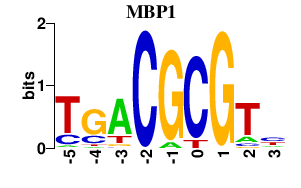
Results for MBP1
Z-value: 2.64
Transcription factors associated with MBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MBP1
|
S000002214 | Transcription factor involved in cell cycle progression from G1 to S |
Activity-expression correlation:
Activity profile of MBP1 motif
Sorted Z-values of MBP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YBL003C | 17.18 |
HTA2
|
One of two nearly identical (see also HTA1) histone H2A subtypes; core histone required for chromatin assembly and chromosome function; DNA damage-dependent phosphorylation by Mec1p facilitates DNA repair; acetylated by Nat4p |
|
| YFR055W | 16.40 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YFR056C | 15.30 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YBL085W | 13.87 |
BOI1
|
Protein implicated in polar growth, functionally redundant with Boi2p; interacts with bud-emergence protein Bem1p; contains an SH3 (src homology 3) domain and a PH (pleckstrin homology) domain |
|
| YNL178W | 13.76 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YER070W | 11.83 |
RNR1
|
One of two large regulatory subunits of ribonucleotide-diphosphate reductase; the RNR complex catalyzes rate-limiting step in dNTP synthesis, regulated by DNA replication and DNA damage checkpoint pathways via localization of small subunits |
|
| YOR315W | 11.82 |
SFG1
|
Nuclear protein, putative transcription factor required for growth of superficial pseudohyphae (which do not invade the agar substrate) but not for invasive pseudohyphal growth; may act together with Phd1p; potential Cdc28p substrate |
|
| YIL053W | 10.93 |
RHR2
|
Constitutively expressed isoform of DL-glycerol-3-phosphatase; involved in glycerol biosynthesis, induced in response to both anaerobic and, along with the Hor2p/Gpp2p isoform, osmotic stress |
|
| YMR199W | 10.80 |
CLN1
|
G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) |
|
| YOR073W-A | 9.58 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially/completely overlaps the verified ORF CDC21/YOR074C; identified by RT-PCR |
||
| YBL002W | 9.51 |
HTB2
|
One of two nearly identical (see HTB1) histone H2B subtypes required for chromatin assembly and chromosome function; Rad6p-Bre1p-Lge1p mediated ubiquitination regulates transcriptional activation, meiotic DSB formation and H3 methylation |
|
| YGR140W | 8.72 |
CBF2
|
Essential kinetochore protein, component of the CBF3 multisubunit complex that binds to the CDEIII region of the centromere; Cbf2p also binds to the CDEII region possibly forming a different multimeric complex, ubiquitinated in vivo |
|
| YDL023C | 8.57 |
Dubious open reading frame, unlikely to encode a protein; not conserved in other Saccharomyces species; overlaps the verified gene GPD1; deletion confers sensitivity to GSAO; deletion in cyr1 mutant results in loss of stress resistance |
||
| YER001W | 8.53 |
MNN1
|
Alpha-1,3-mannosyltransferase, integral membrane glycoprotein of the Golgi complex, required for addition of alpha1,3-mannose linkages to N-linked and O-linked oligosaccharides, one of five S. cerevisiae proteins of the MNN1 family |
|
| YLR154C | 8.17 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YOR074C | 7.77 |
CDC21
|
Thymidylate synthase, required for de novo biosynthesis of pyrimidine deoxyribonucleotides; expression is induced at G1/S |
|
| YJR094W-A | 7.68 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YNL030W | 7.59 |
HHF2
|
One of two identical histone H4 proteins (see also HHF1); core histone required for chromatin assembly and chromosome function; contributes to telomeric silencing; N-terminal domain involved in maintaining genomic integrity |
|
| YNL301C | 7.59 |
RPL18B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl18Ap and has similarity to rat L18 ribosomal protein |
|
| YMR290W-A | 7.58 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; overlaps 5' end of essential HAS1 gene which encodes an ATP-dependent RNA helicase |
||
| YGL147C | 7.58 |
RPL9A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl9Bp and has similarity to E. coli L6 and rat L9 ribosomal proteins |
|
| YDL022W | 7.50 |
GPD1
|
NAD-dependent glycerol-3-phosphate dehydrogenase, key enzyme of glycerol synthesis, essential for growth under osmotic stress; expression regulated by high-osmolarity glycerol response pathway; homolog of Gpd2p |
|
| YJR145C | 7.43 |
RPS4A
|
Protein component of the small (40S) ribosomal subunit; mutation affects 20S pre-rRNA processing; identical to Rps4Bp and has similarity to rat S4 ribosomal protein |
|
| YCL024W | 7.33 |
KCC4
|
Protein kinase of the bud neck involved in the septin checkpoint, associates with septin proteins, negatively regulates Swe1p by phosphorylation, shows structural homology to bud neck kinases Gin4p and Hsl1p |
|
| YMR290C | 7.16 |
HAS1
|
ATP-dependent RNA helicase; localizes to both the nuclear periphery and nucleolus; highly enriched in nuclear pore complex fractions; constituent of 66S pre-ribosomal particles |
|
| YDL003W | 7.14 |
MCD1
|
Essential protein required for sister chromatid cohesion in mitosis and meiosis; subunit of the cohesin complex; expression is cell cycle regulated and peaks in S phase |
|
| YDL047W | 6.87 |
SIT4
|
Type 2A-related serine-threonine phosphatase that functions in the G1/S transition of the mitotic cycle; cytoplasmic and nuclear protein that modulates functions mediated by Pkc1p including cell wall and actin cytoskeleton organization |
|
| YGR138C | 6.81 |
TPO2
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; transcription of TPO2 is regulated by Haa1p; member of the major facilitator superfamily |
|
| YLR349W | 6.59 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified ORF DIC1/YLR348C |
||
| YGR108W | 6.50 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YAR008W | 6.49 |
SEN34
|
Subunit of the tRNA splicing endonuclease, which is composed of Sen2p, Sen15p, Sen34p, and Sen54p; Sen34p contains the active site for tRNA 3' splice site cleavage and has similarity to Sen2p and to Archaeal tRNA splicing endonuclease |
|
| YLR154W-C | 6.42 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YKL063C | 6.25 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the Golgi |
||
| YJL194W | 6.24 |
CDC6
|
Essential ATP-binding protein required for DNA replication, component of the pre-replicative complex (pre-RC) which requires ORC to associate with chromatin and is in turn required for Mcm2-7p DNA association; homologous to S. pombe Cdc18p |
|
| YJL190C | 6.23 |
RPS22A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps22Bp and has similarity to E. coli S8 and rat S15a ribosomal proteins |
|
| YLR348C | 6.22 |
DIC1
|
Mitochondrial dicarboxylate carrier, integral membrane protein, catalyzes a dicarboxylate-phosphate exchange across the inner mitochondrial membrane, transports cytoplasmic dicarboxylates into the mitochondrial matrix |
|
| YLR154W-A | 6.17 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YNR009W | 6.10 |
NRM1
|
Transcriptional co-repressor of MBF (MCB binding factor)-regulated gene expression; Nrm1p associates stably with promoters via MBF to repress transcription upon exit from G1 phase |
|
| YDR507C | 6.00 |
GIN4
|
Protein kinase involved in bud growth and assembly of the septin ring, proposed to have kinase-dependent and kinase-independent activities; undergoes autophosphorylation; similar to Kcc4p and Hsl1p |
|
| YJL115W | 5.97 |
ASF1
|
Nucleosome assembly factor, involved in chromatin assembly and disassembly, anti-silencing protein that causes derepression of silent loci when overexpressed; plays a role in regulating Ty1 transposition |
|
| YLR300W | 5.96 |
EXG1
|
Major exo-1,3-beta-glucanase of the cell wall, involved in cell wall beta-glucan assembly; exists as three differentially glycosylated isoenzymes |
|
| YCL023C | 5.86 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified ORF KCC4 |
||
| YGR139W | 5.84 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR154W-B | 5.80 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YOR313C | 5.75 |
SPS4
|
Protein whose expression is induced during sporulation; not required for sporulation; heterologous expression in E. coli induces the SOS response that senses DNA damage |
|
| YGL201C | 5.69 |
MCM6
|
Protein involved in DNA replication; component of the Mcm2-7 hexameric complex that binds chromatin as a part of the pre-replicative complex |
|
| YBR088C | 5.60 |
POL30
|
Proliferating cell nuclear antigen (PCNA), functions as the sliding clamp for DNA polymerase delta; may function as a docking site for other proteins required for mitotic and meiotic chromosomal DNA replication and for DNA repair |
|
| YOL040C | 5.52 |
RPS15
|
Protein component of the small (40S) ribosomal subunit; has similarity to E. coli S19 and rat S15 ribosomal proteins |
|
| YPL177C | 5.45 |
CUP9
|
Homeodomain-containing transcriptional repressor of PTR2, which encodes a major peptide transporter; imported peptides activate ubiquitin-dependent proteolysis, resulting in degradation of Cup9p and de-repression of PTR2 transcription |
|
| YPL081W | 5.44 |
RPS9A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps9Bp and has similarity to E. coli S4 and rat S9 ribosomal proteins |
|
| YDL101C | 5.40 |
DUN1
|
Cell-cycle checkpoint serine-threonine kinase required for DNA damage-induced transcription of certain target genes, phosphorylation of Rad55p and Sml1p, and transient G2/M arrest after DNA damage; also regulates postreplicative DNA repair |
|
| YOR376W-A | 5.39 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YDL022C-A | 5.35 |
Dubious open reading frame unlikely to encode a protein; partially overlaps the verified gene DIA3; identified by fungal homology and RT-PCR |
||
| YOL039W | 5.34 |
RPP2A
|
Ribosomal protein P2 alpha, a component of the ribosomal stalk, which is involved in the interaction between translational elongation factors and the ribosome; regulates the accumulation of P1 (Rpp1Ap and Rpp1Bp) in the cytoplasm |
|
| YER003C | 5.32 |
PMI40
|
Mannose-6-phosphate isomerase, catalyzes the interconversion of fructose-6-P and mannose-6-P; required for early steps in protein mannosylation |
|
| YMR144W | 5.24 |
Putative protein of unknown function; localized to the nucleus; YMR144W is not an essential gene |
||
| YHR154W | 5.21 |
RTT107
|
Protein implicated in Mms22-dependent DNA repair during S phase, DNA damage induces phosphorylation by Mec1p at one or more SQ/TQ motifs; interacts with Mms22p and Slx4p; has four BRCT domains; has a role in regulation of Ty1 transposition |
|
| YGL179C | 5.17 |
TOS3
|
Protein kinase, related to and functionally redundant with Elm1p and Sak1p for the phosphorylation and activation of Snf1p; functionally orthologous to LKB1, a mammalian kinase associated with Peutz-Jeghers cancer-susceptibility syndrome |
|
| YGL076C | 4.97 |
RPL7A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl7Bp and has similarity to E. coli L30 and rat L7 ribosomal proteins; contains a conserved C-terminal Nucleic acid Binding Domain (NDB2) |
|
| YOR375C | 4.91 |
GDH1
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh3p; expression regulated by nitrogen and carbon sources |
|
| YLR049C | 4.90 |
Putative protein of unknown function |
||
| YDL055C | 4.83 |
PSA1
|
GDP-mannose pyrophosphorylase (mannose-1-phosphate guanyltransferase), synthesizes GDP-mannose from GTP and mannose-1-phosphate in cell wall biosynthesis; required for normal cell wall structure |
|
| YEL001C | 4.79 |
IRC22
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion localizes to the ER; YEL001C is non-essential; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YOR096W | 4.62 |
RPS7A
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps7Bp; interacts with Kti11p; deletion causes hypersensitivity to zymocin; has similarity to rat S7 and Xenopus S8 ribosomal proteins |
|
| YBL063W | 4.61 |
KIP1
|
Kinesin-related motor protein required for mitotic spindle assembly and chromosome segregation; functionally redundant with Cin8p |
|
| YOR314W | 4.61 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YFR054C | 4.60 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR103C | 4.48 |
CDC45
|
DNA replication initiation factor; recruited to MCM pre-RC complexes at replication origins; promotes release of MCM from Mcm10p, recruits elongation machinery; mutants in human homolog may cause velocardiofacial and DiGeorge syndromes |
|
| YLR441C | 4.45 |
RPS1A
|
Ribosomal protein 10 (rp10) of the small (40S) subunit; nearly identical to Rps1Bp and has similarity to rat S3a ribosomal protein |
|
| YOR063W | 4.44 |
RPL3
|
Protein component of the large (60S) ribosomal subunit, has similarity to E. coli L3 and rat L3 ribosomal proteins; involved in the replication and maintenance of killer double stranded RNA virus |
|
| YPL090C | 4.43 |
RPS6A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps6Bp and has similarity to rat S6 ribosomal protein |
|
| YBR081C | 4.39 |
SPT7
|
Subunit of the SAGA transcriptional regulatory complex, involved in proper assembly of the complex; also present as a C-terminally truncated form in the SLIK/SALSA transcriptional regulatory complex |
|
| YLR150W | 4.35 |
STM1
|
Protein that binds G4 quadruplex and purine motif triplex nucleic acid; acts with Cdc13p to maintain telomere structure; interacts with ribosomes and subtelomeric Y' DNA; multicopy suppressor of tom1 and pop2 mutations |
|
| YPL256C | 4.35 |
CLN2
|
G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) |
|
| YML088W | 4.30 |
UFO1
|
F-box receptor protein, subunit of the Skp1-Cdc53-F-box receptor (SCF) E3 ubiquitin ligase complex; binds to phosphorylated Ho endonuclease, allowing its ubiquitylation by SCF and subsequent degradation |
|
| YMR177W | 4.25 |
MMT1
|
Putative metal transporter involved in mitochondrial iron accumulation; closely related to Mmt2p |
|
| YOR254C | 4.25 |
SEC63
|
Essential subunit of Sec63 complex (Sec63p, Sec62p, Sec66p and Sec72p); with Sec61 complex, Kar2p/BiP and Lhs1p forms a channel competent for SRP-dependent and post-translational SRP-independent protein targeting and import into the ER |
|
| YCR065W | 4.17 |
HCM1
|
Forkhead transcription factor that drives S-phase specific expression of genes involved in chromosome segregation, spindle dynamics, and budding; suppressor of calmodulin mutants with specific SPB assembly defects; telomere maintenance role |
|
| YDL164C | 4.17 |
CDC9
|
DNA ligase found in the nucleus and mitochondria, an essential enzyme that joins Okazaki fragments during DNA replication; also acts in nucleotide excision repair, base excision repair, and recombination |
|
| YJR009C | 4.16 |
TDH2
|
Glyceraldehyde-3-phosphate dehydrogenase, isozyme 2, involved in glycolysis and gluconeogenesis; tetramer that catalyzes the reaction of glyceraldehyde-3-phosphate to 1,3 bis-phosphoglycerate; detected in the cytoplasm and cell-wall |
|
| YPL267W | 4.15 |
ACM1
|
Cell cycle regulated protein of unknown function; associated with Cdh1p and may supress the APC/C[Cdh1]-mediated proteolysis of mitotic cyclins |
|
| YHR010W | 4.11 |
RPL27A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl27Bp and has similarity to rat L27 ribosomal protein |
|
| YPR121W | 4.10 |
THI22
|
Protein with similarity to hydroxymethylpyrimidine phosphate kinases; member of a gene family with THI20 and THI21; not required for thiamine biosynthesis |
|
| YDL192W | 4.09 |
ARF1
|
ADP-ribosylation factor, GTPase of the Ras superfamily involved in regulation of coated vesicle formation in intracellular trafficking within the Golgi; functionally interchangeable with Arf2p |
|
| YFL034C-A | 4.06 |
RPL22B
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl22Ap and to rat L22 ribosomal protein |
|
| YPR050C | 4.05 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene MAK3/YPR051W |
||
| YJL173C | 4.03 |
RFA3
|
Subunit of heterotrimeric Replication Protein A (RPA), which is a highly conserved single-stranded DNA binding protein involved in DNA replication, repair, and recombination |
|
| YGL103W | 4.03 |
RPL28
|
Ribosomal protein of the large (60S) ribosomal subunit, has similarity to E. coli L15 and rat L27a ribosomal proteins; may have peptidyl transferase activity; can mutate to cycloheximide resistance |
|
| YPL153C | 4.03 |
RAD53
|
Protein kinase, required for cell-cycle arrest in response to DNA damage; activated by trans autophosphorylation when interacting with hyperphosphorylated Rad9p; also interacts with ARS1 and plays a role in initiation of DNA replication |
|
| YNL031C | 4.01 |
HHT2
|
One of two identical histone H3 proteins (see also HHT1); core histone required for chromatin assembly, involved in heterochromatin-mediated telomeric and HM silencing; regulated by acetylation, methylation, and mitotic phosphorylation |
|
| YLR032W | 4.01 |
RAD5
|
DNA helicase proposed to promote replication fork regression during postreplication repair by template switching; contains RING finger domain |
|
| YPL075W | 4.00 |
GCR1
|
Transcriptional activator of genes involved in glycolysis; DNA-binding protein that interacts and functions with the transcriptional activator Gcr2p |
|
| YJR043C | 3.95 |
POL32
|
Third subunit of DNA polymerase delta, involved in chromosomal DNA replication; required for error-prone DNA synthesis in the presence of DNA damage and processivity; interacts with Hys2p, PCNA (Pol30p), and Pol1p |
|
| YDL048C | 3.92 |
STP4
|
Protein containing a Kruppel-type zinc-finger domain; has similarity to Stp1p, Stp2p, and Stp3p |
|
| YGR152C | 3.90 |
RSR1
|
GTP-binding protein of the ras superfamily required for bud site selection, morphological changes in response to mating pheromone, and efficient cell fusion; localized to the plasma membrane; significantly similar to mammalian Rap GTPases |
|
| YDL211C | 3.87 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YMR006C | 3.86 |
PLB2
|
Phospholipase B (lysophospholipase) involved in phospholipid metabolism; displays transacylase activity in vitro; overproduction confers resistance to lysophosphatidylcholine |
|
| YIL009W | 3.85 |
FAA3
|
Long chain fatty acyl-CoA synthetase, has a preference for C16 and C18 fatty acids; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery |
|
| YKR038C | 3.83 |
KAE1
|
Putative glycoprotease proposed to be in transcription as a component of the EKC protein complex with Bud32p, Cgi121p, Pcc1p, and Gon7p; also identified as a component of the KEOPS protein complex |
|
| YJL136C | 3.78 |
RPS21B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps21Ap and has similarity to rat S21 ribosomal protein |
|
| YOL086C | 3.77 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YKR094C | 3.77 |
RPL40B
|
Fusion protein, identical to Rpl40Ap, that is cleaved to yield ubiquitin and a ribosomal protein of the large (60S) ribosomal subunit with similarity to rat L40; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes |
|
| YMR070W | 3.75 |
MOT3
|
Nuclear transcription factor with two Cys2-His2 zinc fingers; involved in repression of a subset of hypoxic genes by Rox1p, repression of several DAN/TIR genes during aerobic growth, and repression of ergosterol biosynthetic genes |
|
| YHR123W | 3.73 |
EPT1
|
sn-1,2-diacylglycerol ethanolamine- and cholinephosphotranferase; not essential for viability |
|
| YKL110C | 3.72 |
KTI12
|
Protein that plays a role, with Elongator complex, in modification of wobble nucleosides in tRNA; involved in sensitivity to G1 arrest induced by zymocin; interacts with chromatin throughout the genome; also interacts with Cdc19p |
|
| YKL096W-A | 3.72 |
CWP2
|
Covalently linked cell wall mannoprotein, major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored |
|
| YJR006W | 3.72 |
POL31
|
DNA polymerase III (delta) subunit, essential for cell viability; involved in DNA replication and DNA repair |
|
| YJR010W | 3.67 |
MET3
|
ATP sulfurylase, catalyzes the primary step of intracellular sulfate activation, essential for assimilatory reduction of sulfate to sulfide, involved in methionine metabolism |
|
| YGL035C | 3.60 |
MIG1
|
Transcription factor involved in glucose repression; sequence specific DNA binding protein containing two Cys2His2 zinc finger motifs; regulated by the SNF1 kinase and the GLC7 phosphatase |
|
| YNL231C | 3.59 |
PDR16
|
Phosphatidylinositol transfer protein (PITP) controlled by the multiple drug resistance regulator Pdr1p, localizes to lipid particles and microsomes, controls levels of various lipids, may regulate lipid synthesis, homologous to Pdr17p |
|
| YDL085C-A | 3.59 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YNL102W | 3.59 |
POL1
|
Catalytic subunit of the DNA polymerase I alpha-primase complex, required for the initiation of DNA replication during mitotic DNA synthesis and premeiotic DNA synthesis |
|
| YOR327C | 3.58 |
SNC2
|
Vesicle membrane receptor protein (v-SNARE) involved in the fusion between Golgi-derived secretory vesicles with the plasma membrane; member of the synaptobrevin/VAMP family of R-type v-SNARE proteins |
|
| YNL273W | 3.54 |
TOF1
|
Subunit of a replication-pausing checkpoint complex (Tof1p-Mrc1p-Csm3p) that acts at the stalled replication fork to promote sister chromatid cohesion after DNA damage, facilitating gap repair of damaged DNA; interacts with the MCM helicase |
|
| YDR509W | 3.53 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR113C | 3.45 |
PDS1
|
Securin that inhibits anaphase by binding separin Esp1p, also blocks cyclin destruction and mitotic exit, essential for cell cycle arrest in mitosis in the presence of DNA damage or aberrant mitotic spindles; also present in meiotic nuclei |
|
| YBR084W | 3.43 |
MIS1
|
Mitochondrial C1-tetrahydrofolate synthase, involved in interconversion between different oxidation states of tetrahydrofolate (THF); provides activities of formyl-THF synthetase, methenyl-THF cyclohydrolase, and methylene-THF dehydrogenase |
|
| YGR251W | 3.38 |
Putative protein of unknown function; deletion mutant has defects in pre-rRNA processing; green fluorescent protein (GFP)-fusion protein localizes to both the nucleus and the nucleolus; YGR251W is an essential gene |
||
| YCR018C | 3.37 |
SRD1
|
Protein involved in the processing of pre-rRNA to mature rRNA; contains a C2/C2 zinc finger motif; srd1 mutation suppresses defects caused by the rrp1-1 mutation |
|
| YMR142C | 3.34 |
RPL13B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl13Ap; not essential for viability; has similarity to rat L13 ribosomal protein |
|
| YIL159W | 3.32 |
BNR1
|
Formin, nucleates the formation of linear actin filaments, involved in cell processes such as budding and mitotic spindle orientation which require the formation of polarized actin cables, functionally redundant with BNI1 |
|
| YLR143W | 3.32 |
Putative protein of unknown function; green fluorescent protein (GFP)-tagged protein localizes to the cytoplasm; YLR143W is not an essential gene |
||
| YJL181W | 3.30 |
Putative protein of unknown function; expression is cell-cycle regulated as shown by microarray analysis |
||
| YDR508C | 3.29 |
GNP1
|
High-affinity glutamine permease, also transports Leu, Ser, Thr, Cys, Met and Asn; expression is fully dependent on Grr1p and modulated by the Ssy1p-Ptr3p-Ssy5p (SPS) sensor of extracellular amino acids |
|
| YFL008W | 3.26 |
SMC1
|
Subunit of the multiprotein cohesin complex, essential protein involved in chromosome segregation and in double-strand DNA break repair; SMC chromosomal ATPase family member, binds DNA with a preference for DNA with secondary structure |
|
| YGL242C | 3.24 |
Putative protein of unknown function; deletion mutant is viable |
||
| YMR076C | 3.22 |
PDS5
|
Protein required for establishment and maintenance of sister chromatid condensation and cohesion, colocalizes with cohesin on chromosomes in an interdependent manner, may function as a protein-protein interaction scaffold |
|
| YOL080C | 3.20 |
REX4
|
Putative RNA exonuclease possibly involved in pre-rRNA processing and ribosome assembly |
|
| YKL209C | 3.19 |
STE6
|
Plasma membrane ATP-binding cassette (ABC) transporter required for the export of a-factor, catalyzes ATP hydrolysis coupled to a-factor transport; contains 12 transmembrane domains and two ATP binding domains; expressed only in MATa cells |
|
| YMR011W | 3.17 |
HXT2
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YHR153C | 3.17 |
SPO16
|
Protein of unknown function, required for spore formation |
|
| YNL090W | 3.16 |
RHO2
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, involved in the establishment of cell polarity and in microtubule assembly |
|
| YAR007C | 3.16 |
RFA1
|
Subunit of heterotrimeric Replication Protein A (RPA), which is a highly conserved single-stranded DNA binding protein involved in DNA replication, repair, and recombination |
|
| YDR279W | 3.15 |
RNH202
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YPL250W-A | 3.13 |
Identified by fungal homology and RT-PCR |
||
| YHR203C | 3.10 |
RPS4B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps4Ap and has similarity to rat S4 ribosomal protein |
|
| YGR060W | 3.10 |
ERG25
|
C-4 methyl sterol oxidase, catalyzes the first of three steps required to remove two C-4 methyl groups from an intermediate in ergosterol biosynthesis; mutants accumulate the sterol intermediate 4,4-dimethylzymosterol |
|
| YJL122W | 3.09 |
ALB1
|
Shuttling pre-60S factor; involved in the biogenesis of ribosomal large subunit; interacts directly with Arx1p; responsible for Tif6p recycling defects in absence of Rei1p |
|
| YKL062W | 3.06 |
MSN4
|
Transcriptional activator related to Msn2p; activated in stress conditions, which results in translocation from the cytoplasm to the nucleus; binds DNA at stress response elements of responsive genes, inducing gene expression |
|
| YJR005C-A | 3.05 |
Putative protein of unknown function, originally identified as a syntenic homolog of an Ashbya gossypii gene |
||
| YHR149C | 3.03 |
SKG6
|
Integral membrane protein that localizes primarily to growing sites such as the bud tip or the cell periphery; potential Cdc28p substrate; Skg6p interacts with Zds1p and Zds2p |
|
| YPL019C | 3.03 |
VTC3
|
Vacuolar membrane protein involved in vacuolar polyphosphate accumulation; functions as a regulator of vacuolar H+-ATPase activity and vacuolar transporter chaperones; involved in non-autophagic vacuolar fusion |
|
| YBR252W | 3.02 |
DUT1
|
dUTPase, catalyzes hydrolysis of dUTP to dUMP and PPi, thereby preventing incorporation of uracil into DNA during replication; critical for the maintenance of genetic stability |
|
| YOL079W | 3.02 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YLR372W | 2.99 |
SUR4
|
Elongase, involved in fatty acid and sphingolipid biosynthesis; synthesizes very long chain 20-26-carbon fatty acids from C18-CoA primers; involved in regulation of sphingolipid biosynthesis |
|
| YNL166C | 2.98 |
BNI5
|
Protein involved in organization of septins at the mother-bud neck, may interact directly with the Cdc11p septin, localizes to bud neck in a septin-dependent manner |
|
| YJL196C | 2.97 |
ELO1
|
Elongase I, medium-chain acyl elongase, catalyzes carboxy-terminal elongation of unsaturated C12-C16 fatty acyl-CoAs to C16-C18 fatty acids |
|
| YJR094C | 2.96 |
IME1
|
Master regulator of meiosis that is active only during meiotic events, activates transcription of early meiotic genes through interaction with Ume6p, degraded by the 26S proteasome following phosphorylation by Ime2p |
|
| YMR143W | 2.96 |
RPS16A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps16Bp and has similarity to E. coli S9 and rat S16 ribosomal proteins |
|
| YER118C | 2.95 |
SHO1
|
Transmembrane osmosensor, participates in activation of both the Cdc42p- and MAP kinase-dependent filamentous growth pathway and the high-osmolarity glycerol response pathway |
|
| YDR502C | 2.94 |
SAM2
|
S-adenosylmethionine synthetase, catalyzes transfer of the adenosyl group of ATP to the sulfur atom of methionine; one of two differentially regulated isozymes (Sam1p and Sam2p) |
|
| YOL017W | 2.94 |
ESC8
|
Protein involved in telomeric and mating-type locus silencing, interacts with Sir2p and also interacts with the Gal11p, which is a component of the RNA pol II mediator complex |
|
| YDL163W | 2.92 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the verified gene CDC9/YDL164C encoding DNA ligase |
||
| YMR083W | 2.92 |
ADH3
|
Mitochondrial alcohol dehydrogenase isozyme III; involved in the shuttling of mitochondrial NADH to the cytosol under anaerobic conditions and ethanol production |
|
| YPR010C | 2.89 |
RPA135
|
RNA polymerase I subunit A135 |
|
| YOR115C | 2.85 |
TRS33
|
One of 10 subunits of the transport protein particle (TRAPP) complex of the cis-Golgi which mediates vesicle docking and fusion; involved in endoplasmic reticulum (ER) to Golgi membrane traffic |
|
| YOR092W | 2.80 |
ECM3
|
Non-essential protein of unknown function; involved in signal transduction and the genotoxic response; induced rapidly in response to treatment with 8-methoxypsoralen and UVA irradiation |
|
| YNL024C | 2.79 |
Putative protein of unknown function with seven beta-strand methyltransferase motif; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YNL024C is not an essential gene |
||
| YKL128C | 2.79 |
PMU1
|
Putative phosphomutase, contains a region homologous to the active site of phosphomutases; overexpression suppresses the histidine auxotrophy of an ade3 ade16 ade17 triple mutant and the temperature sensitivity of a tps2 mutant |
|
| YMR296C | 2.79 |
LCB1
|
Component of serine palmitoyltransferase, responsible along with Lcb2p for the first committed step in sphingolipid synthesis, which is the condensation of serine with palmitoyl-CoA to form 3-ketosphinganine |
|
| YBR106W | 2.77 |
PHO88
|
Probable membrane protein, involved in phosphate transport; pho88 pho86 double null mutant exhibits enhanced synthesis of repressible acid phosphatase at high inorganic phosphate concentrations |
|
| YJR022W | 2.76 |
LSM8
|
Lsm (Like Sm) protein; forms heteroheptameric complex (with Lsm2p, Lsm3p, Lsm4p, Lsm5p, Lsm6p, and Lsm7p) that is part of spliceosomal U6 snRNP and is also implicated in processing of pre-tRNA, pre-snoRNA, and pre-rRNA |
|
| YLR313C | 2.74 |
SPH1
|
Protein involved in shmoo formation and bipolar bud site selection; homologous to Spa2p, localizes to sites of polarized growth in a cell cycle dependent- and Spa2p-dependent manner, interacts with MAPKKs Mkk1p, Mkk2p, and Ste7p |
|
| YLL021W | 2.73 |
SPA2
|
Component of the polarisome, which functions in actin cytoskeletal organization during polarized growth; acts as a scaffold for Mkk1p and Mpk1p cell wall integrity signaling components; potential Cdc28p substrate |
|
| YIL127C | 2.73 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus |
||
| YIL169C | 2.71 |
Putative protein of unknown function; serine/threonine rich and highly similar to YOL155C, a putative glucan alpha-1,4-glucosidase; transcript is induced in both high and low pH environments; YIL169C is a non-essential gene |
||
| YNL233W | 2.71 |
BNI4
|
Targeting subunit for Glc7p protein phosphatase, localized to the bud neck, required for localization of chitin synthase III to the bud neck via interaction with the chitin synthase III regulatory subunit Skt5p |
|
| YMR141W-A | 2.67 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene RPL13B/YMR142C |
||
| YKL101W | 2.67 |
HSL1
|
Nim1p-related protein kinase that regulates the morphogenesis and septin checkpoints; associates with the assembled septin filament; required along with Hsl7p for bud neck recruitment, phosphorylation, and degradation of Swe1p |
|
| YNR021W | 2.65 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; YNR021W is not an essential gene |
||
| YML063W | 2.64 |
RPS1B
|
Ribosomal protein 10 (rp10) of the small (40S) subunit; nearly identical to Rps1Ap and has similarity to rat S3a ribosomal protein |
|
| YMR277W | 2.62 |
FCP1
|
Carboxy-terminal domain (CTD) phosphatase, essential for dephosphorylation of the repeated C-terminal domain of the RNA polymerase II large subunit (Rpo21p) |
|
| YDR510W | 2.62 |
SMT3
|
Ubiquitin-like protein of the SUMO family, conjugated to lysine residues of target proteins; regulates chromatid cohesion, chromosome segregation, APC-mediated proteolysis, DNA replication and septin ring dynamics |
|
| YGR249W | 2.61 |
MGA1
|
Protein similar to heat shock transcription factor; multicopy suppressor of pseudohyphal growth defects of ammonium permease mutants |
|
| YJR054W | 2.60 |
Vacuolar protein of unknown function; potential Cdc28p substrate |
||
| YHR045W | 2.60 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum |
||
| YPL062W | 2.60 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YPL062W is not an essential gene; homozygous diploid mutant shows a decrease in glycogen accumulation |
||
| YPL254W | 2.59 |
HFI1
|
Adaptor protein required for structural integrity of the SAGA complex, a histone acetyltransferase-coactivator complex that is involved in global regulation of gene expression through acetylation and transcription functions |
|
| YMR194C-A | 2.57 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YKL152C | 2.53 |
GPM1
|
Tetrameric phosphoglycerate mutase, mediates the conversion of 3-phosphoglycerate to 2-phosphoglycerate during glycolysis and the reverse reaction during gluconeogenesis |
|
| YPR018W | 2.53 |
RLF2
|
Largest subunit (p90) of the Chromatin Assembly Complex (CAF-1) with Cac2p and Msi1p that assembles newly synthesized histones onto recently replicated DNA; involved in the maintenance of transcriptionally silent chromatin |
|
| YOR130C | 2.52 |
ORT1
|
Ornithine transporter of the mitochondrial inner membrane, exports ornithine from mitochondria as part of arginine biosynthesis; human ortholog is associated with hyperammonaemia-hyperornithinaemia-homocitrullinuria (HHH) syndrome |
|
| YKR043C | 2.51 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YJL187C | 2.51 |
SWE1
|
Protein kinase that regulates the G2/M transition by inhibition of Cdc28p kinase activity; localizes to the nucleus and to the daughter side of the mother-bud neck; homolog of S. pombe Wee1p; potential Cdc28p substrate |
|
| YBR126W-A | 2.51 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR126W-B; identified by gene-trapping, microarray analysis, and genome-wide homology searches |
||
| YDR500C | 2.49 |
RPL37B
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl37Ap and to rat L37 ribosomal protein |
|
| YLR321C | 2.48 |
SFH1
|
Component of the RSC chromatin remodeling complex; essential gene required for cell cycle progression and maintenance of proper ploidy; phosphorylated in the G1 phase of the cell cycle; Snf5p paralog |
|
| YDL103C | 2.47 |
QRI1
|
UDP-N-acetylglucosamine pyrophosphorylase, catalyzes the formation of UDP-N-acetylglucosamine (UDP-GlcNAc), which is important in cell wall biosynthesis, protein N-glycosylation, and GPI anchor biosynthesis |
|
| YDR133C | 2.46 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR134C |
||
| YKR039W | 2.45 |
GAP1
|
General amino acid permease; localization to the plasma membrane is regulated by nitrogen source |
|
| YGL177W | 2.44 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YKL042W | 2.43 |
SPC42
|
Central plaque component of spindle pole body (SPB); involved in SPB duplication, may facilitate attachment of the SPB to the nuclear membrane |
|
| YFL015W-A | 2.42 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR025W | 2.42 |
RPS11A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps11Bp and has similarity to E. coli S17 and rat S11 ribosomal proteins |
|
| YBR078W | 2.41 |
ECM33
|
GPI-anchored protein of unknown function, has a possible role in apical bud growth; GPI-anchoring on the plasma membrane crucial to function; phosphorylated in mitochondria; similar to Sps2p and Pst1p |
|
| YPR175W | 2.41 |
DPB2
|
Second largest subunit of DNA polymerase II (DNA polymerase epsilon), required for normal yeast chromosomal replication; expression peaks at the G1/S phase boundary; potential Cdc28p substrate |
|
| YDL219W | 2.41 |
DTD1
|
D-Tyr-tRNA(Tyr) deacylase, functions in protein translation, may affect nonsense suppression via alteration of the protein synthesis machinery; ubiquitous among eukaryotes |
|
| YGR090W | 2.39 |
UTP22
|
Possible U3 snoRNP protein involved in maturation of pre-18S rRNA, based on computational analysis of large-scale protein-protein interaction data |
|
| YBR143C | 2.38 |
SUP45
|
Polypeptide release factor involved in translation termination; mutant form acts as a recessive omnipotent suppressor |
|
| YDR278C | 2.38 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
Network of associatons between targets according to the STRING database.
First level regulatory network of MBP1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 4.5 | 13.6 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 4.0 | 16.0 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 3.6 | 10.9 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 3.4 | 10.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 3.1 | 21.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 2.5 | 7.5 | GO:0035952 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 2.4 | 4.8 | GO:0030541 | plasmid partitioning(GO:0030541) |
| 2.1 | 10.6 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 2.0 | 40.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 2.0 | 9.8 | GO:0060277 | obsolete negative regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0060277) |
| 1.9 | 11.5 | GO:0048478 | replication fork protection(GO:0048478) |
| 1.9 | 7.5 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 1.9 | 7.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 1.7 | 7.0 | GO:0000296 | spermine transport(GO:0000296) |
| 1.7 | 5.1 | GO:1904667 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 1.7 | 15.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.7 | 3.3 | GO:0009219 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) pyrimidine deoxyribonucleotide metabolic process(GO:0009219) pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) 2'-deoxyribonucleotide biosynthetic process(GO:0009265) 2'-deoxyribonucleotide metabolic process(GO:0009394) deoxyribose phosphate biosynthetic process(GO:0046385) |
| 1.6 | 4.9 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 1.6 | 4.9 | GO:0019379 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 1.6 | 6.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 1.5 | 6.2 | GO:1902931 | negative regulation of alcohol biosynthetic process(GO:1902931) |
| 1.5 | 16.4 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) |
| 1.5 | 8.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 1.5 | 4.4 | GO:0061422 | positive regulation of transcription from RNA polymerase II promoter in response to alkaline pH(GO:0061422) |
| 1.5 | 5.9 | GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000461) |
| 1.5 | 4.4 | GO:0060548 | negative regulation of apoptotic process(GO:0043066) negative regulation of programmed cell death(GO:0043069) negative regulation of cell death(GO:0060548) |
| 1.4 | 14.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 1.4 | 15.6 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 1.4 | 22.5 | GO:0006407 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 1.3 | 4.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 1.2 | 2.5 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 1.2 | 2.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.1 | 4.5 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) barbed-end actin filament capping(GO:0051016) actin filament capping(GO:0051693) |
| 1.1 | 2.2 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 1.1 | 3.3 | GO:0031384 | regulation of initiation of mating projection growth(GO:0031384) |
| 1.1 | 4.3 | GO:0071406 | response to methylmercury(GO:0051597) cellular response to methylmercury(GO:0071406) |
| 1.1 | 2.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 1.0 | 11.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 1.0 | 4.0 | GO:0014070 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 1.0 | 9.0 | GO:0010695 | regulation of spindle pole body separation(GO:0010695) |
| 1.0 | 6.0 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.9 | 8.4 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.9 | 3.7 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.9 | 1.9 | GO:0009847 | spore germination(GO:0009847) |
| 0.9 | 126.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.9 | 5.5 | GO:0000084 | mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.9 | 2.7 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.9 | 1.8 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.9 | 4.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.8 | 3.2 | GO:0009262 | deoxyribonucleotide metabolic process(GO:0009262) deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.8 | 4.0 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.8 | 2.4 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.8 | 2.3 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.8 | 1.5 | GO:0031684 | heterotrimeric G-protein complex cycle(GO:0031684) |
| 0.7 | 0.7 | GO:0007232 | osmosensory signaling pathway via Sho1 osmosensor(GO:0007232) |
| 0.7 | 5.9 | GO:0001402 | signal transduction involved in filamentous growth(GO:0001402) |
| 0.7 | 3.7 | GO:0046656 | folic acid metabolic process(GO:0046655) folic acid biosynthetic process(GO:0046656) |
| 0.7 | 1.5 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.7 | 2.9 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.7 | 8.7 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.7 | 14.4 | GO:0007020 | microtubule nucleation(GO:0007020) microtubule polymerization(GO:0046785) |
| 0.7 | 4.9 | GO:0045014 | negative regulation of transcription from RNA polymerase II promoter by glucose(GO:0000433) negative regulation of transcription by glucose(GO:0045014) |
| 0.7 | 17.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.7 | 8.3 | GO:0010383 | cell wall polysaccharide metabolic process(GO:0010383) |
| 0.7 | 2.6 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.7 | 2.0 | GO:0045128 | negative regulation of reciprocal meiotic recombination(GO:0045128) |
| 0.7 | 2.6 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.7 | 2.6 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.6 | 2.5 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.6 | 3.6 | GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity(GO:0051090) |
| 0.6 | 4.2 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.6 | 1.2 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.6 | 1.2 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.6 | 7.0 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.6 | 1.7 | GO:0031382 | mating projection assembly(GO:0031382) |
| 0.5 | 2.2 | GO:0050667 | transsulfuration(GO:0019346) homocysteine metabolic process(GO:0050667) |
| 0.5 | 1.6 | GO:0010994 | free ubiquitin chain polymerization(GO:0010994) |
| 0.5 | 2.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.5 | 1.6 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.5 | 4.2 | GO:0045860 | positive regulation of protein kinase activity(GO:0045860) |
| 0.5 | 2.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.5 | 1.5 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 0.5 | 3.0 | GO:0010529 | negative regulation of transposition, RNA-mediated(GO:0010526) negative regulation of transposition(GO:0010529) |
| 0.5 | 11.0 | GO:0002098 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.5 | 2.0 | GO:0048313 | Golgi inheritance(GO:0048313) |
| 0.5 | 2.0 | GO:0015883 | FAD transport(GO:0015883) |
| 0.5 | 2.0 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.5 | 1.4 | GO:0033241 | regulation of cellular amine catabolic process(GO:0033241) |
| 0.5 | 5.7 | GO:0006415 | translational termination(GO:0006415) |
| 0.5 | 0.9 | GO:0031292 | gene conversion at mating-type locus, DNA double-strand break processing(GO:0031292) |
| 0.5 | 4.1 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.5 | 3.2 | GO:0045005 | DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.5 | 0.9 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.5 | 1.8 | GO:0072396 | cellular response to biotic stimulus(GO:0071216) response to cell cycle checkpoint signaling(GO:0072396) response to mitotic cell cycle checkpoint signaling(GO:0072414) response to spindle checkpoint signaling(GO:0072417) response to mitotic spindle checkpoint signaling(GO:0072476) response to mitotic cell cycle spindle assembly checkpoint signaling(GO:0072479) response to spindle assembly checkpoint signaling(GO:0072485) negative regulation of protein import into nucleus during spindle assembly checkpoint(GO:1901925) |
| 0.4 | 2.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.4 | 4.0 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.4 | 1.3 | GO:0043419 | urea catabolic process(GO:0043419) |
| 0.4 | 2.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.4 | 4.4 | GO:0015833 | peptide transport(GO:0015833) |
| 0.4 | 1.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.4 | 1.3 | GO:0001111 | promoter clearance during DNA-templated transcription(GO:0001109) promoter clearance from RNA polymerase II promoter(GO:0001111) |
| 0.4 | 1.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 1.2 | GO:0044819 | mitotic G1 DNA damage checkpoint(GO:0031571) mitotic G1/S transition checkpoint(GO:0044819) |
| 0.4 | 2.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.4 | 3.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.4 | 2.0 | GO:0072668 | obsolete tubulin complex biogenesis(GO:0072668) |
| 0.4 | 1.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.4 | 3.7 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.4 | 1.8 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.4 | 3.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.4 | 3.2 | GO:0055070 | cellular copper ion homeostasis(GO:0006878) copper ion homeostasis(GO:0055070) |
| 0.3 | 1.4 | GO:0043007 | maintenance of rDNA(GO:0043007) |
| 0.3 | 5.0 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.3 | 8.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.3 | 2.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.3 | 1.3 | GO:0017157 | regulation of exocytosis(GO:0017157) regulation of secretion(GO:0051046) regulation of formin-nucleated actin cable assembly(GO:0090337) positive regulation of formin-nucleated actin cable assembly(GO:0090338) regulation of secretion by cell(GO:1903530) |
| 0.3 | 3.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.3 | 0.6 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.3 | 1.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.3 | 1.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.3 | 2.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) snRNA modification(GO:0040031) |
| 0.3 | 1.8 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.3 | 1.5 | GO:0042435 | tryptophan biosynthetic process(GO:0000162) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.3 | 0.9 | GO:0007535 | donor selection(GO:0007535) |
| 0.3 | 0.9 | GO:0070623 | regulation of vitamin metabolic process(GO:0030656) positive regulation of vitamin metabolic process(GO:0046136) regulation of thiamine biosynthetic process(GO:0070623) positive regulation of thiamine biosynthetic process(GO:0090180) |
| 0.3 | 3.8 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.3 | 1.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.3 | 1.7 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.3 | 0.3 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.3 | 2.5 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.3 | 0.8 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.3 | 0.5 | GO:0071044 | histone mRNA metabolic process(GO:0008334) histone mRNA catabolic process(GO:0071044) |
| 0.3 | 0.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.3 | 1.0 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.3 | 2.9 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) regulation of vacuole organization(GO:0044088) |
| 0.3 | 1.0 | GO:0060188 | regulation of protein desumoylation(GO:0060188) |
| 0.3 | 1.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 8.9 | GO:0006694 | steroid biosynthetic process(GO:0006694) sterol biosynthetic process(GO:0016126) |
| 0.2 | 1.0 | GO:0007157 | agglutination involved in conjugation with cellular fusion(GO:0000752) agglutination involved in conjugation(GO:0000771) heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) cell-cell adhesion(GO:0098609) adhesion between unicellular organisms(GO:0098610) multi organism cell adhesion(GO:0098740) cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.2 | 1.4 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.2 | 5.4 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.2 | 1.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 1.7 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.2 | 3.0 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.2 | 0.6 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.2 | 1.3 | GO:0000755 | cytogamy(GO:0000755) |
| 0.2 | 0.6 | GO:0034310 | ethanol catabolic process(GO:0006068) acetate biosynthetic process(GO:0019413) primary alcohol catabolic process(GO:0034310) |
| 0.2 | 2.6 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.2 | 1.5 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.2 | 0.6 | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) |
| 0.2 | 1.7 | GO:0006797 | polyphosphate metabolic process(GO:0006797) |
| 0.2 | 2.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 4.0 | GO:0098876 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.2 | 0.8 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.2 | 8.1 | GO:0007124 | pseudohyphal growth(GO:0007124) |
| 0.2 | 0.9 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 0.5 | GO:0038032 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 0.9 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 0.5 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) maintenance of chromatin silencing at telomere(GO:0035392) |
| 0.2 | 0.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 0.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 3.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.2 | 1.5 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 1.5 | GO:0042991 | transcription factor import into nucleus(GO:0042991) |
| 0.2 | 0.5 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.2 | 2.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 0.7 | GO:0031135 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.2 | 4.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.2 | 0.6 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 0.9 | GO:0090158 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) endoplasmic reticulum membrane organization(GO:0090158) |
| 0.2 | 1.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 0.6 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.1 | 0.9 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.5 | GO:0000921 | septin ring assembly(GO:0000921) |
| 0.1 | 1.0 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 1.9 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 1.9 | GO:0018208 | protein peptidyl-prolyl isomerization(GO:0000413) peptidyl-proline modification(GO:0018208) |
| 0.1 | 0.3 | GO:2000197 | regulation of mRNA export from nucleus(GO:0010793) regulation of nucleobase-containing compound transport(GO:0032239) regulation of RNA export from nucleus(GO:0046831) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.1 | 6.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 0.5 | GO:0043489 | RNA stabilization(GO:0043489) mRNA stabilization(GO:0048255) |
| 0.1 | 0.3 | GO:1901983 | regulation of histone acetylation(GO:0035065) regulation of protein acetylation(GO:1901983) regulation of peptidyl-lysine acetylation(GO:2000756) |
| 0.1 | 17.3 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.1 | 0.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.5 | GO:0009423 | chorismate biosynthetic process(GO:0009423) |
| 0.1 | 2.9 | GO:0006885 | regulation of pH(GO:0006885) |
| 0.1 | 5.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 2.3 | GO:0009304 | tRNA transcription(GO:0009304) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 2.2 | GO:0000725 | recombinational repair(GO:0000725) |
| 0.1 | 0.4 | GO:0051258 | protein polymerization(GO:0051258) |
| 0.1 | 1.3 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.1 | 0.6 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.1 | 0.2 | GO:0031106 | septin ring organization(GO:0031106) |
| 0.1 | 0.6 | GO:0071267 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 0.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.6 | GO:0071027 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 0.4 | GO:0009306 | protein secretion(GO:0009306) |
| 0.1 | 0.3 | GO:0006784 | heme a biosynthetic process(GO:0006784) heme a metabolic process(GO:0046160) |
| 0.1 | 1.6 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.1 | 0.8 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 2.1 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 0.2 | GO:0070893 | DNA integration(GO:0015074) transposon integration(GO:0070893) |
| 0.1 | 0.8 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.6 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 2.7 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 1.1 | GO:0000750 | pheromone-dependent signal transduction involved in conjugation with cellular fusion(GO:0000750) signal transduction involved in conjugation with cellular fusion(GO:0032005) |
| 0.1 | 0.4 | GO:0033014 | porphyrin-containing compound metabolic process(GO:0006778) porphyrin-containing compound biosynthetic process(GO:0006779) heme biosynthetic process(GO:0006783) tetrapyrrole metabolic process(GO:0033013) tetrapyrrole biosynthetic process(GO:0033014) heme metabolic process(GO:0042168) |
| 0.1 | 0.3 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) regulation of cell cycle G2/M phase transition(GO:1902749) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 0.9 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.4 | GO:0019365 | nicotinate nucleotide biosynthetic process(GO:0019357) nicotinate nucleotide salvage(GO:0019358) pyridine nucleotide salvage(GO:0019365) nicotinate nucleotide metabolic process(GO:0046497) |
| 0.1 | 4.0 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.1 | 0.7 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 0.4 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.1 | 0.6 | GO:0051666 | actin cortical patch localization(GO:0051666) |
| 0.1 | 1.4 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 0.2 | GO:0045862 | positive regulation of proteolysis(GO:0045862) |
| 0.1 | 0.2 | GO:0000389 | mRNA branch site recognition(GO:0000348) mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 1.0 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.2 | GO:0000709 | meiotic joint molecule formation(GO:0000709) |
| 0.1 | 0.2 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.1 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.2 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) |
| 0.1 | 0.2 | GO:2001252 | positive regulation of chromosome organization(GO:2001252) |
| 0.1 | 1.5 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 0.3 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 0.8 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 1.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 0.3 | GO:0048017 | inositol lipid-mediated signaling(GO:0048017) |
| 0.1 | 1.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.9 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.8 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.0 | 0.2 | GO:0000376 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.0 | 1.3 | GO:0046474 | glycerophospholipid biosynthetic process(GO:0046474) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:0000742 | karyogamy involved in conjugation with cellular fusion(GO:0000742) |
| 0.0 | 0.1 | GO:0060237 | regulation of fungal-type cell wall organization(GO:0060237) |
| 0.0 | 0.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.3 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.3 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.2 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0019932 | second-messenger-mediated signaling(GO:0019932) |
| 0.0 | 0.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.0 | GO:0035690 | cellular response to drug(GO:0035690) regulation of response to drug(GO:2001023) positive regulation of response to drug(GO:2001025) regulation of cellular response to drug(GO:2001038) positive regulation of cellular response to drug(GO:2001040) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:2000241 | regulation of reproductive process(GO:2000241) |
| 0.0 | 0.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 3.4 | 13.6 | GO:0034990 | mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) |
| 3.2 | 38.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 2.8 | 8.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 2.5 | 7.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 2.5 | 7.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 2.0 | 8.1 | GO:0031518 | CBF3 complex(GO:0031518) |
| 1.6 | 6.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 1.5 | 6.2 | GO:0030907 | MBF transcription complex(GO:0030907) |
| 1.4 | 2.9 | GO:0008278 | nuclear cohesin complex(GO:0000798) cohesin complex(GO:0008278) |
| 1.4 | 8.4 | GO:0042555 | MCM complex(GO:0042555) |
| 1.3 | 82.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 1.3 | 5.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.3 | 20.0 | GO:0005940 | septin ring(GO:0005940) |
| 1.2 | 16.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 1.1 | 3.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.1 | 3.3 | GO:0008275 | gamma-tubulin small complex, spindle pole body(GO:0000928) gamma-tubulin complex(GO:0000930) gamma-tubulin small complex(GO:0008275) |
| 1.0 | 5.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.0 | 1.0 | GO:0005822 | inner plaque of spindle pole body(GO:0005822) |
| 1.0 | 5.2 | GO:0035361 | Cul8-RING ubiquitin ligase complex(GO:0035361) |
| 1.0 | 3.1 | GO:0044697 | HICS complex(GO:0044697) |
| 1.0 | 3.8 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 1.0 | 79.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.9 | 8.7 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.8 | 4.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.8 | 4.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.8 | 3.2 | GO:0033254 | vacuolar transporter chaperone complex(GO:0033254) |
| 0.8 | 2.4 | GO:0071261 | Ssh1 translocon complex(GO:0071261) |
| 0.8 | 3.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.7 | 3.0 | GO:0005823 | central plaque of spindle pole body(GO:0005823) |
| 0.7 | 2.0 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.7 | 2.6 | GO:0031207 | endoplasmic reticulum Sec complex(GO:0031205) Sec62/Sec63 complex(GO:0031207) |
| 0.6 | 3.9 | GO:0000133 | polarisome(GO:0000133) |
| 0.6 | 3.1 | GO:0031391 | Elg1 RFC-like complex(GO:0031391) |
| 0.6 | 3.6 | GO:0000796 | condensin complex(GO:0000796) nuclear condensin complex(GO:0000799) |
| 0.6 | 4.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.6 | 2.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.6 | 1.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.6 | 9.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.5 | 2.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.5 | 4.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.5 | 2.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.5 | 4.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.5 | 2.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.5 | 7.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.5 | 8.9 | GO:0016586 | RSC complex(GO:0016586) |
| 0.5 | 0.5 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.5 | 2.4 | GO:0032545 | CURI complex(GO:0032545) |
| 0.5 | 3.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.5 | 1.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.5 | 7.7 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.4 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.4 | 0.9 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.4 | 1.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.4 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 2.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.4 | 2.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.4 | 4.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.4 | 6.7 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.4 | 1.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.3 | 1.4 | GO:0097196 | Shu complex(GO:0097196) |
| 0.3 | 1.4 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.3 | 1.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 3.6 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.3 | 1.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.3 | 1.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.3 | 1.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.3 | 1.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.3 | 13.3 | GO:0000131 | incipient cellular bud site(GO:0000131) |
| 0.3 | 0.6 | GO:0005824 | outer plaque of spindle pole body(GO:0005824) |
| 0.3 | 10.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.3 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.3 | 1.7 | GO:0032126 | eisosome(GO:0032126) |
| 0.3 | 1.9 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.3 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 0.8 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 14.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.3 | 1.5 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.3 | 0.8 | GO:0070274 | RES complex(GO:0070274) |
| 0.3 | 1.0 | GO:0000444 | MIS12/MIND type complex(GO:0000444) nuclear MIS12/MIND complex(GO:0000818) |
| 0.2 | 2.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 23.4 | GO:0005935 | cellular bud neck(GO:0005935) |
| 0.2 | 2.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 0.7 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.2 | 0.7 | GO:0005880 | polar microtubule(GO:0005827) nuclear microtubule(GO:0005880) tubulin complex(GO:0045298) |
| 0.2 | 3.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 0.9 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.2 | 7.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 1.4 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.2 | 0.9 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 0.8 | GO:0034044 | exomer complex(GO:0034044) |
| 0.2 | 2.9 | GO:0070210 | Rpd3L-Expanded complex(GO:0070210) |
| 0.2 | 0.5 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.2 | 0.2 | GO:0033309 | SBF transcription complex(GO:0033309) |
| 0.2 | 12.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.2 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 0.6 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.1 | 0.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) nuclear telomeric heterochromatin(GO:0005724) telomeric heterochromatin(GO:0031933) |
| 0.1 | 6.6 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 0.9 | GO:0033180 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.6 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.1 | 0.6 | GO:0072380 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 0.1 | 0.6 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 1.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.9 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.3 | GO:0000417 | HIR complex(GO:0000417) |
| 0.1 | 0.5 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.1 | 0.6 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 0.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 2.9 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 6.7 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 4.4 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.1 | 1.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.2 | GO:0044462 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 0.1 | 1.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.8 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0033176 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.3 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0034099 | luminal surveillance complex(GO:0034099) |
| 0.0 | 0.3 | GO:0090544 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.0 | 30.8 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.2 | GO:0005934 | cellular bud tip(GO:0005934) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.6 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 2.7 | 10.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 2.5 | 7.6 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 2.5 | 15.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 2.5 | 7.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 2.4 | 14.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 2.2 | 8.9 | GO:0008310 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) single-stranded DNA exodeoxyribonuclease activity(GO:0008297) single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 1.9 | 5.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 1.7 | 7.0 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 1.7 | 8.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 1.7 | 8.5 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 1.6 | 4.8 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 1.6 | 6.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 1.4 | 4.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.4 | 5.4 | GO:0005034 | osmosensor activity(GO:0005034) |
| 1.3 | 5.3 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.3 | 4.0 | GO:0008902 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 1.3 | 9.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 1.3 | 3.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 1.3 | 37.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 1.2 | 4.9 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 1.2 | 4.7 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 1.2 | 3.5 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 1.1 | 4.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 1.1 | 3.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.0 | 4.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 1.0 | 5.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 1.0 | 2.0 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 1.0 | 15.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 1.0 | 2.9 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 1.0 | 2.9 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.9 | 23.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.9 | 3.5 | GO:0004338 | glucan exo-1,3-beta-glucosidase activity(GO:0004338) |
| 0.8 | 37.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.8 | 2.5 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.8 | 3.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.8 | 6.1 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.7 | 3.7 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.7 | 2.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.7 | 2.1 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 0.7 | 2.0 | GO:0004000 | adenosine deaminase activity(GO:0004000) tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.6 | 6.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.6 | 4.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.6 | 1.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.6 | 4.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.6 | 102.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.6 | 2.3 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.5 | 1.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.5 | 4.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.5 | 1.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.5 | 2.7 | GO:0043141 | 5'-3' DNA helicase activity(GO:0043139) ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.5 | 2.6 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.5 | 9.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.5 | 2.0 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.5 | 2.9 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.5 | 12.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 2.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.5 | 1.4 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.5 | 7.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.5 | 2.3 | GO:0048256 | 5'-flap endonuclease activity(GO:0017108) flap endonuclease activity(GO:0048256) |
| 0.5 | 1.4 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) |
| 0.4 | 4.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.4 | 1.7 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 0.4 | 15.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.4 | 1.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 4.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.4 | 13.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.4 | 2.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 4.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.3 | 1.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 4.8 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.3 | 2.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.3 | 1.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 1.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.3 | 19.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.3 | 9.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.3 | 1.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 1.8 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.3 | 0.9 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.3 | 0.3 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.3 | 0.3 | GO:1904680 | peptide transmembrane transporter activity(GO:1904680) |
| 0.3 | 0.9 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.3 | 1.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.3 | 0.3 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.3 | 0.5 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.3 | 7.6 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.3 | 1.6 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.3 | 1.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 5.0 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.2 | 2.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 0.7 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.2 | 1.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 4.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 1.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 1.1 | GO:0001139 | transcription factor activity, core RNA polymerase II recruiting(GO:0001139) |
| 0.2 | 20.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.2 | 3.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 2.4 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 23.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.2 | 0.6 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.2 | 8.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.2 | 6.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.2 | 11.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.2 | 0.6 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.2 | 0.6 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.2 | 1.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.2 | 2.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 0.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 1.3 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
| 0.2 | 0.5 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 4.4 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.2 | 1.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.2 | 0.6 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.2 | 0.8 | GO:0032934 | steroid binding(GO:0005496) sterol binding(GO:0032934) |
| 0.2 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 1.2 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 1.9 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 0.3 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 2.0 | GO:0019001 | GTP binding(GO:0005525) guanyl nucleotide binding(GO:0019001) guanyl ribonucleotide binding(GO:0032561) |
| 0.1 | 1.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.3 | GO:0019207 | kinase regulator activity(GO:0019207) |
| 0.1 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.1 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.7 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 1.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 9.0 | GO:0008289 | lipid binding(GO:0008289) |
| 0.1 | 6.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.6 | GO:0004030 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 4.1 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.1 | 0.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.6 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.1 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.1 | 0.5 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 1.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.3 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 6.5 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.1 | 4.7 | GO:0016791 | phosphatase activity(GO:0016791) |
| 0.1 | 1.1 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.1 | 0.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 2.8 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 0.1 | GO:0015645 | long-chain fatty acid-CoA ligase activity(GO:0004467) fatty acid ligase activity(GO:0015645) |
| 0.1 | 1.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 0.5 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 0.5 | GO:0043566 | structure-specific DNA binding(GO:0043566) |
| 0.1 | 1.4 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.1 | 0.3 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.7 | GO:0030234 | enzyme regulator activity(GO:0030234) |
| 0.0 | 0.1 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.2 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.1 | PID E2F PATHWAY | E2F transcription factor network |
| 1.4 | 4.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 1.0 | 3.0 | PID ATM PATHWAY | ATM pathway |
| 0.6 | 1.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.6 | 1.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.5 | 4.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.4 | 0.9 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.4 | 0.4 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.4 | 0.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.3 | 1.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.3 | 1.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.3 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 2.8 | 11.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 2.7 | 21.8 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 1.5 | 23.8 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 1.4 | 8.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.4 | 4.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.6 | 1.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.6 | 3.7 | REACTOME LAGGING STRAND SYNTHESIS | Genes involved in Lagging Strand Synthesis |
| 0.6 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.5 | 0.5 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.5 | 0.5 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.4 | 4.4 | REACTOME G2 M CHECKPOINTS | Genes involved in G2/M Checkpoints |
| 0.4 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.4 | 1.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.3 | 1.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 3.6 | REACTOME MITOTIC M M G1 PHASES | Genes involved in Mitotic M-M/G1 phases |
| 0.2 | 1.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 0.2 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.2 | 0.4 | REACTOME CHROMOSOME MAINTENANCE | Genes involved in Chromosome Maintenance |
| 0.2 | 1.7 | REACTOME METABOLISM OF MRNA | Genes involved in Metabolism of mRNA |
| 0.2 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 2.0 | REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | Genes involved in Class I MHC mediated antigen processing & presentation |
| 0.1 | 2.5 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 0.4 | REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | Genes involved in Platelet activation, signaling and aggregation |
| 0.1 | 0.5 | REACTOME SIGNALING BY GPCR | Genes involved in Signaling by GPCR |
| 0.0 | 0.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |