFIIIC); part of the TauA globular domain of TFIIIC that binds DNA at the BoxA promoter sites of tRNA and similar genes; human homolog is TFIIIC-63 |
|
| YMR295C | 0.18 |
Protein of unknown function that associates with ribosomes; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery and bud; YMR295C is not an essential gene |
||
| YPR151C | 0.18 |
SUE1
|
Mitochondrial protein required for degradation of unstable forms of cytochrome c |
|
| YKR073C | 0.18 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR171W | 0.18 |
HSP42
|
Small heat shock protein (sHSP) with chaperone activity; forms barrel-shaped oligomers that suppress unfolded protein aggregation; involved in cytoskeleton reorganization after heat shock |
|
| YDR389W | 0.18 |
SAC7
|
GTPase activating protein (GAP) for Rho1p, involved in signaling to the actin cytoskeleton, null mutations suppress tor2 mutations and temperature sensitive mutations in actin; potential Cdc28p substrate |
|
| YHR030C | 0.18 |
SLT2
|
Serine/threonine MAP kinase involved in regulating the maintenance of cell wall integrity and progression through the cell cycle; regulated by the PKC1-mediated signaling pathway |
|
| YDL234C | 0.17 |
GYP7
|
GTPase-activating protein for yeast Rab family members including: Ypt7p (most effective), Ypt1p, Ypt31p, and Ypt32p (in vitro); involved in vesicle mediated protein trafficking |
|
| YOL035C | 0.17 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNL143C | 0.17 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR394W | 0.17 |
RPT3
|
One of six ATPases of the 19S regulatory particle of the 26S proteasome involved in the degradation of ubiquitinated substrates; substrate of N-acetyltransferase B |
|
| YDL037C | 0.17 |
BSC1
|
Protein of unconfirmed function, similar to cell surface flocculin Muc1p; ORF exhibits genomic organization compatible with a translational readthrough-dependent mode of expression |
|
| YDR477W | 0.17 |
SNF1
|
AMP-activated serine/threonine protein kinase found in a complex containing Snf4p and members of the Sip1p/Sip2p/Gal83p family; required for transcription of glucose-repressed genes, thermotolerance, sporulation, and peroxisome biogenesis |
|
| YJL188C | 0.17 |
BUD19
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 88% of ORF overlaps the verified gene RPL39; diploid mutant displays a weak budding pattern phenotype in a systematic assay |
|
| YNR058W | 0.17 |
BIO3
|
7,8-diamino-pelargonic acid aminotransferase (DAPA), catalyzes the second step in the biotin biosynthesis pathway; BIO3 is in a cluster of 3 genes (BIO3, BIO4, and BIO5) that mediate biotin synthesis |
|
| YLR176C | 0.17 |
RFX1
|
Major transcriptional repressor of DNA-damage-regulated genes, recruits repressors Tup1p and Cyc8p to their promoters; involved in DNA damage and replication checkpoint pathway; similar to a family of mammalian DNA binding RFX1-4 proteins |
|
| YKL190W | 0.17 |
CNB1
|
Calcineurin B; the regulatory subunit of calcineurin, a Ca++/calmodulin-regulated type 2B protein phosphatase which regulates Crz1p (a stress-response transcription factor), the other calcineurin subunit is encoded by CNA1 and/or CMP1 |
|
| YDR001C | 0.17 |
NTH1
|
Neutral trehalase, degrades trehalose; required for thermotolerance and may mediate resistance to other cellular stresses; may be phosphorylated by Cdc28p |
|
| YER098W | 0.16 |
UBP9
|
Ubiquitin carboxyl-terminal hydrolase, ubiquitin-specific protease that cleaves ubiquitin-protein fusions |
|
| YAR053W | 0.16 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR192C-C | 0.16 |
Putative protein of unknown function; identified by expression profiling and mass spectrometry |
||
| YLR108C | 0.16 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus; YLR108C is not an esssential gene |
||
| YIL045W | 0.16 |
PIG2
|
Putative type-1 protein phosphatase targeting subunit that tethers Glc7p type-1 protein phosphatase to Gsy2p glycogen synthase |
|
| YDR515W | 0.16 |
SLF1
|
RNA binding protein that associates with polysomes; proposed to be involved in regulating mRNA translation; involved in the copper-dependent mineralization of copper sulfide complexes on cell surface in cells cultured in copper salts |
|
| YIL100W | 0.16 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps the dubious ORF YIL100C-A |
||
| YGL156W | 0.16 |
AMS1
|
Vacuolar alpha mannosidase, involved in free oligosaccharide (fOS) degradation; delivered to the vacuole in a novel pathway separate from the secretory pathway |
|
| YOL118C | 0.16 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YDR193W | 0.16 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR090W | 0.16 |
Putative protein of unknown function with similarity to DTDP-glucose 4,6-dehydratases; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YMR090W is not an essential gene |
||
| YAL044C | 0.16 |
GCV3
|
H subunit of the mitochondrial glycine decarboxylase complex, required for the catabolism of glycine to 5,10-methylene-THF; expression is regulated by levels of levels of 5,10-methylene-THF in the cytoplasm |
|
| YER014C-A | 0.16 |
BUD25
|
Protein involved in bud-site selection; diploid mutants display a random budding pattern instead of the wild-type bipolar pattern |
|
| YBR203W | 0.16 |
COS111
|
Protein required for resistance to the antifungal drug ciclopirox olamine; not related to the subtelomerically-encoded COS family; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YPL222C-A | 0.16 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
Network of associatons between targets according to the STRING database.
First level regulatory network of NHP6A
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0090295 | nitrogen catabolite repression of transcription(GO:0090295) |
| 0.3 | 0.9 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.2 | 0.2 | GO:0071472 | cellular response to salt stress(GO:0071472) |
| 0.2 | 0.5 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.2 | 0.5 | GO:0009732 | detection of chemical stimulus(GO:0009593) detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) detection of stimulus(GO:0051606) |
| 0.2 | 0.2 | GO:0042710 | biofilm formation(GO:0042710) |
| 0.2 | 0.7 | GO:0009415 | response to water(GO:0009415) cellular response to water stimulus(GO:0071462) |
| 0.2 | 1.0 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.2 | 0.5 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 0.6 | GO:0001323 | age-dependent general metabolic decline involved in chronological cell aging(GO:0001323) age-dependent response to oxidative stress involved in chronological cell aging(GO:0001324) |
| 0.1 | 0.1 | GO:0072353 | cellular age-dependent response to reactive oxygen species(GO:0072353) |
| 0.1 | 0.5 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.1 | 0.4 | GO:0045895 | positive regulation of mating-type specific transcription, DNA-templated(GO:0045895) |
| 0.1 | 0.4 | GO:0030656 | regulation of vitamin metabolic process(GO:0030656) positive regulation of vitamin metabolic process(GO:0046136) regulation of thiamine biosynthetic process(GO:0070623) positive regulation of thiamine biosynthetic process(GO:0090180) |
| 0.1 | 0.3 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.4 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.8 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.1 | 0.6 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.1 | 0.4 | GO:0019419 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.1 | 0.5 | GO:1903313 | positive regulation of mRNA metabolic process(GO:1903313) |
| 0.1 | 0.5 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) thiamine diphosphate metabolic process(GO:0042357) |
| 0.1 | 0.3 | GO:0052547 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 0.1 | 0.3 | GO:0043419 | urea catabolic process(GO:0043419) |
| 0.1 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.1 | 0.4 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.1 | 0.4 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 1.2 | GO:0000753 | cell morphogenesis involved in conjugation with cellular fusion(GO:0000753) |
| 0.1 | 0.4 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.1 | GO:0019627 | urea metabolic process(GO:0019627) |
| 0.1 | 0.4 | GO:1901072 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.1 | 0.1 | GO:0000736 | double-strand break repair via single-strand annealing, removal of nonhomologous ends(GO:0000736) |
| 0.1 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.3 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.1 | 0.4 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.1 | 0.2 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.1 | 0.2 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 0.1 | 0.4 | GO:0010388 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.1 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.6 | GO:0015688 | iron chelate transport(GO:0015688) |
| 0.1 | 0.2 | GO:0000092 | mitotic anaphase B(GO:0000092) |
| 0.1 | 0.2 | GO:2000058 | regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.1 | 0.3 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.3 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.1 | 0.3 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glycogen metabolic process(GO:0070873) |
| 0.1 | 0.1 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.1 | 0.4 | GO:0009743 | response to carbohydrate(GO:0009743) |
| 0.1 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.2 | GO:0070054 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) IRE1-mediated unfolded protein response(GO:0036498) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) |
| 0.1 | 0.3 | GO:0045597 | positive regulation of cell fate commitment(GO:0010455) positive regulation of mating type switching(GO:0031496) positive regulation of cell differentiation(GO:0045597) |
| 0.1 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.1 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.1 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) positive regulation of cellular protein catabolic process(GO:1903364) |
| 0.0 | 0.1 | GO:0061395 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) positive regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061395) cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0035950 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 0.0 | 0.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) peptidyl-serine modification(GO:0018209) |
| 0.0 | 0.4 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0001041 | transcription from RNA polymerase III type 2 promoter(GO:0001009) transcription from a RNA polymerase III hybrid type promoter(GO:0001041) |
| 0.0 | 0.2 | GO:0045033 | peroxisome inheritance(GO:0045033) |
| 0.0 | 0.2 | GO:0055075 | cellular potassium ion homeostasis(GO:0030007) potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.3 | GO:0051346 | negative regulation of hydrolase activity(GO:0051346) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) mitotic G1/S transition checkpoint(GO:0044819) |
| 0.0 | 0.1 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.1 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.0 | 0.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0042173 | regulation of sporulation resulting in formation of a cellular spore(GO:0042173) |
| 0.0 | 0.1 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 0.0 | 0.2 | GO:0007234 | osmosensory signaling via phosphorelay pathway(GO:0007234) |
| 0.0 | 0.2 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) |
| 0.0 | 0.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0030491 | heteroduplex formation(GO:0030491) |
| 0.0 | 0.1 | GO:0006827 | high-affinity iron ion transmembrane transport(GO:0006827) |
| 0.0 | 0.2 | GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000461) |
| 0.0 | 0.1 | GO:0043388 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.0 | 0.1 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:0000730 | meiotic DNA recombinase assembly(GO:0000707) DNA recombinase assembly(GO:0000730) |
| 0.0 | 0.2 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.1 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0019626 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 0.0 | 0.1 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0015883 | FAD transport(GO:0015883) |
| 0.0 | 0.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.1 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:1904667 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.1 | GO:1901654 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) response to ketone(GO:1901654) |
| 0.0 | 0.1 | GO:1900544 | positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.0 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.0 | 0.6 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.1 | GO:0006037 | cell wall chitin metabolic process(GO:0006037) |
| 0.0 | 0.1 | GO:0032443 | regulation of steroid metabolic process(GO:0019218) regulation of ergosterol biosynthetic process(GO:0032443) regulation of steroid biosynthetic process(GO:0050810) |
| 0.0 | 0.3 | GO:0031578 | mitotic spindle orientation checkpoint(GO:0031578) |
| 0.0 | 0.4 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0061392 | regulation of transcription from RNA polymerase II promoter in response to osmotic stress(GO:0061392) |
| 0.0 | 0.0 | GO:0061013 | regulation of mRNA catabolic process(GO:0061013) |
| 0.0 | 0.1 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:0007534 | gene conversion at mating-type locus(GO:0007534) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.9 | GO:0051304 | chromosome separation(GO:0051304) |
| 0.0 | 0.1 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0048313 | Golgi inheritance(GO:0048313) |
| 0.0 | 0.0 | GO:0071050 | snoRNA polyadenylation(GO:0071050) |
| 0.0 | 0.1 | GO:0000729 | meiotic DNA double-strand break processing(GO:0000706) DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.2 | GO:2000144 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.0 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.0 | 0.7 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0044091 | ascospore-type prospore membrane assembly(GO:0032120) membrane biogenesis(GO:0044091) membrane assembly(GO:0071709) |
| 0.0 | 0.2 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.0 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:1900436 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0019932 | second-messenger-mediated signaling(GO:0019932) |
| 0.0 | 0.0 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.0 | GO:0000279 | mitotic M phase(GO:0000087) M phase(GO:0000279) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0051046 | regulation of exocytosis(GO:0017157) regulation of secretion(GO:0051046) regulation of secretion by cell(GO:1903530) |
| 0.0 | 0.2 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.1 | GO:0000078 | obsolete cytokinesis after mitosis checkpoint(GO:0000078) positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.0 | GO:0043937 | regulation of sporulation(GO:0043937) |
| 0.0 | 0.7 | GO:0070590 | ascospore wall assembly(GO:0030476) spore wall assembly(GO:0042244) spore wall biogenesis(GO:0070590) ascospore wall biogenesis(GO:0070591) fungal-type cell wall assembly(GO:0071940) |
| 0.0 | 0.1 | GO:0032231 | regulation of actin filament bundle assembly(GO:0032231) positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.3 | GO:0030474 | spindle pole body duplication(GO:0030474) |
| 0.0 | 0.0 | GO:0043270 | positive regulation of ion transport(GO:0043270) |
| 0.0 | 0.1 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.0 | 0.1 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.0 | GO:0071361 | positive regulation of transcription from RNA polymerase II promoter in response to ethanol(GO:0061410) cellular response to ethanol(GO:0071361) cellular response to alcohol(GO:0097306) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.0 | 0.1 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.1 | GO:0035392 | maintenance of chromatin silencing(GO:0006344) maintenance of chromatin silencing at telomere(GO:0035392) |
| 0.0 | 0.2 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.0 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:2000284 | positive regulation of cellular amino acid metabolic process(GO:0045764) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 0.1 | GO:0015758 | glucose transport(GO:0015758) |
| 0.0 | 0.0 | GO:0001315 | age-dependent response to reactive oxygen species(GO:0001315) |
| 0.0 | 0.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:1900062 | regulation of cell aging(GO:0090342) regulation of replicative cell aging(GO:1900062) |
| 0.0 | 0.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.7 | GO:0018393 | internal protein amino acid acetylation(GO:0006475) histone acetylation(GO:0016573) internal peptidyl-lysine acetylation(GO:0018393) peptidyl-lysine acetylation(GO:0018394) |
| 0.0 | 0.1 | GO:1903726 | negative regulation of phospholipid biosynthetic process(GO:0071072) negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.0 | 0.0 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.1 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:0043200 | response to amino acid(GO:0043200) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0055070 | cellular copper ion homeostasis(GO:0006878) copper ion homeostasis(GO:0055070) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) |
| 0.0 | 0.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.1 | GO:0009371 | positive regulation of transcription from RNA polymerase II promoter by pheromones(GO:0007329) positive regulation of transcription by pheromones(GO:0009371) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.0 | GO:0016077 | snRNA catabolic process(GO:0016076) snoRNA catabolic process(GO:0016077) nuclear polyadenylation-dependent snoRNA catabolic process(GO:0071036) nuclear polyadenylation-dependent snRNA catabolic process(GO:0071037) |
| 0.0 | 0.0 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.2 | GO:0007118 | budding cell apical bud growth(GO:0007118) |
| 0.0 | 0.1 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.0 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.1 | GO:0043954 | cellular component maintenance(GO:0043954) |
| 0.0 | 0.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.0 | GO:0097271 | protein localization to bud neck(GO:0097271) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) negative regulation of DNA-dependent DNA replication(GO:2000104) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.0 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.0 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0000743 | nuclear migration involved in conjugation with cellular fusion(GO:0000743) |
| 0.0 | 0.1 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.1 | GO:0000084 | mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.0 | 0.3 | GO:0009272 | fungal-type cell wall biogenesis(GO:0009272) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0045991 | carbon catabolite activation of transcription(GO:0045991) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.0 | GO:0009847 | spore germination(GO:0009847) |
| 0.0 | 0.0 | GO:0034310 | ethanol catabolic process(GO:0006068) primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.0 | GO:0000272 | polysaccharide catabolic process(GO:0000272) |
| 0.0 | 0.3 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.0 | GO:0015691 | cadmium ion transport(GO:0015691) |
| 0.0 | 0.1 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.1 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.0 | 0.2 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.4 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.3 | GO:0034990 | mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) |
| 0.1 | 0.7 | GO:0005619 | ascospore wall(GO:0005619) |
| 0.1 | 0.3 | GO:0001400 | mating projection base(GO:0001400) |
| 0.1 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 0.1 | 0.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.3 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.1 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.2 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.1 | 0.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0072379 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 0.0 | 0.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0005823 | central plaque of spindle pole body(GO:0005823) |
| 0.0 | 0.1 | GO:0034099 | luminal surveillance complex(GO:0034099) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.0 | 0.1 | GO:0008278 | nuclear cohesin complex(GO:0000798) cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0031518 | CBF3 complex(GO:0031518) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.4 | GO:0005940 | septin ring(GO:0005940) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.4 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.0 | 0.1 | GO:0033597 | mitotic checkpoint complex(GO:0033597) |
| 0.0 | 0.1 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.0 | 0.4 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0032126 | eisosome(GO:0032126) |
| 0.0 | 0.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.0 | GO:0000111 | nucleotide-excision repair factor 2 complex(GO:0000111) |
| 0.0 | 0.0 | GO:0045240 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial oxoglutarate dehydrogenase complex(GO:0009353) dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0090544 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.0 | 0.1 | GO:0032221 | Rpd3S complex(GO:0032221) |
| 0.0 | 0.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.0 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.4 | GO:0005628 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.0 | 0.1 | GO:0000446 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) nucleoplasmic THO complex(GO:0000446) |
| 0.0 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.0 | GO:0070211 | Snt2C complex(GO:0070211) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0000133 | polarisome(GO:0000133) |
| 0.0 | 0.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.0 | 0.1 | GO:0032806 | carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 0.0 | GO:0070274 | RES complex(GO:0070274) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.0 | GO:0044697 | HICS complex(GO:0044697) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0000772 | mating pheromone activity(GO:0000772) pheromone activity(GO:0005186) |
| 0.2 | 0.7 | GO:0015603 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.2 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 0.5 | GO:0005536 | glucose binding(GO:0005536) |
| 0.2 | 0.6 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.1 | 0.5 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 0.1 | 0.5 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.4 | GO:0008972 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.1 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.3 | GO:0030414 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 0.1 | 0.4 | GO:0016642 | glycine dehydrogenase (decarboxylating) activity(GO:0004375) oxidoreductase activity, acting on the CH-NH2 group of donors, disulfide as acceptor(GO:0016642) |
| 0.1 | 0.4 | GO:0004338 | glucan exo-1,3-beta-glucosidase activity(GO:0004338) |
| 0.1 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.5 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.1 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.1 | 0.2 | GO:0015489 | putrescine transmembrane transporter activity(GO:0015489) |
| 0.1 | 0.2 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.1 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.3 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.1 | 0.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.1 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) phosphatase binding(GO:0019902) protein phosphatase binding(GO:0019903) |
| 0.1 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.4 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.5 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.0 | 2.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.6 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.2 | GO:0019212 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.3 | GO:0001008 | RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) transcription factor activity, RNA polymerase III promoter sequence-specific binding, TFIIIB recruiting(GO:0001004) transcription factor activity, RNA polymerase III type 1 promoter sequence-specific binding, TFIIIB recruiting(GO:0001005) transcription factor activity, RNA polymerase III type 2 promoter sequence-specific binding, TFIIIB recruiting(GO:0001008) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.1 | GO:0005034 | osmosensor activity(GO:0005034) |
| 0.0 | 0.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0048256 | 5'-flap endonuclease activity(GO:0017108) flap endonuclease activity(GO:0048256) |
| 0.0 | 0.7 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.4 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.6 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.6 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.1 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0042736 | NAD+ kinase activity(GO:0003951) NADH kinase activity(GO:0042736) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.4 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.1 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 2.1 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.0 | 0.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0015079 | potassium ion transmembrane transporter activity(GO:0015079) |
| 0.0 | 0.2 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.3 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.0 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.0 | 0.9 | GO:0000988 | transcription factor activity, protein binding(GO:0000988) |
| 0.0 | 0.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 1.1 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.8 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0045182 | translation regulator activity(GO:0045182) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |