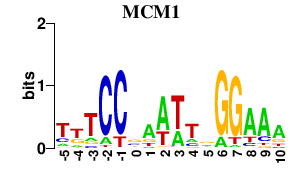
Results for MCM1
Z-value: 1.79
Transcription factors associated with MCM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MCM1
|
S000004646 | Transcription factor involved in both repression and activation |
Activity-expression correlation:
Activity profile of MCM1 motif
Sorted Z-values of MCM1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YFR055W | 10.43 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YPR119W | 9.55 |
CLB2
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YFR056C | 9.26 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YGR279C | 8.93 |
SCW4
|
Cell wall protein with similarity to glucanases; scw4 scw10 double mutants exhibit defects in mating |
|
| YMR032W | 8.86 |
HOF1
|
Bud neck-localized, SH3 domain-containing protein required for cytokinesis; regulates actomyosin ring dynamics and septin localization; interacts with the formins, Bni1p and Bnr1p, and with Cyk3p, Vrp1p, and Bni5p |
|
| YHR092C | 8.61 |
HXT4
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YNL160W | 8.51 |
YGP1
|
Cell wall-related secretory glycoprotein; induced by nutrient deprivation-associated growth arrest and upon entry into stationary phase; may be involved in adaptation prior to stationary phase entry; has similarity to Sps100p |
|
| YDR133C | 8.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR134C |
||
| YBR067C | 7.58 |
TIP1
|
Major cell wall mannoprotein with possible lipase activity; transcription is induced by heat- and cold-shock; member of the Srp1p/Tip1p family of serine-alanine-rich proteins |
|
| YBR092C | 7.55 |
PHO3
|
Constitutively expressed acid phosphatase similar to Pho5p; brought to the cell surface by transport vesicles; hydrolyzes thiamin phosphates in the periplasmic space, increasing cellular thiamin uptake; expression is repressed by thiamin |
|
| YIL158W | 7.54 |
AIM20
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; null mutant displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YEL040W | 7.02 |
UTR2
|
Cell wall protein that functions in the transfer of chitin to beta(1-6)glucan; putative chitin transglycosidase; glycosylphosphatidylinositol (GPI)-anchored protein localized to the bud neck; has a role in cell wall maintenance |
|
| YLR110C | 6.71 |
CCW12
|
Cell wall mannoprotein, mutants are defective in mating and agglutination, expression is downregulated by alpha-factor |
|
| YOR315W | 6.38 |
SFG1
|
Nuclear protein, putative transcription factor required for growth of superficial pseudohyphae (which do not invade the agar substrate) but not for invasive pseudohyphal growth; may act together with Phd1p; potential Cdc28p substrate |
|
| YPR157W | 6.22 |
Putative protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YNL327W | 5.44 |
EGT2
|
Glycosylphosphatidylinositol (GPI)-anchored cell wall endoglucanase required for proper cell separation after cytokinesis, expression is activated by Swi5p and tightly regulated in a cell cycle-dependent manner |
|
| YNR067C | 5.27 |
DSE4
|
Daughter cell-specific secreted protein with similarity to glucanases, degrades cell wall from the daughter side causing daughter to separate from mother |
|
| YAL038W | 5.19 |
CDC19
|
Pyruvate kinase, functions as a homotetramer in glycolysis to convert phosphoenolpyruvate to pyruvate, the input for aerobic (TCA cycle) or anaerobic (glucose fermentation) respiration |
|
| YBR158W | 5.17 |
AMN1
|
Protein required for daughter cell separation, multiple mitotic checkpoints, and chromosome stability; contains 12 degenerate leucine-rich repeat motifs; expression is induced by the Mitotic Exit Network (MEN) |
|
| YJR094W-A | 5.06 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YJL159W | 4.84 |
HSP150
|
O-mannosylated heat shock protein that is secreted and covalently attached to the cell wall via beta-1,3-glucan and disulfide bridges; required for cell wall stability; induced by heat shock, oxidative stress, and nitrogen limitation |
|
| YNL289W | 4.74 |
PCL1
|
Pho85 cyclin of the Pcl1,2-like subfamily, involved in entry into the mitotic cell cycle and regulation of morphogenesis, localizes to sites of polarized cell growth |
|
| YKL096W-A | 4.65 |
CWP2
|
Covalently linked cell wall mannoprotein, major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored |
|
| YML052W | 4.61 |
SUR7
|
Putative integral membrane protein; component of eisosomes; associated with endocytosis, along with Pil1p and Lsp1p; sporulation and plasma membrane sphingolipid content are altered in mutants |
|
| YDR044W | 4.54 |
HEM13
|
Coproporphyrinogen III oxidase, an oxygen requiring enzyme that catalyzes the sixth step in the heme biosynthetic pathway; localizes to the mitochondrial inner membrane; transcription is repressed by oxygen and heme (via Rox1p and Hap1p) |
|
| YFL026W | 4.50 |
STE2
|
Receptor for alpha-factor pheromone; seven transmembrane-domain GPCR that interacts with both pheromone and a heterotrimeric G protein to initiate the signaling response that leads to mating between haploid a and alpha cells |
|
| YKL164C | 4.50 |
PIR1
|
O-glycosylated protein required for cell wall stability; attached to the cell wall via beta-1,3-glucan; mediates mitochondrial translocation of Apn1p; expression regulated by the cell integrity pathway and by Swi5p during the cell cycle |
|
| YMR251W-A | 4.48 |
HOR7
|
Protein of unknown function; overexpression suppresses Ca2+ sensitivity of mutants lacking inositol phosphorylceramide mannosyltransferases Csg1p and Csh1p; transcription is induced under hyperosmotic stress and repressed by alpha factor |
|
| YJL196C | 4.21 |
ELO1
|
Elongase I, medium-chain acyl elongase, catalyzes carboxy-terminal elongation of unsaturated C12-C16 fatty acyl-CoAs to C16-C18 fatty acids |
|
| YOL154W | 4.21 |
ZPS1
|
Putative GPI-anchored protein; transcription is induced under low-zinc conditions, as mediated by the Zap1p transcription factor, and at alkaline pH |
|
| YDR033W | 4.11 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YPL256C | 3.84 |
CLN2
|
G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) |
|
| YOR314W | 3.79 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHL028W | 3.62 |
WSC4
|
ER membrane protein involved in the translocation of soluble secretory proteins and insertion of membrane proteins into the ER membrane; may also have a role in the stress response but has only partial functional overlap with WSC1-3 |
|
| YMR122W-A | 3.33 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and endoplasmic reticulum |
||
| YGR108W | 3.33 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YPR113W | 3.29 |
PIS1
|
Phosphatidylinositol synthase, required for biosynthesis of phosphatidylinositol, which is a precursor for polyphosphoinositides, sphingolipids, and glycolipid anchors for some of the plasma membrane proteins |
|
| YMR305C | 3.27 |
SCW10
|
Cell wall protein with similarity to glucanases; may play a role in conjugation during mating based on mutant phenotype and its regulation by Ste12p |
|
| YGR086C | 3.22 |
PIL1
|
Primary component of eisosomes, which are large immobile cell cortex structures associated with endocytosis; null mutants show activation of Pkc1p/Ypk1p stress resistance pathways; detected in phosphorylated state in mitochondria |
|
| YDR345C | 3.07 |
HXT3
|
Low affinity glucose transporter of the major facilitator superfamily, expression is induced in low or high glucose conditions |
|
| YJL079C | 2.96 |
PRY1
|
Protein of unknown function, has similarity to Pry2p and Pry3p and to the plant PR-1 class of pathogen related proteins |
|
| YNR001W-A | 2.89 |
Dubious open reading frame unlikely to encode a functional protein; identified by homology |
||
| YIR021W | 2.87 |
MRS1
|
Protein required for the splicing of two mitochondrial group I introns (BI3 in COB and AI5beta in COX1); forms a splicing complex, containing four subunits of Mrs1p and two subunits of the BI3-encoded maturase, that binds to the BI3 RNA |
|
| YML132W | 2.80 |
COS3
|
Protein involved in salt resistance; interacts with sodium:hydrogen antiporter Nha1p; member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YOL155C | 2.74 |
HPF1
|
Haze-protective mannoprotein that reduces the particle size of aggregated proteins in white wines |
|
| YNL336W | 2.68 |
COS1
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YDL236W | 2.68 |
PHO13
|
Alkaline phosphatase specific for p-nitrophenyl phosphate, involved in dephosphorylation of histone II-A and casein |
|
| YKL185W | 2.66 |
ASH1
|
Zinc-finger inhibitor of HO transcription; mRNA is localized and translated in the distal tip of anaphase cells, resulting in accumulation of Ash1p in daughter cell nuclei and inhibition of HO expression; potential Cdc28p substrate |
|
| YNL300W | 2.62 |
Glycosylphosphatidylinositol-dependent cell wall protein, expression is periodic and decreases in respone to ergosterol perturbation or upon entry into stationary phase; depletion increases resistance to lactic acid |
||
| YJL157C | 2.58 |
FAR1
|
Cyclin-dependent kinase inhibitor that mediates cell cycle arrest in response to pheromone; also forms a complex with Cdc24p, Ste4p, and Ste18p that may specify the direction of polarized growth during mating; potential Cdc28p substrate |
|
| YIL123W | 2.51 |
SIM1
|
Protein of the SUN family (Sim1p, Uth1p, Nca3p, Sun4p) that may participate in DNA replication, promoter contains SCB regulation box at -300 bp indicating that expression may be cell cycle-regulated |
|
| YKL152C | 2.49 |
GPM1
|
Tetrameric phosphoglycerate mutase, mediates the conversion of 3-phosphoglycerate to 2-phosphoglycerate during glycolysis and the reverse reaction during gluconeogenesis |
|
| YER131W | 2.46 |
RPS26B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps26Ap and has similarity to rat S26 ribosomal protein |
|
| YIL015W | 2.39 |
BAR1
|
Aspartyl protease secreted into the periplasmic space of mating type a cells, helps cells find mating partners, cleaves and inactivates alpha factor allowing cells to recover from alpha-factor-induced cell cycle arrest |
|
| YER070W | 2.38 |
RNR1
|
One of two large regulatory subunits of ribonucleotide-diphosphate reductase; the RNR complex catalyzes rate-limiting step in dNTP synthesis, regulated by DNA replication and DNA damage checkpoint pathways via localization of small subunits |
|
| YKR013W | 2.35 |
PRY2
|
Protein of unknown function, has similarity to Pry1p and Pry3p and to the plant PR-1 class of pathogen related proteins |
|
| YBR053C | 2.34 |
Putative protein of unknown function; induced by cell wall perturbation |
||
| YOR247W | 2.33 |
SRL1
|
Mannoprotein that exhibits a tight association with the cell wall, required for cell wall stability in the absence of GPI-anchored mannoproteins; has a high serine-threonine content; expression is induced in cell wall mutants |
|
| YIL114C | 2.30 |
POR2
|
Putative mitochondrial porin (voltage-dependent anion channel), related to Por1p but not required for mitochondrial membrane permeability or mitochondrial osmotic stability |
|
| YNL338W | 2.24 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps TEL14L-XC, which is Telomeric X element Core sequence on the left arm of Chromosome XIV |
||
| YPL014W | 2.14 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and to the nucleus |
||
| YKL209C | 2.10 |
STE6
|
Plasma membrane ATP-binding cassette (ABC) transporter required for the export of a-factor, catalyzes ATP hydrolysis coupled to a-factor transport; contains 12 transmembrane domains and two ATP binding domains; expressed only in MATa cells |
|
| YMR001C-A | 2.09 |
Putative protein of unknown function |
||
| YPR149W | 2.09 |
NCE102
|
Protein of unknown function; contains transmembrane domains; involved in secretion of proteins that lack classical secretory signal sequences; component of the detergent-insoluble glycolipid-enriched complexes (DIGs) |
|
| YBR009C | 2.09 |
HHF1
|
One of two identical histone H4 proteins (see also HHF2); core histone required for chromatin assembly and chromosome function; contributes to telomeric silencing; N-terminal domain involved in maintaining genomic integrity |
|
| YCR102W-A | 2.07 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOL130W | 2.05 |
ALR1
|
Plasma membrane Mg(2+) transporter, expression and turnover are regulated by Mg(2+) concentration; overexpression confers increased tolerance to Al(3+) and Ga(3+) ions |
|
| YFR054C | 2.02 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHR141C | 1.87 |
RPL42B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl42Ap and has similarity to rat L44; required for propagation of the killer toxin-encoding M1 double-stranded RNA satellite of the L-A double-stranded RNA virus |
|
| YEL034C-A | 1.85 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; completely overlaps HYP2/YEL034W, a verified gene that encodes eiF-5A |
||
| YKR012C | 1.84 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene PRY2 |
||
| YLR113W | 1.79 |
HOG1
|
Mitogen-activated protein kinase involved in osmoregulation via three independent osmosensors; mediates the recruitment and activation of RNA Pol II at Hot1p-dependent promoters; localization regulated by Ptp2p and Ptp3p |
|
| YKL216W | 1.77 |
URA1
|
Dihydroorotate dehydrogenase, catalyzes the fourth enzymatic step in the de novo biosynthesis of pyrimidines, converting dihydroorotic acid into orotic acid |
|
| YLR190W | 1.74 |
MMR1
|
Phosphorylated protein of the mitochondrial outer membrane, localizes only to mitochondria of the bud; interacts with Myo2p to mediate mitochondrial distribution to buds; mRNA is targeted to the bud via the transport system involving She2p |
|
| YOR313C | 1.74 |
SPS4
|
Protein whose expression is induced during sporulation; not required for sporulation; heterologous expression in E. coli induces the SOS response that senses DNA damage |
|
| YLR028C | 1.73 |
ADE16
|
Enzyme of 'de novo' purine biosynthesis containing both 5-aminoimidazole-4-carboxamide ribonucleotide transformylase and inosine monophosphate cyclohydrolase activities, isozyme of Ade17p; ade16 ade17 mutants require adenine and histidine |
|
| YPR069C | 1.70 |
SPE3
|
Spermidine synthase, involved in biosynthesis of spermidine and also in biosynthesis of pantothenic acid; spermidine is required for growth of wild-type cells |
|
| YFL027C | 1.65 |
GYP8
|
GTPase-activating protein for yeast Rab family members; Ypt1p is the preferred in vitro substrate but also acts on Sec4p, Ypt31p and Ypt32p; involved in the regulation of ER to Golgi vesicle transport |
|
| YKL080W | 1.64 |
VMA5
|
Subunit C of the eight-subunit V1 peripheral membrane domain of vacuolar H+-ATPase (V-ATPase), an electrogenic proton pump found throughout the endomembrane system; required for the V1 domain to assemble onto the vacuolar membrane |
|
| YHR149C | 1.64 |
SKG6
|
Integral membrane protein that localizes primarily to growing sites such as the bud tip or the cell periphery; potential Cdc28p substrate; Skg6p interacts with Zds1p and Zds2p |
|
| YBR093C | 1.62 |
PHO5
|
Repressible acid phosphatase (1 of 3) that also mediates extracellular nucleotide-derived phosphate hydrolysis; secretory pathway derived cell surface glycoprotein; induced by phosphate starvation and coordinately regulated by PHO4 and PHO2 |
|
| YJL158C | 1.62 |
CIS3
|
Mannose-containing glycoprotein constituent of the cell wall; member of the PIR (proteins with internal repeats) family |
|
| YNL066W | 1.60 |
SUN4
|
Cell wall protein related to glucanases, possibly involved in cell wall septation; member of the SUN family |
|
| YPR148C | 1.60 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
||
| YEL066W | 1.59 |
HPA3
|
D-Amino acid N-acetyltransferase, catalyzes N-acetylation of D-amino acids through ordered bi-bi mechanism in which acetyl-CoA is first substrate bound and CoA is last product liberated; similar to Hpa2p, acetylates histones weakly in vitro |
|
| YOR103C | 1.59 |
OST2
|
Epsilon subunit of the oligosaccharyltransferase complex of the ER lumen, which catalyzes asparagine-linked glycosylation of newly synthesized proteins |
|
| YMR122C | 1.58 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHR007C | 1.54 |
ERG11
|
Lanosterol 14-alpha-demethylase, catalyzes the C-14 demethylation of lanosterol to form 4,4''-dimethyl cholesta-8,14,24-triene-3-beta-ol in the ergosterol biosynthesis pathway; member of the cytochrome P450 family |
|
| YAL040C | 1.53 |
CLN3
|
G1 cyclin involved in cell cycle progression; activates Cdc28p kinase to promote the G1 to S phase transition; plays a role in regulating transcription of the other G1 cyclins, CLN1 and CLN2; regulated by phosphorylation and proteolysis |
|
| YDR465C | 1.53 |
RMT2
|
Arginine methyltransferase; ribosomal protein L12 is a substrate |
|
| YIL009C-A | 1.52 |
EST3
|
Component of the telomerase holoenzyme, involved in telomere replication |
|
| YKR092C | 1.51 |
SRP40
|
Nucleolar, serine-rich protein with a role in preribosome assembly or transport; may function as a chaperone of small nucleolar ribonucleoprotein particles (snoRNPs); immunologically and structurally to rat Nopp140 |
|
| YOR226C | 1.51 |
ISU2
|
Conserved protein of the mitochondrial matrix, required for synthesis of mitochondrial and cytosolic iron-sulfur proteins, performs a scaffolding function in mitochondria during Fe/S cluster assembly; isu1 isu2 double mutant is inviable |
|
| YIR020C | 1.51 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR014W | 1.49 |
MSB2
|
Mucin family member involved in the Cdc42p- and MAP kinase-dependent filamentous growth signaling pathway; also functions as an osmosensor in parallel to the Sho1p-mediated pathway; potential Cdc28p substrate |
|
| YGL256W | 1.49 |
ADH4
|
Alcohol dehydrogenase isoenzyme type IV, dimeric enzyme demonstrated to be zinc-dependent despite sequence similarity to iron-activated alcohol dehydrogenases; transcription is induced in response to zinc deficiency |
|
| YDR309C | 1.48 |
GIC2
|
Redundant rho-like GTPase Cdc42p effector; homolog of Gic1p; involved in initiation of budding and cellular polarization; interacts with Cdc42p via the Cdc42/Rac-interactive binding (CRIB) domain and with PI(4,5)P2 via a polybasic region |
|
| YCL049C | 1.48 |
Protein of unknown function; localizes to membrane fraction; YCL049C is not an essential gene; |
||
| YGR078C | 1.48 |
PAC10
|
Part of the heteromeric co-chaperone GimC/prefoldin complex, which promotes efficient protein folding |
|
| YPR156C | 1.46 |
TPO3
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; member of the major facilitator superfamily |
|
| YKR038C | 1.44 |
KAE1
|
Putative glycoprotease proposed to be in transcription as a component of the EKC protein complex with Bud32p, Cgi121p, Pcc1p, and Gon7p; also identified as a component of the KEOPS protein complex |
|
| YHR005C-A | 1.44 |
MRS11
|
Essential protein of the mitochondrial intermembrane space, forms a complex with Tim9p (TIM10 complex) that mediates insertion of hydrophobic proteins at the inner membrane, has homology to Mrs5p, which is also involved in this process |
|
| YLR432W | 1.44 |
IMD3
|
Inosine monophosphate dehydrogenase, catalyzes the first step of GMP biosynthesis, member of a four-gene family in S. cerevisiae, constitutively expressed |
|
| YIL009W | 1.43 |
FAA3
|
Long chain fatty acyl-CoA synthetase, has a preference for C16 and C18 fatty acids; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery |
|
| YCL048W-A | 1.43 |
Putative protein of unknown function |
||
| YLR112W | 1.40 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR451C | 1.39 |
YHP1
|
One of two homeobox transcriptional repressors (see also Yox1p), that bind to Mcm1p and to early cell cycle box (ECB) elements of cell cycle regulated genes, thereby restricting ECB-mediated transcription to the M/G1 interval |
|
| YMR121C | 1.38 |
RPL15B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl15Ap and has similarity to rat L15 ribosomal protein; binds to 5.8 S rRNA |
|
| YIR013C | 1.34 |
GAT4
|
Protein containing GATA family zinc finger motifs |
|
| YIR020W-A | 1.33 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YJR145C | 1.33 |
RPS4A
|
Protein component of the small (40S) ribosomal subunit; mutation affects 20S pre-rRNA processing; identical to Rps4Bp and has similarity to rat S4 ribosomal protein |
|
| YHR005C | 1.32 |
GPA1
|
GTP-binding alpha subunit of the heterotrimeric G protein that couples to pheromone receptors; negatively regulates the mating pathway by sequestering G(beta)gamma and by triggering an adaptive response; activates Vps34p at the endosome |
|
| YFR015C | 1.32 |
GSY1
|
Glycogen synthase with similarity to Gsy2p, the more highly expressed yeast homolog; expression induced by glucose limitation, nitrogen starvation, environmental stress, and entry into stationary phase |
|
| YMR199W | 1.32 |
CLN1
|
G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) |
|
| YIL169C | 1.31 |
Putative protein of unknown function; serine/threonine rich and highly similar to YOL155C, a putative glucan alpha-1,4-glucosidase; transcript is induced in both high and low pH environments; YIL169C is a non-essential gene |
||
| YJL145W | 1.29 |
SFH5
|
Putative phosphatidylinositol transfer protein (PITP), exhibits phosphatidylinositol- but not phosphatidylcholine-transfer activity, localized to cytosol and microsomes, similar to Sec14p; may be PITP regulator rather than actual PITP |
|
| YNL058C | 1.29 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YNL058C is not an essential gene |
||
| YMR266W | 1.29 |
RSN1
|
Membrane protein of unknown function; overexpression suppresses NaCl sensitivity of sro7 mutant |
|
| YNL078W | 1.29 |
NIS1
|
Protein localized in the bud neck at G2/M phase; physically interacts with septins; possibly involved in a mitotic signaling network |
|
| YOR273C | 1.29 |
TPO4
|
Polyamine transport protein, recognizes spermine, putrescine, and spermidine; localizes to the plasma membrane; member of the major facilitator superfamily |
|
| YDR032C | 1.28 |
PST2
|
Protein with similarity to members of a family of flavodoxin-like proteins; induced by oxidative stress in a Yap1p dependent manner; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YHR094C | 1.25 |
HXT1
|
Low-affinity glucose transporter of the major facilitator superfamily, expression is induced by Hxk2p in the presence of glucose and repressed by Rgt1p when glucose is limiting |
|
| YLR196W | 1.24 |
PWP1
|
Protein with WD-40 repeats involved in rRNA processing; associates with trans-acting ribosome biogenesis factors; similar to beta-transducin superfamily |
|
| YNL301C | 1.23 |
RPL18B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl18Ap and has similarity to rat L18 ribosomal protein |
|
| YLR257W | 1.22 |
Putative protein of unknown function |
||
| YLR120C | 1.21 |
YPS1
|
Aspartic protease, attached to the plasma membrane via a glycosylphosphatidylinositol (GPI) anchor |
|
| YNL079C | 1.19 |
TPM1
|
Major isoform of tropomyosin; binds to and stabilizes actin cables and filaments, which direct polarized cell growth and the distribution of several organelles; acetylated by the NatB complex and acetylated form binds actin most efficiently |
|
| YBR010W | 1.18 |
HHT1
|
One of two identical histone H3 proteins (see also HHT2); core histone required for chromatin assembly, involved in heterochromatin-mediated telomeric and HM silencing; regulated by acetylation, methylation, and mitotic phosphorylation |
|
| YCL050C | 1.18 |
APA1
|
Diadenosine 5',5''-P1,P4-tetraphosphate phosphorylase I (AP4A phosphorylase), involved in catabolism of bis(5'-nucleosidyl) tetraphosphates; has similarity to Apa2p |
|
| YLL064C | 1.18 |
PAU18
|
Hypothetical protein |
|
| YGL032C | 1.17 |
AGA2
|
Adhesion subunit of a-agglutinin of a-cells, C-terminal sequence acts as a ligand for alpha-agglutinin (Sag1p) during agglutination, modified with O-linked oligomannosyl chains, linked to anchorage subunit Aga1p via two disulfide bonds |
|
| YDR399W | 1.15 |
HPT1
|
Dimeric hypoxanthine-guanine phosphoribosyltransferase, catalyzes the formation of both inosine monophosphate and guanosine monophosphate; mutations in the human homolog HPRT1 can cause Lesch-Nyhan syndrome and Kelley-Seegmiller syndrome |
|
| YGL257C | 1.13 |
MNT2
|
Mannosyltransferase involved in adding the 4th and 5th mannose residues of O-linked glycans |
|
| YML119W | 1.11 |
Putative protein of unknown funtion; YML119W is not an essential gene; potential Cdc28p substrate |
||
| YBR164C | 1.11 |
ARL1
|
Soluble GTPase with a role in regulation of membrane traffic; regulates potassium influx; G protein of the Ras superfamily, similar to ADP-ribosylation factor |
|
| YOL007C | 1.10 |
CSI2
|
Protein of unknown function; green fluorescent protein (GFP)- fusion protein localizes to the mother side of the bud neck and the vacuole; YOL007C is not an essential gene |
|
| YER091C | 1.10 |
MET6
|
Cobalamin-independent methionine synthase, involved in amino acid biosynthesis; requires a minimum of two glutamates on the methyltetrahydrofolate substrate, similar to bacterial metE homologs |
|
| YDR276C | 1.10 |
PMP3
|
Small plasma membrane protein related to a family of plant polypeptides that are overexpressed under high salt concentration or low temperature, not essential for viability, deletion causes hyperpolarization of the plasma membrane potential |
|
| YAL022C | 1.09 |
FUN26
|
Nucleoside transporter with broad nucleoside selectivity; localized to intracellular membranes |
|
| YCR104W | 1.08 |
PAU3
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YGL039W | 1.07 |
Oxidoreductase, catalyzes NADPH-dependent reduction of the bicyclic diketone bicyclo[2.2.2]octane-2,6-dione (BCO2,6D) to the chiral ketoalcohol (1R,4S,6S)-6-hydroxybicyclo[2.2.2]octane-2-one (BCO2one6ol) |
||
| YBR054W | 1.06 |
YRO2
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in a phosphorylated state in highly purified mitochondria in high-throughput studies; transcriptionally regulated by Haa1p |
|
| YLR154C | 1.04 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YOL127W | 1.03 |
RPL25
|
Primary rRNA-binding ribosomal protein component of the large (60S) ribosomal subunit, has similarity to E. coli L23 and rat L23a ribosomal proteins; binds to 26S rRNA via a conserved C-terminal motif |
|
| YNL057W | 1.03 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YKL165C | 1.03 |
MCD4
|
Protein involved in glycosylphosphatidylinositol (GPI) anchor synthesis; multimembrane-spanning protein that localizes to the endoplasmic reticulum; highly conserved among eukaryotes |
|
| YGL201C | 1.03 |
MCM6
|
Protein involved in DNA replication; component of the Mcm2-7 hexameric complex that binds chromatin as a part of the pre-replicative complex |
|
| YGR164W | 1.02 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNL178W | 1.01 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YER031C | 1.01 |
YPT31
|
GTPase of the Ypt/Rab family, very similar to Ypt32p; involved in the exocytic pathway; mediates intra-Golgi traffic or the budding of post-Golgi vesicles from the trans-Golgi |
|
| YNL337W | 1.00 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YML058W | 1.00 |
SML1
|
Ribonucleotide reductase inhibitor involved in regulating dNTP production; regulated by Mec1p and Rad53p during DNA damage and S phase |
|
| YGL040C | 0.99 |
HEM2
|
Delta-aminolevulinate dehydratase, a homo-octameric enzyme, catalyzes the conversion of delta-aminolevulinic acid to porphobilinogen, the second step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus |
|
| YFR005C | 0.99 |
SAD1
|
Conserved zinc-finger domain protein involved in pre-mRNA splicing, required for assembly of U4 snRNA into the U4/U6 particle |
|
| YGL258W | 0.97 |
VEL1
|
Protein of unknown function; highly induced in zinc-depleted conditions and has increased expression in NAP1 deletion mutants |
|
| YPR063C | 0.97 |
ER-localized protein of unknown function |
||
| YBL092W | 0.96 |
RPL32
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L32 ribosomal protein; overexpression disrupts telomeric silencing |
|
| YGL088W | 0.95 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps snR10, a snoRNA required for preRNA processing |
||
| YOR222W | 0.95 |
ODC2
|
Mitochondrial inner membrane transporter, exports 2-oxoadipate and 2-oxoglutarate from the mitochondrial matrix to the cytosol for use in lysine and glutamate biosynthesis and in lysine catabolism |
|
| YGL021W | 0.95 |
ALK1
|
Protein kinase; accumulation and phosphorylation are periodic during the cell cycle; phosphorylated in response to DNA damage; contains characteristic motifs for degradation via the APC pathway; similar to Alk2p and to mammalian haspins |
|
| YDL037C | 0.94 |
BSC1
|
Protein of unconfirmed function, similar to cell surface flocculin Muc1p; ORF exhibits genomic organization compatible with a translational readthrough-dependent mode of expression |
|
| YCR018C | 0.93 |
SRD1
|
Protein involved in the processing of pre-rRNA to mature rRNA; contains a C2/C2 zinc finger motif; srd1 mutation suppresses defects caused by the rrp1-1 mutation |
|
| YER130C | 0.93 |
Hypothetical protein |
||
| YEL068C | 0.93 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YAR020C | 0.93 |
PAU7
|
Part of 23-member seripauperin multigene family, active during alcoholic fermentation, regulated by anaerobiosis, inhibited by oxygen, repressed by heme |
|
| YLR341W | 0.92 |
SPO77
|
Meiosis-specific protein of unknown function, required for spore wall formation during sporulation; dispensable for both nuclear divisions during meiosis |
|
| YDR341C | 0.92 |
Arginyl-tRNA synthetase, proposed to be cytoplasmic but the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YFL062W | 0.92 |
COS4
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YJL170C | 0.92 |
ASG7
|
Protein that regulates signaling from a G protein beta subunit Ste4p and its relocalization within the cell; specific to a-cells and induced by alpha-factor |
|
| YHL018W | 0.91 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to mitochondria and is induced in response to the DNA-damaging agent MMS |
||
| YMR009W | 0.91 |
ADI1
|
Acireductone dioxygenease involved in the methionine salvage pathway; ortholog of human MTCBP-1; transcribed with YMR010W and regulated post-transcriptionally by RNase III (Rnt1p) cleavage; ADI1 mRNA is induced in heat shock conditions |
|
| YAR071W | 0.89 |
PHO11
|
One of three repressible acid phosphatases, a glycoprotein that is transported to the cell surface by the secretory pathway; induced by phosphate starvation and coordinately regulated by PHO4 and PHO2 |
|
| YPR196W | 0.89 |
Putative maltose activator |
||
| YFL015W-A | 0.88 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR214W | 0.88 |
RPS0A
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps0Bp; required for maturation of 18S rRNA along with Rps0Bp; deletion of either RPS0 gene reduces growth rate, deletion of both genes is lethal |
|
| YLR183C | 0.87 |
TOS4
|
Transcription factor that binds to a number of promoter regions, particularly promoters of some genes involved in pheromone response and cell cycle; potential Cdc28p substrate; expression is induced in G1 by bound SBF |
|
| YLR154W-B | 0.86 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YGR284C | 0.86 |
ERV29
|
Protein localized to COPII-coated vesicles, involved in vesicle formation and incorporation of specific secretory cargo |
|
| YGR189C | 0.85 |
CRH1
|
Putative chitin transglycosidase, cell wall protein that functions in the transfer of chitin to beta(1-6)glucan; localizes to sites of polarized growth; expression is induced under cell wall stress conditions |
|
| YPL274W | 0.85 |
SAM3
|
High-affinity S-adenosylmethionine permease, required for utilization of S-adenosylmethionine as a sulfur source; has similarity to S-methylmethionine permease Mmp1p |
|
| YIL069C | 0.85 |
RPS24B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps24Ap and has similarity to rat S24 ribosomal protein |
|
| YIL076W | 0.84 |
SEC28
|
Epsilon-COP subunit of the coatomer; regulates retrograde Golgi-to-ER protein traffic; stabilizes Cop1p, the alpha-COP and the coatomer complex; non-essential for cell growth |
|
| YDR225W | 0.84 |
HTA1
|
One of two nearly identical (see also HTA2) histone H2A subtypes; core histone required for chromatin assembly and chromosome function; DNA damage-dependent phosphorylation by Mec1p facilitates DNA repair; acetylated by Nat4p |
|
| YIL118W | 0.84 |
RHO3
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins involved in the establishment of cell polarity; GTPase activity positively regulated by the GTPase activating protein (GAP) Rgd1p |
|
| YPL079W | 0.83 |
RPL21B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl21Ap and has similarity to rat L21 ribosomal protein |
|
| YGR169C-A | 0.83 |
Putative protein of unknown function |
||
| YMR006C | 0.82 |
PLB2
|
Phospholipase B (lysophospholipase) involved in phospholipid metabolism; displays transacylase activity in vitro; overproduction confers resistance to lysophosphatidylcholine |
|
| YHR144C | 0.82 |
DCD1
|
Deoxycytidine monophosphate (dCMP) deaminase required for dCTP and dTTP synthesis; expression is NOT cell cycle regulated |
|
| YCR102C | 0.82 |
Putative protein of unknown function; involved in copper metabolism; similar to C.carbonum toxD gene; YCR102C is not an essential gene |
||
| YEL017W | 0.82 |
GTT3
|
Protein of unknown function with a possible role in glutathione metabolism, as suggested by computational analysis of large-scale protein-protein interaction data; GFP-fusion protein localizes to the nuclear periphery |
|
| YHR013C | 0.80 |
ARD1
|
Subunit of the N-terminal acetyltransferase NatA (Nat1p, Ard1p, Nat5p); N-terminally acetylates many proteins, which influences multiple processes such as the cell cycle, heat-shock resistance, mating, sporulation, and telomeric silencing |
|
| YJR099W | 0.80 |
YUH1
|
Ubiquitin C-terminal hydrolase that cleaves ubiquitin-protein fusions to generate monomeric ubiquitin; hydrolyzes the peptide bond at the C-terminus of ubiquitin; also the major processing enzyme for the ubiquitin-like protein Rub1p |
|
| YGR177C | 0.80 |
ATF2
|
Alcohol acetyltransferase, may play a role in steroid detoxification; forms volatile esters during fermentation, which is important in brewing |
|
| YKR093W | 0.80 |
PTR2
|
Integral membrane peptide transporter, mediates transport of di- and tri-peptides; conserved protein that contains 12 transmembrane domains; PTR2 expression is regulated by the N-end rule pathway via repression by Cup9p |
|
| YGR106C | 0.79 |
VOA1
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuolar memebrane |
|
| YOR011W | 0.79 |
AUS1
|
Transporter of the ATP-binding cassette family, involved in uptake of sterols and anaerobic growth |
|
| YCR024C-B | 0.78 |
Putative protein of unknown function; identified by expression profiling and mass spectrometry |
||
| YGL202W | 0.78 |
ARO8
|
Aromatic aminotransferase I, expression is regulated by general control of amino acid biosynthesis |
|
| YFL015C | 0.78 |
Dubious open reading frame unlikely to encode a protein; partially overlaps dubious ORF YFL015W-A; YFL015C is not an essential gene |
||
| YGR107W | 0.78 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
Network of associatons between targets according to the STRING database.
First level regulatory network of MCM1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 1.8 | 5.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 1.7 | 11.8 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 1.7 | 6.6 | GO:0098742 | agglutination involved in conjugation with cellular fusion(GO:0000752) agglutination involved in conjugation(GO:0000771) heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) cell-cell adhesion(GO:0098609) adhesion between unicellular organisms(GO:0098610) multi organism cell adhesion(GO:0098740) cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 1.2 | 5.9 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 1.1 | 4.2 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.9 | 11.2 | GO:0090529 | barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.9 | 2.8 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.9 | 2.7 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) |
| 0.9 | 2.6 | GO:0071850 | mitotic cell cycle arrest in response to pheromone(GO:0000751) mitotic cell cycle arrest(GO:0071850) |
| 0.8 | 5.5 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.8 | 3.1 | GO:0000296 | spermine transport(GO:0000296) |
| 0.8 | 4.5 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.7 | 2.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.7 | 2.7 | GO:0046898 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 0.6 | 3.6 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.6 | 2.4 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.6 | 8.9 | GO:0006037 | cell wall chitin metabolic process(GO:0006037) |
| 0.6 | 1.7 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.5 | 1.6 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.5 | 2.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.5 | 1.5 | GO:1902223 | L-phenylalanine biosynthetic process(GO:0009094) erythrose 4-phosphate/phosphoenolpyruvate family amino acid biosynthetic process(GO:1902223) |
| 0.5 | 1.5 | GO:0018195 | peptidyl-arginine modification(GO:0018195) peptidyl-arginine methylation(GO:0018216) |
| 0.5 | 1.5 | GO:0031684 | heterotrimeric G-protein complex cycle(GO:0031684) |
| 0.4 | 0.8 | GO:0015848 | spermidine transport(GO:0015848) |
| 0.4 | 5.7 | GO:0007009 | plasma membrane organization(GO:0007009) |
| 0.4 | 1.6 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) |
| 0.4 | 2.0 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.4 | 0.4 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.4 | 1.1 | GO:0009177 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.4 | 2.9 | GO:0000372 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.3 | 5.2 | GO:0001100 | negative regulation of exit from mitosis(GO:0001100) |
| 0.3 | 2.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.3 | 1.6 | GO:0060277 | obsolete negative regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0060277) |
| 0.3 | 7.7 | GO:0008645 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.3 | 5.5 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.3 | 0.9 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.3 | 5.6 | GO:0042723 | thiamine-containing compound metabolic process(GO:0042723) |
| 0.3 | 0.9 | GO:0090110 | cargo loading into vesicle(GO:0035459) cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.3 | 1.4 | GO:0007232 | osmosensory signaling pathway via Sho1 osmosensor(GO:0007232) |
| 0.3 | 6.9 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.3 | 4.0 | GO:0015833 | peptide transport(GO:0015833) |
| 0.3 | 1.8 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.3 | 1.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.3 | 1.0 | GO:0015867 | ATP transport(GO:0015867) |
| 0.3 | 6.5 | GO:0006885 | regulation of pH(GO:0006885) |
| 0.2 | 0.7 | GO:0016094 | polyprenol metabolic process(GO:0016093) polyprenol biosynthetic process(GO:0016094) dolichol metabolic process(GO:0019348) dolichol biosynthetic process(GO:0019408) |
| 0.2 | 5.8 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.2 | 1.4 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.2 | 0.5 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 0.7 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.2 | 3.8 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.2 | 1.9 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.2 | 0.8 | GO:0051046 | regulation of exocytosis(GO:0017157) regulation of secretion(GO:0051046) regulation of formin-nucleated actin cable assembly(GO:0090337) positive regulation of formin-nucleated actin cable assembly(GO:0090338) regulation of secretion by cell(GO:1903530) |
| 0.2 | 0.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.2 | 0.6 | GO:0007535 | donor selection(GO:0007535) |
| 0.2 | 1.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 0.6 | GO:1901073 | aminoglycan biosynthetic process(GO:0006023) chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.2 | 1.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 0.9 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.2 | 0.9 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.2 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 0.5 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.2 | 0.7 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 0.2 | 1.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 2.2 | GO:0006113 | fermentation(GO:0006113) |
| 0.2 | 1.0 | GO:0009164 | nucleoside catabolic process(GO:0009164) glycosyl compound catabolic process(GO:1901658) |
| 0.2 | 0.8 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.2 | 1.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 1.2 | GO:0061572 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.1 | 3.1 | GO:0015918 | sterol transport(GO:0015918) |
| 0.1 | 0.7 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 2.0 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.1 | 0.7 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 0.4 | GO:0009090 | homoserine biosynthetic process(GO:0009090) |
| 0.1 | 1.3 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 17.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 1.2 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.1 | 4.0 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.1 | 3.1 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) phytosteroid biosynthetic process(GO:0016129) cellular alcohol biosynthetic process(GO:0044108) cellular lipid biosynthetic process(GO:0097384) |
| 0.1 | 0.2 | GO:0019673 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 0.5 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 1.2 | GO:0052803 | histidine biosynthetic process(GO:0000105) histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.1 | 0.5 | GO:0046417 | chorismate metabolic process(GO:0046417) |
| 0.1 | 0.3 | GO:0042992 | regulation of transcription factor import into nucleus(GO:0042990) negative regulation of transcription factor import into nucleus(GO:0042992) |
| 0.1 | 1.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.3 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 1.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.7 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.1 | 1.0 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 0.4 | GO:0010674 | negative regulation of transcription from RNA polymerase II promoter involved in meiotic cell cycle(GO:0010674) |
| 0.1 | 0.4 | GO:0015883 | FAD transport(GO:0015883) |
| 0.1 | 0.7 | GO:0009262 | deoxyribonucleotide metabolic process(GO:0009262) deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 2.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 1.5 | GO:0008202 | steroid metabolic process(GO:0008202) sterol metabolic process(GO:0016125) |
| 0.1 | 0.5 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.1 | 0.3 | GO:0000092 | mitotic anaphase B(GO:0000092) |
| 0.1 | 3.7 | GO:0070726 | cell wall assembly(GO:0070726) |
| 0.1 | 0.4 | GO:0034227 | tRNA wobble position uridine thiolation(GO:0002143) tRNA thio-modification(GO:0034227) |
| 0.1 | 0.6 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 2.1 | GO:0048311 | mitochondrion inheritance(GO:0000001) mitochondrion distribution(GO:0048311) |
| 0.1 | 1.2 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 0.2 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.3 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.3 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 3.0 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.1 | 4.3 | GO:0048468 | ascospore formation(GO:0030437) cell development(GO:0048468) |
| 0.1 | 0.3 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.1 | 0.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.8 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 0.7 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.1 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.7 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 1.5 | GO:0002098 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.3 | GO:0033241 | regulation of cellular amine catabolic process(GO:0033241) |
| 0.1 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.6 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 0.2 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.3 | GO:0045010 | positive regulation of actin filament polymerization(GO:0030838) actin nucleation(GO:0045010) |
| 0.1 | 0.3 | GO:0006551 | leucine metabolic process(GO:0006551) leucine biosynthetic process(GO:0009098) |
| 0.1 | 1.1 | GO:0098876 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.1 | 0.3 | GO:0060304 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.3 | GO:0070096 | mitochondrial outer membrane translocase complex assembly(GO:0070096) |
| 0.1 | 0.4 | GO:0000743 | nuclear migration involved in conjugation with cellular fusion(GO:0000743) |
| 0.1 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.7 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.1 | 0.2 | GO:0046495 | nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) |
| 0.1 | 0.4 | GO:0009272 | fungal-type cell wall biogenesis(GO:0009272) |
| 0.1 | 0.2 | GO:0000709 | meiotic joint molecule formation(GO:0000709) |
| 0.0 | 0.2 | GO:0007329 | positive regulation of transcription from RNA polymerase II promoter by pheromones(GO:0007329) positive regulation of transcription by pheromones(GO:0009371) |
| 0.0 | 0.1 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.2 | GO:0048279 | vesicle fusion with endoplasmic reticulum(GO:0048279) |
| 0.0 | 0.8 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.1 | GO:0040031 | snRNA pseudouridine synthesis(GO:0031120) snRNA modification(GO:0040031) |
| 0.0 | 0.2 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.4 | GO:0071039 | nuclear polyadenylation-dependent CUT catabolic process(GO:0071039) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 0.5 | GO:0042181 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) ketone biosynthetic process(GO:0042181) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.0 | 0.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.3 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 0.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of metal ion(GO:0051238) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0043487 | regulation of RNA stability(GO:0043487) regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.5 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0046219 | tryptophan biosynthetic process(GO:0000162) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.0 | 1.2 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.7 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.2 | GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity(GO:0051090) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.5 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.3 | GO:0033967 | box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) |
| 0.0 | 0.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 1.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0015940 | pantothenate metabolic process(GO:0015939) pantothenate biosynthetic process(GO:0015940) |
| 0.0 | 0.2 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.2 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.0 | 0.0 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.6 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 0.0 | GO:0035753 | maintenance of DNA trinucleotide repeats(GO:0035753) |
| 0.0 | 0.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0030428 | cell septum(GO:0030428) |
| 2.2 | 8.9 | GO:0044697 | HICS complex(GO:0044697) |
| 2.2 | 8.8 | GO:0030287 | cell wall-bounded periplasmic space(GO:0030287) |
| 1.3 | 7.7 | GO:0032126 | eisosome(GO:0032126) |
| 0.9 | 7.2 | GO:0000144 | cellular bud neck septin ring(GO:0000144) |
| 0.8 | 2.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.6 | 10.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.5 | 33.8 | GO:0005576 | extracellular region(GO:0005576) |
| 0.4 | 4.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.4 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 1.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 1.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 1.4 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.3 | 2.2 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.3 | 1.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.3 | 0.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 1.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 1.8 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 6.5 | GO:0009277 | fungal-type cell wall(GO:0009277) |
| 0.2 | 1.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 6.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.2 | 17.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 1.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 0.8 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.2 | 0.7 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.2 | 0.7 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 0.5 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.2 | 1.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.2 | 2.9 | GO:0070210 | Rpd3L-Expanded complex(GO:0070210) |
| 0.2 | 9.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.7 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.6 | GO:0005940 | septin ring(GO:0005940) |
| 0.1 | 2.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 8.7 | GO:0005815 | microtubule organizing center(GO:0005815) spindle pole body(GO:0005816) |
| 0.1 | 0.6 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.1 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 3.6 | GO:0000131 | incipient cellular bud site(GO:0000131) |
| 0.1 | 0.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 13.5 | GO:0000324 | storage vacuole(GO:0000322) lytic vacuole(GO:0000323) fungal-type vacuole(GO:0000324) |
| 0.1 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.3 | GO:0033597 | mitotic checkpoint complex(GO:0033597) |
| 0.1 | 0.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 0.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 0.2 | GO:0071261 | Ssh1 translocon complex(GO:0071261) |
| 0.1 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) mitochondrial chromosome(GO:0000262) |
| 0.0 | 0.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 15.2 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.2 | GO:0030907 | MBF transcription complex(GO:0030907) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 8.3 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 0.0 | GO:0034456 | UTP-C complex(GO:0034456) |
| 0.0 | 2.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0031933 | nuclear heterochromatin(GO:0005720) nuclear telomeric heterochromatin(GO:0005724) telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.0 | 0.1 | GO:0035327 | Cdc73/Paf1 complex(GO:0016593) transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.4 | GO:0044445 | cytosolic part(GO:0044445) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.6 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 1.7 | 10.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 1.6 | 6.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 1.5 | 4.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 1.4 | 8.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 1.4 | 4.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 1.3 | 5.3 | GO:0042973 | glucan endo-1,3-beta-D-glucosidase activity(GO:0042973) |
| 1.0 | 17.8 | GO:0015926 | glucosidase activity(GO:0015926) |
| 1.0 | 24.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.9 | 2.6 | GO:0005253 | anion channel activity(GO:0005253) voltage-gated anion channel activity(GO:0008308) |
| 0.8 | 2.5 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 0.8 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.8 | 3.1 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.7 | 3.6 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.7 | 13.0 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.7 | 4.9 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.7 | 2.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.6 | 7.6 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.5 | 5.3 | GO:0005496 | steroid binding(GO:0005496) sterol binding(GO:0032934) |
| 0.5 | 1.5 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.5 | 2.0 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.5 | 1.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 1.9 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.5 | 3.7 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.5 | 1.4 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.4 | 0.8 | GO:0015606 | spermidine transmembrane transporter activity(GO:0015606) |
| 0.4 | 1.3 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.4 | 2.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.4 | 1.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.4 | 1.5 | GO:0005034 | osmosensor activity(GO:0005034) |
| 0.4 | 1.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 1.3 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.3 | 1.6 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.3 | 1.5 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.3 | 1.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.3 | 1.4 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 1.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.3 | 1.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 1.6 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.3 | 1.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.7 | GO:0019238 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) cyclohydrolase activity(GO:0019238) |
| 0.2 | 0.7 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.2 | 1.5 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.2 | 0.6 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.2 | 0.6 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.2 | 0.8 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 1.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 0.5 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.2 | 0.5 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.2 | 0.7 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 0.7 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 1.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.2 | 1.6 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.2 | 0.5 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 0.6 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.7 | GO:0004026 | alcohol O-acetyltransferase activity(GO:0004026) alcohol O-acyltransferase activity(GO:0034318) |
| 0.1 | 4.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 1.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.7 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.5 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.1 | 0.7 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 4.2 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.1 | 3.2 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.1 | 10.6 | GO:0008092 | cytoskeletal protein binding(GO:0008092) |
| 0.1 | 0.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 24.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.8 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.5 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 0.7 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.1 | 0.4 | GO:0015085 | calcium ion transmembrane transporter activity(GO:0015085) potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.1 | 1.9 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 0.4 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.1 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 2.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.6 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.9 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.1 | 1.0 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.1 | 0.5 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 2.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.3 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 0.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.4 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.1 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 1.3 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.1 | 0.3 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.1 | 0.3 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.1 | 0.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.1 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 2.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.7 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.1 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 1.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) thiol oxidase activity(GO:0016972) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.7 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.8 | GO:0016410 | N-acyltransferase activity(GO:0016410) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.5 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0008144 | drug binding(GO:0008144) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.0 | GO:0035591 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.0 | 0.2 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 0.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.3 | 0.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.2 | 0.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 0.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 1.1 | REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | Genes involved in NFkB and MAP kinases activation mediated by TLR4 signaling repertoire |
| 0.3 | 2.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 1.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 3.2 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 0.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 0.3 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.1 | 2.6 | REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | Genes involved in Metabolism of lipids and lipoproteins |
| 0.1 | 0.4 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 0.5 | REACTOME APOPTOSIS | Genes involved in Apoptosis |
| 0.1 | 0.4 | REACTOME INTERFERON SIGNALING | Genes involved in Interferon Signaling |
| 0.1 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.1 | REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | Genes involved in Platelet activation, signaling and aggregation |