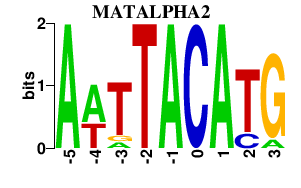
Results for MATALPHA2
Z-value: 0.60
Transcription factors associated with MATALPHA2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MATALPHA2
|
S000000635 | Homeobox-domain protein that represses a-specific genes in haploids |
Activity-expression correlation:
Activity profile of MATALPHA2 motif
Sorted Z-values of MATALPHA2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YFL026W | 2.77 |
STE2
|
Receptor for alpha-factor pheromone; seven transmembrane-domain GPCR that interacts with both pheromone and a heterotrimeric G protein to initiate the signaling response that leads to mating between haploid a and alpha cells |
|
| YKL209C | 2.73 |
STE6
|
Plasma membrane ATP-binding cassette (ABC) transporter required for the export of a-factor, catalyzes ATP hydrolysis coupled to a-factor transport; contains 12 transmembrane domains and two ATP binding domains; expressed only in MATa cells |
|
| YBR092C | 2.40 |
PHO3
|
Constitutively expressed acid phosphatase similar to Pho5p; brought to the cell surface by transport vesicles; hydrolyzes thiamin phosphates in the periplasmic space, increasing cellular thiamin uptake; expression is repressed by thiamin |
|
| YIL015W | 2.36 |
BAR1
|
Aspartyl protease secreted into the periplasmic space of mating type a cells, helps cells find mating partners, cleaves and inactivates alpha factor allowing cells to recover from alpha-factor-induced cell cycle arrest |
|
| YGL135W | 2.26 |
RPL1B
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, nearly identical to Rpl1Ap and has similarity to E. coli L1 and rat L10a ribosomal proteins; rpl1a rpl1b double null mutation is lethal |
|
| YML056C | 2.24 |
IMD4
|
Inosine monophosphate dehydrogenase, catalyzes the first step of GMP biosynthesis, member of a four-gene family in S. cerevisiae, constitutively expressed |
|
| YDR033W | 1.96 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YBR158W | 1.83 |
AMN1
|
Protein required for daughter cell separation, multiple mitotic checkpoints, and chromosome stability; contains 12 degenerate leucine-rich repeat motifs; expression is induced by the Mitotic Exit Network (MEN) |
|
| YGR148C | 1.71 |
RPL24B
|
Ribosomal protein L30 of the large (60S) ribosomal subunit, nearly identical to Rpl24Ap and has similarity to rat L24 ribosomal protein; not essential for translation but may be required for normal translation rate |
|
| YKL165C | 1.66 |
MCD4
|
Protein involved in glycosylphosphatidylinositol (GPI) anchor synthesis; multimembrane-spanning protein that localizes to the endoplasmic reticulum; highly conserved among eukaryotes |
|
| YBR244W | 1.65 |
GPX2
|
Phospholipid hydroperoxide glutathione peroxidase induced by glucose starvation that protects cells from phospholipid hydroperoxides and nonphospholipid peroxides during oxidative stress |
|
| YPL131W | 1.64 |
RPL5
|
Protein component of the large (60S) ribosomal subunit with similarity to E. coli L18 and rat L5 ribosomal proteins; binds 5S rRNA and is required for 60S subunit assembly |
|
| YDR098C | 1.62 |
GRX3
|
Hydroperoxide and superoxide-radical responsive glutathione-dependent oxidoreductase; monothiol glutaredoxin subfamily member along with Grx4p and Grx5p; protects cells from oxidative damage |
|
| YPR157W | 1.48 |
Putative protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YCR018C | 1.46 |
SRD1
|
Protein involved in the processing of pre-rRNA to mature rRNA; contains a C2/C2 zinc finger motif; srd1 mutation suppresses defects caused by the rrp1-1 mutation |
|
| YER026C | 1.45 |
CHO1
|
Phosphatidylserine synthase, functions in phospholipid biosynthesis; catalyzes the reaction CDP-diaclyglycerol + L-serine = CMP + L-1-phosphatidylserine, transcriptionally repressed by myo-inositol and choline |
|
| YGR108W | 1.40 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YFL027C | 1.37 |
GYP8
|
GTPase-activating protein for yeast Rab family members; Ypt1p is the preferred in vitro substrate but also acts on Sec4p, Ypt31p and Ypt32p; involved in the regulation of ER to Golgi vesicle transport |
|
| YHR063C | 1.37 |
PAN5
|
2-dehydropantoate 2-reductase, part of the pantothenic acid pathway, structurally homologous to E. coli panE |
|
| YEL053W-A | 1.35 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YEL054C |
||
| YEL020C-B | 1.30 |
Dubious open reading frame unlikely to encode a protein; partially overlaps verified gene YEL020W-A; identified by fungal homology and RT-PCR |
||
| YHR216W | 1.28 |
IMD2
|
Inosine monophosphate dehydrogenase, catalyzes the first step of GMP biosynthesis, expression is induced by mycophenolic acid resulting in resistance to the drug, expression is repressed by nutrient limitation |
|
| YLR432W | 1.28 |
IMD3
|
Inosine monophosphate dehydrogenase, catalyzes the first step of GMP biosynthesis, member of a four-gene family in S. cerevisiae, constitutively expressed |
|
| YPR119W | 1.26 |
CLB2
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YNR001W-A | 1.24 |
Dubious open reading frame unlikely to encode a functional protein; identified by homology |
||
| YMR003W | 1.24 |
AIM34
|
Protein of unknown function; GFP-fusion protein localizes to the mitochondria; null mutant is viable and displays decreased frequency of mitochondrial genome loss (petite formation) and severe growth defect in minimal glycerol media |
|
| YHR063W-A | 1.23 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER055C | 1.20 |
HIS1
|
ATP phosphoribosyltransferase, a hexameric enzyme, catalyzes the first step in histidine biosynthesis; mutations cause histidine auxotrophy and sensitivity to Cu, Co, and Ni salts; transcription is regulated by general amino acid control |
|
| YHR094C | 1.19 |
HXT1
|
Low-affinity glucose transporter of the major facilitator superfamily, expression is induced by Hxk2p in the presence of glucose and repressed by Rgt1p when glucose is limiting |
|
| YGL032C | 1.14 |
AGA2
|
Adhesion subunit of a-agglutinin of a-cells, C-terminal sequence acts as a ligand for alpha-agglutinin (Sag1p) during agglutination, modified with O-linked oligomannosyl chains, linked to anchorage subunit Aga1p via two disulfide bonds |
|
| YLR406C | 1.13 |
RPL31B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl31Ap and has similarity to rat L31 ribosomal protein; associates with the karyopherin Sxm1p |
|
| YOL101C | 1.12 |
IZH4
|
Membrane protein involved in zinc metabolism, member of the four-protein IZH family, expression induced by fatty acids and altered zinc levels; deletion reduces sensitivity to excess zinc; possible role in sterol metabolism |
|
| YNL111C | 1.12 |
CYB5
|
Cytochrome b5, involved in the sterol and lipid biosynthesis pathways; acts as an electron donor to support sterol C5-6 desaturation |
|
| YAR071W | 1.10 |
PHO11
|
One of three repressible acid phosphatases, a glycoprotein that is transported to the cell surface by the secretory pathway; induced by phosphate starvation and coordinately regulated by PHO4 and PHO2 |
|
| YIL133C | 1.10 |
RPL16A
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, binds to 5.8 S rRNA; has similarity to Rpl16Bp, E. coli L13 and rat L13a ribosomal proteins; transcriptionally regulated by Rap1p |
|
| YPL177C | 1.07 |
CUP9
|
Homeodomain-containing transcriptional repressor of PTR2, which encodes a major peptide transporter; imported peptides activate ubiquitin-dependent proteolysis, resulting in degradation of Cup9p and de-repression of PTR2 transcription |
|
| YPR048W | 1.06 |
TAH18
|
Protein with a potential role in DNA replication; displays synthetic lethal genetic interaction with the pol3-13 allele of POL3, which encodes DNA polymerase delta |
|
| YMR102C | 1.06 |
Protein of unknown function; transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in multidrug resistance; mutant shows increased resistance to azoles; YMR102C is not an essential gene |
||
| YMR049C | 1.06 |
ERB1
|
Protein required for maturation of the 25S and 5.8S ribosomal RNAs; constituent of 66S pre-ribosomal particles; homologous to mammalian Bop1 |
|
| YOR248W | 1.03 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YIL169C | 1.02 |
Putative protein of unknown function; serine/threonine rich and highly similar to YOL155C, a putative glucan alpha-1,4-glucosidase; transcript is induced in both high and low pH environments; YIL169C is a non-essential gene |
||
| YLR367W | 1.02 |
RPS22B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps22Ap and has similarity to E. coli S8 and rat S15a ribosomal proteins |
|
| YGR052W | 1.01 |
FMP48
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YBR177C | 1.01 |
EHT1
|
Acyl-coenzymeA:ethanol O-acyltransferase that plays a minor role in medium-chain fatty acid ethyl ester biosynthesis; possesses short-chain esterase activity; localizes to lipid particles and the mitochondrial outer membrane |
|
| YOR369C | 1.01 |
RPS12
|
Protein component of the small (40S) ribosomal subunit; has similarity to rat ribosomal protein S12 |
|
| YCR102W-A | 0.99 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOL049W | 0.97 |
GSH2
|
Glutathione synthetase, catalyzes the ATP-dependent synthesis of glutathione (GSH) from gamma-glutamylcysteine and glycine; induced by oxidative stress and heat shock |
|
| YKL008C | 0.95 |
LAC1
|
Ceramide synthase component, involved in synthesis of ceramide from C26(acyl)-coenzyme A and dihydrosphingosine or phytosphingosine, functionally equivalent to Lag1p |
|
| YOR176W | 0.95 |
HEM15
|
Ferrochelatase, a mitochondrial inner membrane protein, catalyzes the insertion of ferrous iron into protoporphyrin IX, the eighth and final step in the heme biosynthetic pathway |
|
| YHR123W | 0.94 |
EPT1
|
sn-1,2-diacylglycerol ethanolamine- and cholinephosphotranferase; not essential for viability |
|
| YDR297W | 0.94 |
SUR2
|
Sphinganine C4-hydroxylase, catalyses the conversion of sphinganine to phytosphingosine in sphingolipid biosyntheis |
|
| YGR036C | 0.93 |
CAX4
|
Dolichyl pyrophosphate (Dol-P-P) phosphatase with a luminally oriented active site in the ER, cleaves the anhydride linkage in Dol-P-P, required for Dol-P-P-linked oligosaccharide intermediate synthesis and protein N-glycosylation |
|
| YHR143W | 0.92 |
DSE2
|
Daughter cell-specific secreted protein with similarity to glucanases, degrades cell wall from the daughter side causing daughter to separate from mother; expression is repressed by cAMP |
|
| YBR067C | 0.92 |
TIP1
|
Major cell wall mannoprotein with possible lipase activity; transcription is induced by heat- and cold-shock; member of the Srp1p/Tip1p family of serine-alanine-rich proteins |
|
| YGL158W | 0.91 |
RCK1
|
Protein kinase involved in the response to oxidative stress; identified as suppressor of S. pombe cell cycle checkpoint mutations |
|
| YBR252W | 0.91 |
DUT1
|
dUTPase, catalyzes hydrolysis of dUTP to dUMP and PPi, thereby preventing incorporation of uracil into DNA during replication; critical for the maintenance of genetic stability |
|
| YCR034W | 0.91 |
FEN1
|
Fatty acid elongase, involved in sphingolipid biosynthesis; acts on fatty acids of up to 24 carbons in length; mutations have regulatory effects on 1,3-beta-glucan synthase, vacuolar ATPase, and the secretory pathway |
|
| YEL027W | 0.90 |
CUP5
|
Proteolipid subunit of the vacuolar H(+)-ATPase V0 sector (subunit c; dicyclohexylcarbodiimide binding subunit); required for vacuolar acidification and important for copper and iron metal ion homeostasis |
|
| YIL158W | 0.90 |
AIM20
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; null mutant displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YER049W | 0.89 |
TPA1
|
Protein of unknown function; interacts with Sup45p (eRF1), Sup35p (eRF3) and Pab1p; has a role in translation termination efficiency, mRNA poly(A) tail length and mRNA stability |
|
| YOL086C | 0.89 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YLR167W | 0.88 |
RPS31
|
Fusion protein that is cleaved to yield a ribosomal protein of the small (40S) subunit and ubiquitin; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes; interacts genetically with translation factor eIF2B |
|
| YGL067W | 0.87 |
NPY1
|
NADH diphosphatase (pyrophosphatase), hydrolyzes the pyrophosphate linkage in NADH and related nucleotides; localizes to peroxisomes |
|
| YAL059W | 0.85 |
ECM1
|
Protein of unknown function, localized in the nucleoplasm and the nucleolus, genetically interacts with MTR2 in 60S ribosomal protein subunit export |
|
| YJL011C | 0.85 |
RPC17
|
RNA polymerase III subunit C17; physically interacts with C31, C11, and TFIIIB70; may be involved in the recruitment of pol III by the preinitiation complex |
|
| YGL029W | 0.85 |
CGR1
|
Protein involved in nucleolar integrity and processing of the pre-rRNA for the 60S ribosome subunit; transcript is induced in response to cytotoxic stress but not genotoxic stress |
|
| YGR192C | 0.84 |
TDH3
|
Glyceraldehyde-3-phosphate dehydrogenase, isozyme 3, involved in glycolysis and gluconeogenesis; tetramer that catalyzes the reaction of glyceraldehyde-3-phosphate to 1,3 bis-phosphoglycerate; detected in the cytoplasm and cell-wall |
|
| YFR032C | 0.84 |
Putative protein of unknown function; transcribed during sporulation; YFR032C is not an essential gene |
||
| YGL201C | 0.81 |
MCM6
|
Protein involved in DNA replication; component of the Mcm2-7 hexameric complex that binds chromatin as a part of the pre-replicative complex |
|
| YOR175C | 0.81 |
ALE1
|
Lysophospholipid acyltransferase, partially redundant with Slc1p; part of MBOAT family of membrane-bound O-acyltransferases; key component of Lands cycle; may have role in fatty acid exchange at sn-2 position of mature glycerophospholipids |
|
| YNL148C | 0.81 |
ALF1
|
Alpha-tubulin folding protein, similar to mammalian cofactor B; Alf1p-GFP localizes to cytoplasmic microtubules; required for the folding of alpha-tubulin and may play an additional role in microtubule maintenance |
|
| YGL209W | 0.81 |
MIG2
|
Protein containing zinc fingers, involved in repression, along with Mig1p, of SUC2 (invertase) expression by high levels of glucose; binds to Mig1p-binding sites in SUC2 promoter |
|
| YJR009C | 0.80 |
TDH2
|
Glyceraldehyde-3-phosphate dehydrogenase, isozyme 2, involved in glycolysis and gluconeogenesis; tetramer that catalyzes the reaction of glyceraldehyde-3-phosphate to 1,3 bis-phosphoglycerate; detected in the cytoplasm and cell-wall |
|
| YGR050C | 0.80 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YIR020C-B | 0.80 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified ORF MRS1 |
||
| YJR070C | 0.79 |
LIA1
|
Deoxyhypusine hydroxylase, a HEAT-repeat containing metalloenzyme that catalyses hypusine formation; binds to and is required for the modification of Hyp2p (eIF5A); complements S. pombe mmd1 mutants defective in mitochondrial positioning |
|
| YHL003C | 0.79 |
LAG1
|
Ceramide synthase component, involved in synthesis of ceramide from C26(acyl)-coenzyme A and dihydrosphingosine or phytosphingosine, functionally equivalent to Lac1p |
|
| YPR041W | 0.79 |
TIF5
|
Translation initiation factor eIF-5; N-terminal domain functions as a GTPase-activating protein to mediate hydrolysis of ribosome-bound GTP; C-terminal domain is the core of ribosomal preinitiation complex formation |
|
| YPL227C | 0.78 |
ALG5
|
UDP-glucose:dolichyl-phosphate glucosyltransferase, involved in asparagine-linked glycosylation in the endoplasmic reticulum |
|
| YJR071W | 0.76 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR249W | 0.76 |
YEF3
|
Translational elongation factor 3, stimulates the binding of aminoacyl-tRNA (AA-tRNA) to ribosomes by releasing EF-1 alpha from the ribosomal complex; contains two ABC cassettes; binds and hydrolyses ATP |
|
| YBR031W | 0.75 |
RPL4A
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, nearly identical to Rpl4Bp and has similarity to E. coli L4 and rat L4 ribosomal proteins |
|
| YER137C | 0.74 |
Putative protein of unknown function |
||
| YMR143W | 0.74 |
RPS16A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps16Bp and has similarity to E. coli S9 and rat S16 ribosomal proteins |
|
| YPR010C | 0.74 |
RPA135
|
RNA polymerase I subunit A135 |
|
| YGR040W | 0.72 |
KSS1
|
Mitogen-activated protein kinase (MAPK) involved in signal transduction pathways that control filamentous growth and pheromone response; the KSS1 gene is nonfunctional in S288C strains and functional in W303 strains |
|
| YGL097W | 0.72 |
SRM1
|
Nucleotide exchange factor for Gsp1p, localizes to the nucleus, required for nucleocytoplasmic trafficking of macromolecules; suppressor of the pheromone response pathway; potentially phosphorylated by Cdc28p |
|
| YDR215C | 0.72 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; null mutant displays elevated sensitivity to expression of a mutant huntingtin fragment or of alpha-synuclein |
||
| YHR141C | 0.71 |
RPL42B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl42Ap and has similarity to rat L44; required for propagation of the killer toxin-encoding M1 double-stranded RNA satellite of the L-A double-stranded RNA virus |
|
| YJL097W | 0.71 |
PHS1
|
Essential 3-hydroxyacyl-CoA dehydratase of the ER membrane, involved in elongation of very long-chain fatty acids; evolutionarily conserved, similar to mammalian PTPLA and PTPLB; involved in sphingolipid biosynthesis and protein trafficking |
|
| YJR094W-A | 0.71 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YDR101C | 0.71 |
ARX1
|
Shuttling pre-60S factor; involved in the biogenesis of ribosomal large subunit biogenesis; interacts directly with Alb1; responsible for Tif6 recycling defects in absence of Rei1; associated with the ribosomal export complex |
|
| YKL216W | 0.71 |
URA1
|
Dihydroorotate dehydrogenase, catalyzes the fourth enzymatic step in the de novo biosynthesis of pyrimidines, converting dihydroorotic acid into orotic acid |
|
| YEL029C | 0.71 |
BUD16
|
Putative pyridoxal kinase, key enzyme in vitamin B6 metabolism; involved in maintaining levels of pyridoxal 5'-phosphate, the active form of vitamin B6; required for genome integrity; homolog of E. coli PdxK; involved in bud-site selection |
|
| YPR029C | 0.71 |
APL4
|
Gamma-adaptin, large subunit of the clathrin-associated protein (AP-1) complex; binds clathrin; involved in vesicle mediated transport |
|
| YML124C | 0.71 |
TUB3
|
Alpha-tubulin; associates with beta-tubulin (Tub2p) to form tubulin dimer, which polymerizes to form microtubules; expressed at lower level than Tub1p |
|
| YIL029C | 0.71 |
Putative protein of unknown function; deletion confers sensitivity to 4-(N-(S-glutathionylacetyl)amino) phenylarsenoxide (GSAO) |
||
| YGL120C | 0.70 |
PRP43
|
RNA helicase in the DEAH-box family, functions in both RNA polymerase I and polymerase II transcript metabolism, involved in release of the lariat-intron from the spliceosome |
|
| YCR104W | 0.68 |
PAU3
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YGR060W | 0.67 |
ERG25
|
C-4 methyl sterol oxidase, catalyzes the first of three steps required to remove two C-4 methyl groups from an intermediate in ergosterol biosynthesis; mutants accumulate the sterol intermediate 4,4-dimethylzymosterol |
|
| YCR022C | 0.67 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YCR022C is not an essential gene |
||
| YPL041C | 0.67 |
Protein of unknown function involved in maintenance of proper telomere length |
||
| YLR411W | 0.67 |
CTR3
|
High-affinity copper transporter of the plasma membrane, acts as a trimer; gene is disrupted by a Ty2 transposon insertion in many laboratory strains of S. cerevisiae |
|
| YPR146C | 0.66 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR293W | 0.66 |
RPS10A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps10Bp and has similarity to rat ribosomal protein S10 |
|
| YMR208W | 0.66 |
ERG12
|
Mevalonate kinase, acts in the biosynthesis of isoprenoids and sterols, including ergosterol, from mevalonate |
|
| YDR413C | 0.66 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the essential uncharacterized ORF YDR412W that is implicated in rRNA processing |
||
| YDL055C | 0.66 |
PSA1
|
GDP-mannose pyrophosphorylase (mannose-1-phosphate guanyltransferase), synthesizes GDP-mannose from GTP and mannose-1-phosphate in cell wall biosynthesis; required for normal cell wall structure |
|
| YIL096C | 0.66 |
Putative protein of unknown function; associates with precursors of the 60S ribosomal subunit |
||
| YDL240W | 0.65 |
LRG1
|
Putative GTPase-activating protein (GAP) involved in the Pkc1p-mediated signaling pathway that controls cell wall integrity; appears to specifically regulate 1,3-beta-glucan synthesis |
|
| YMR142C | 0.65 |
RPL13B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl13Ap; not essential for viability; has similarity to rat L13 ribosomal protein |
|
| YCR009C | 0.65 |
RVS161
|
Amphiphysin-like lipid raft protein; subunit of a complex (Rvs161p-Rvs167p) that regulates polarization of the actin cytoskeleton, endocytosis, cell polarity, cell fusion and viability following starvation or osmotic stress |
|
| YOL103W | 0.65 |
ITR2
|
Myo-inositol transporter with strong similarity to the major myo-inositol transporter Itr1p, member of the sugar transporter superfamily; expressed constitutively |
|
| YLR257W | 0.65 |
Putative protein of unknown function |
||
| YGL039W | 0.65 |
Oxidoreductase, catalyzes NADPH-dependent reduction of the bicyclic diketone bicyclo[2.2.2]octane-2,6-dione (BCO2,6D) to the chiral ketoalcohol (1R,4S,6S)-6-hydroxybicyclo[2.2.2]octane-2-one (BCO2one6ol) |
||
| YDR037W | 0.64 |
KRS1
|
Lysyl-tRNA synthetase; also identified as a negative regulator of general control of amino acid biosynthesis |
|
| YHR060W | 0.64 |
VMA22
|
Peripheral membrane protein that is required for vacuolar H+-ATPase (V-ATPase) function, although not an actual component of the V-ATPase complex; functions in the assembly of the V-ATPase; localized to the yeast endoplasmic reticulum (ER) |
|
| YKL219W | 0.64 |
COS9
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YNL300W | 0.63 |
Glycosylphosphatidylinositol-dependent cell wall protein, expression is periodic and decreases in respone to ergosterol perturbation or upon entry into stationary phase; depletion increases resistance to lactic acid |
||
| YCR012W | 0.63 |
PGK1
|
3-phosphoglycerate kinase, catalyzes transfer of high-energy phosphoryl groups from the acyl phosphate of 1,3-bisphosphoglycerate to ADP to produce ATP; key enzyme in glycolysis and gluconeogenesis |
|
| YKR003W | 0.63 |
OSH6
|
Member of an oxysterol-binding protein family with overlapping, redundant functions in sterol metabolism and which collectively perform a function essential for viability; GFP-fusion protein localizes to the cell periphery |
|
| YHR031C | 0.62 |
RRM3
|
DNA helicase involved in rDNA replication and Ty1 transposition; relieves replication fork pauses at telomeric regions; structurally and functionally related to Pif1p |
|
| YHR045W | 0.62 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum |
||
| YDL111C | 0.62 |
RRP42
|
Protein involved in rRNA processing; component of the exosome 3->5 exonuclease complex with Rrp4p, Rrp41p, Rrp43p and Dis3p |
|
| YNL066W | 0.61 |
SUN4
|
Cell wall protein related to glucanases, possibly involved in cell wall septation; member of the SUN family |
|
| YBR066C | 0.61 |
NRG2
|
Transcriptional repressor that mediates glucose repression and negatively regulates filamentous growth; has similarity to Nrg1p |
|
| YGL076C | 0.61 |
RPL7A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl7Bp and has similarity to E. coli L30 and rat L7 ribosomal proteins; contains a conserved C-terminal Nucleic acid Binding Domain (NDB2) |
|
| YJL200C | 0.61 |
ACO2
|
Putative mitochondrial aconitase isozyme; similarity to Aco1p, an aconitase required for the TCA cycle; expression induced during growth on glucose, by amino acid starvation via Gcn4p, and repressed on ethanol |
|
| YBR022W | 0.60 |
POA1
|
Phosphatase that is highly specific for ADP-ribose 1''-phosphate, a tRNA splicing metabolite; may have a role in regulation of tRNA splicing |
|
| YLR328W | 0.60 |
NMA1
|
Nicotinic acid mononucleotide adenylyltransferase, involved in pathways of NAD biosynthesis, including the de novo, NAD(+) salvage, and nicotinamide riboside salvage pathways |
|
| YPR170W-A | 0.60 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; identified by expression profiling and mass spectrometry |
||
| YLR154C | 0.60 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YPL225W | 0.59 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YER012W | 0.58 |
PRE1
|
Beta 4 subunit of the 20S proteasome; localizes to the nucleus throughout the cell cycle |
|
| YKL120W | 0.58 |
OAC1
|
Mitochondrial inner membrane transporter, transports oxaloacetate, sulfate, and thiosulfate; member of the mitochondrial carrier family |
|
| YNL301C | 0.57 |
RPL18B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl18Ap and has similarity to rat L18 ribosomal protein |
|
| YDR509W | 0.57 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR273C | 0.57 |
TPO4
|
Polyamine transport protein, recognizes spermine, putrescine, and spermidine; localizes to the plasma membrane; member of the major facilitator superfamily |
|
| YNR067C | 0.57 |
DSE4
|
Daughter cell-specific secreted protein with similarity to glucanases, degrades cell wall from the daughter side causing daughter to separate from mother |
|
| YLR022C | 0.57 |
SDO1
|
Essential protein involved in 60S ribosome maturation; ortholog of the human protein (SBDS) responsible for autosomal recessive Shwachman-Bodian-Diamond Syndrome; highly conserved across archae and eukaryotes |
|
| YDR492W | 0.56 |
IZH1
|
Membrane protein involved in zinc metabolism, member of the four-protein IZH family; transcription is regulated directly by Zap1p, expression induced by zinc deficiency and fatty acids; deletion increases sensitivity to elevated zinc |
|
| YDR185C | 0.56 |
Mitochondrial protein of unknown function; has similarity to Ups1p, which is involved in regulation of alternative topogenesis of the dynamin-related GTPase Mgm1p |
||
| YLR196W | 0.56 |
PWP1
|
Protein with WD-40 repeats involved in rRNA processing; associates with trans-acting ribosome biogenesis factors; similar to beta-transducin superfamily |
|
| YDL201W | 0.56 |
TRM8
|
Subunit of a tRNA methyltransferase complex composed of Trm8p and Trm82p that catalyzes 7-methylguanosine modification of tRNA |
|
| YER056C-A | 0.55 |
RPL34A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl34Bp and has similarity to rat L34 ribosomal protein |
|
| YDR491C | 0.55 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJL153C | 0.55 |
INO1
|
Inositol 1-phosphate synthase, involved in synthesis of inositol phosphates and inositol-containing phospholipids; transcription is coregulated with other phospholipid biosynthetic genes by Ino2p and Ino4p, which bind the UASINO DNA element |
|
| YDR353W | 0.54 |
TRR1
|
Cytoplasmic thioredoxin reductase, key regulatory enzyme that determines the redox state of the thioredoxin system, which acts as a disulfide reductase system and protects cells against both oxidative and reductive stress |
|
| YKL096W-A | 0.54 |
CWP2
|
Covalently linked cell wall mannoprotein, major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored |
|
| YIL011W | 0.54 |
TIR3
|
Cell wall mannoprotein of the Srp1p/Tip1p family of serine-alanine-rich proteins; expressed under anaerobic conditions and required for anaerobic growth |
|
| YDR508C | 0.54 |
GNP1
|
High-affinity glutamine permease, also transports Leu, Ser, Thr, Cys, Met and Asn; expression is fully dependent on Grr1p and modulated by the Ssy1p-Ptr3p-Ssy5p (SPS) sensor of extracellular amino acids |
|
| YKL063C | 0.54 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the Golgi |
||
| YDR447C | 0.54 |
RPS17B
|
Ribosomal protein 51 (rp51) of the small (40s) subunit; nearly identical to Rps17Ap and has similarity to rat S17 ribosomal protein |
|
| YNR054C | 0.54 |
ESF2
|
Essential nucleolar protein involved in pre-18S rRNA processing; binds to RNA and stimulates ATPase activity of Dbp8; involved in assembly of the small subunit (SSU) processome |
|
| YKL110C | 0.53 |
KTI12
|
Protein that plays a role, with Elongator complex, in modification of wobble nucleosides in tRNA; involved in sensitivity to G1 arrest induced by zymocin; interacts with chromatin throughout the genome; also interacts with Cdc19p |
|
| YGR164W | 0.53 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPR170W-B | 0.53 |
Putative protein of unknown function, conserved in fungi; partially overlaps the dubious genes YPR169W-A, YPR170W-A and YRP170C |
||
| YPL241C | 0.53 |
CIN2
|
GTPase-activating protein (GAP) for Cin4p; tubulin folding factor C involved in beta-tubulin (Tub2p) folding; mutants display increased chromosome loss and benomyl sensitivity; deletion complemented by human GAP, retinitis pigmentosa 2 |
|
| YBR243C | 0.52 |
ALG7
|
UDP-N-acetyl-glucosamine-1-P transferase, transfers Glc-Nac-P from UDP-GlcNac to Dol-P in the ER in the first step of the dolichol pathway of protein asparagine-linked glycosylation; inhibited by tunicamycin |
|
| YJL091C | 0.52 |
GWT1
|
Protein involved in the inositol acylation of glucosaminyl phosphatidylinositol (GlcN-PI) to form glucosaminyl(acyl)phosphatidylinositol (GlcN(acyl)PI), an intermediate in the biosynthesis of glycosylphosphatidylinositol (GPI) anchors |
|
| YLR222C | 0.52 |
UTP13
|
Nucleolar protein, component of the small subunit (SSU) processome containing the U3 snoRNA that is involved in processing of pre-18S rRNA |
|
| YFR039C | 0.52 |
Putative protein of unknown function; may be involved in response to high salt and changes in carbon source |
||
| YCR045C | 0.52 |
Putative protein of unknown function |
||
| YGL069C | 0.51 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 92% of ORF overlaps the uncharacterized ORF YGL068W; deletion in cyr1 mutant results in loss of stress resistance |
||
| YER118C | 0.51 |
SHO1
|
Transmembrane osmosensor, participates in activation of both the Cdc42p- and MAP kinase-dependent filamentous growth pathway and the high-osmolarity glycerol response pathway |
|
| YCL024W | 0.51 |
KCC4
|
Protein kinase of the bud neck involved in the septin checkpoint, associates with septin proteins, negatively regulates Swe1p by phosphorylation, shows structural homology to bud neck kinases Gin4p and Hsl1p |
|
| YNL178W | 0.51 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YGL028C | 0.51 |
SCW11
|
Cell wall protein with similarity to glucanases; may play a role in conjugation during mating based on its regulation by Ste12p |
|
| YOR101W | 0.51 |
RAS1
|
GTPase involved in G-protein signaling in the adenylate cyclase activating pathway, plays a role in cell proliferation; localized to the plasma membrane; homolog of mammalian RAS proto-oncogenes |
|
| YDR434W | 0.51 |
GPI17
|
Transmembrane protein subunit of the glycosylphosphatidylinositol transamidase complex that adds GPIs to newly synthesized proteins; human PIG-Sp homolog |
|
| YMR108W | 0.51 |
ILV2
|
Acetolactate synthase, catalyses the first common step in isoleucine and valine biosynthesis and is the target of several classes of inhibitors, localizes to the mitochondria; expression of the gene is under general amino acid control |
|
| YLR132C | 0.51 |
Protein of unknown function; green fluorescent protein (GFP)-tagged protein localizes to mitochondria and nucleus; YLR132C is an essential gene |
||
| YDR212W | 0.51 |
TCP1
|
Alpha subunit of chaperonin-containing T-complex, which mediates protein folding in the cytosol; involved in maintenance of actin cytoskeleton; homolog to Drosophila melanogaster and mouse tailless complex polypeptide |
|
| YPL235W | 0.50 |
RVB2
|
Essential protein involved in transcription regulation; component of chromatin remodeling complexes; required for assembly and function of the INO80 complex; member of the RUVB-like protein family |
|
| YER134C | 0.50 |
Putative protein of unknown function; non-essential gene |
||
| YMR319C | 0.49 |
FET4
|
Low-affinity Fe(II) transporter of the plasma membrane |
|
| YGL040C | 0.49 |
HEM2
|
Delta-aminolevulinate dehydratase, a homo-octameric enzyme, catalyzes the conversion of delta-aminolevulinic acid to porphobilinogen, the second step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus |
|
| YOR108C-A | 0.49 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YOL057W | 0.49 |
Putative metalloprotease |
||
| YNL028W | 0.48 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YML119W | 0.48 |
Putative protein of unknown funtion; YML119W is not an essential gene; potential Cdc28p substrate |
||
| YGR075C | 0.48 |
PRP38
|
Unique component of the U4/U6.U5 tri-snRNP particle, required for conformational changes which result in the catalytic activation of the spliceosome; dispensible for spliceosome assembly |
|
| YBR121C | 0.48 |
GRS1
|
Cytoplasmic and mitochondrial glycyl-tRNA synthase that ligates glycine to the cognate anticodon bearing tRNA; transcription termination factor that may interact with the 3'-end of pre-mRNA to promote 3'-end formation |
|
| YJL169W | 0.48 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YJL168C/SET2 |
||
| YNL147W | 0.47 |
LSM7
|
Lsm (Like Sm) protein; part of heteroheptameric complexes (Lsm2p-7p and either Lsm1p or 8p): cytoplasmic Lsm1p complex involved in mRNA decay; nuclear Lsm8p complex part of U6 snRNP and possibly involved in processing tRNA, snoRNA, and rRNA |
|
| YDR284C | 0.47 |
DPP1
|
Diacylglycerol pyrophosphate (DGPP) phosphatase, zinc-regulated vacuolar membrane-associated lipid phosphatase, dephosphorylates DGPP to phosphatidate (PA) and Pi, then PA to diacylglycerol; involved in lipid signaling and cell metabolism |
|
| YDL235C | 0.47 |
YPD1
|
Phosphorelay intermediate protein, phosphorylated by the plasma membrane sensor Sln1p in response to osmotic stress and then in turn phosphorylates the response regulators Ssk1p in the cytosol and Skn7p in the nucleus |
|
| YBL077W | 0.47 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene ILS1/YBL076C |
||
| YCR102C | 0.46 |
Putative protein of unknown function; involved in copper metabolism; similar to C.carbonum toxD gene; YCR102C is not an essential gene |
||
| YGR157W | 0.46 |
CHO2
|
Phosphatidylethanolamine methyltransferase (PEMT), catalyzes the first step in the conversion of phosphatidylethanolamine to phosphatidylcholine during the methylation pathway of phosphatidylcholine biosynthesis |
|
| YBR048W | 0.46 |
RPS11B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps11Ap and has similarity to E. coli S17 and rat S11 ribosomal proteins |
|
| YDR539W | 0.46 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YDR539W is not an essential gene; homolog of E. coli UbiD |
||
| YJR047C | 0.46 |
ANB1
|
Translation initiation factor eIF-5A, promotes formation of the first peptide bond; similar to and functionally redundant with Hyp2p; undergoes an essential hypusination modification; expressed under anaerobic conditions |
|
| YBL087C | 0.46 |
RPL23A
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl23Bp and has similarity to E. coli L14 and rat L23 ribosomal proteins |
|
| YOL056W | 0.46 |
GPM3
|
Homolog of Gpm1p phosphoglycerate mutase, which converts 3-phosphoglycerate to 2-phosphoglycerate in glycolysis; may be non-functional derivative of a gene duplication event |
|
| YBR210W | 0.45 |
ERV15
|
Protein involved in export of proteins from the endoplasmic reticulum, has similarity to Erv14p |
|
| YER003C | 0.45 |
PMI40
|
Mannose-6-phosphate isomerase, catalyzes the interconversion of fructose-6-P and mannose-6-P; required for early steps in protein mannosylation |
|
| YMR119W | 0.45 |
ASI1
|
Putative integral membrane E3 ubiquitin ligase; acts with Asi2p and Asi3p to ensure the fidelity of SPS-sensor signalling by maintaining the dormant repressed state of gene expression in the absence of inducing signals |
|
| YOL088C | 0.45 |
MPD2
|
Member of the protein disulfide isomerase (PDI) family, exhibits chaperone activity; overexpression suppresses the lethality of a pdi1 deletion but does not complement all Pdi1p functions; undergoes oxidation by Ero1p |
|
| YPL143W | 0.44 |
RPL33A
|
N-terminally acetylated ribosomal protein L37 of the large (60S) ribosomal subunit, nearly identical to Rpl33Bp and has similarity to rat L35a; rpl33a null mutant exhibits slow growth while rpl33a rpl33b double null mutant is inviable |
Network of associatons between targets according to the STRING database.
First level regulatory network of MATALPHA2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.5 | 1.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.4 | 1.7 | GO:0015867 | ATP transport(GO:0015867) |
| 0.4 | 2.7 | GO:0030030 | cell projection organization(GO:0030030) cell projection assembly(GO:0030031) |
| 0.4 | 1.1 | GO:0035955 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 0.4 | 1.1 | GO:0019673 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.4 | 1.1 | GO:0009177 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.3 | 2.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.3 | 1.4 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.3 | 2.3 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 0.3 | 1.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.3 | 1.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.3 | 1.2 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.3 | 1.2 | GO:2001057 | reactive nitrogen species metabolic process(GO:2001057) |
| 0.3 | 0.8 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.3 | 1.8 | GO:0006672 | ceramide metabolic process(GO:0006672) ceramide biosynthetic process(GO:0046513) |
| 0.3 | 1.0 | GO:0000296 | spermine transport(GO:0000296) |
| 0.2 | 1.0 | GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000461) |
| 0.2 | 0.7 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.2 | 3.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 0.9 | GO:0014070 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 0.2 | 2.5 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.2 | 0.9 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 | 0.8 | GO:0098610 | agglutination involved in conjugation with cellular fusion(GO:0000752) agglutination involved in conjugation(GO:0000771) heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) cell-cell adhesion(GO:0098609) adhesion between unicellular organisms(GO:0098610) multi organism cell adhesion(GO:0098740) cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.2 | 3.3 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.2 | 0.6 | GO:0060988 | lipid tube assembly(GO:0060988) protein-lipid complex assembly(GO:0065005) protein-lipid complex subunit organization(GO:0071825) |
| 0.2 | 2.3 | GO:0015833 | peptide transport(GO:0015833) |
| 0.2 | 0.8 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 1.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) pantothenate biosynthetic process(GO:0015940) |
| 0.2 | 0.7 | GO:0019184 | glutathione biosynthetic process(GO:0006750) nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.2 | 0.3 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.2 | 1.7 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.2 | 1.7 | GO:0006547 | histidine biosynthetic process(GO:0000105) histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.2 | 21.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 0.5 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.2 | 4.0 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.2 | 0.5 | GO:0046901 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.2 | 0.6 | GO:1900436 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.2 | 0.5 | GO:0000388 | spliceosome conformational change to release U4 (or U4atac) and U1 (or U11)(GO:0000388) |
| 0.2 | 1.9 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 0.5 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.2 | 0.6 | GO:0009423 | chorismate biosynthetic process(GO:0009423) |
| 0.1 | 0.7 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.1 | 0.6 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 0.1 | 0.6 | GO:0046655 | folic acid metabolic process(GO:0046655) folic acid biosynthetic process(GO:0046656) |
| 0.1 | 0.4 | GO:0090338 | regulation of formin-nucleated actin cable assembly(GO:0090337) positive regulation of formin-nucleated actin cable assembly(GO:0090338) |
| 0.1 | 0.4 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.7 | GO:0072668 | obsolete tubulin complex biogenesis(GO:0072668) |
| 0.1 | 1.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.5 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.7 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 0.1 | GO:0009135 | purine nucleoside diphosphate metabolic process(GO:0009135) purine ribonucleoside diphosphate metabolic process(GO:0009179) ribonucleoside diphosphate metabolic process(GO:0009185) ADP metabolic process(GO:0046031) |
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 1.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.1 | 3.5 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.1 | 0.5 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.4 | GO:0006798 | polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.5 | GO:0043097 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.1 | 0.6 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.8 | GO:0000743 | nuclear migration involved in conjugation with cellular fusion(GO:0000743) |
| 0.1 | 0.4 | GO:0030541 | plasmid partitioning(GO:0030541) |
| 0.1 | 0.8 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.1 | 1.8 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.1 | 0.6 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 1.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 3.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.3 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.1 | 0.8 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.2 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) |
| 0.1 | 0.3 | GO:0071478 | cellular response to radiation(GO:0071478) cellular response to light stimulus(GO:0071482) |
| 0.1 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.4 | GO:0009164 | nucleoside catabolic process(GO:0009164) glycosyl compound catabolic process(GO:1901658) |
| 0.1 | 0.5 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.1 | GO:0061422 | response to cold(GO:0009409) positive regulation of transcription from RNA polymerase II promoter in response to alkaline pH(GO:0061422) cellular response to cold(GO:0070417) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:0009312 | trehalose biosynthetic process(GO:0005992) oligosaccharide biosynthetic process(GO:0009312) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.1 | GO:0043065 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.1 | 0.6 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.4 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.1 | 0.8 | GO:0006783 | porphyrin-containing compound biosynthetic process(GO:0006779) heme biosynthetic process(GO:0006783) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.1 | 0.6 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 0.8 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.1 | 1.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.1 | GO:0007535 | donor selection(GO:0007535) |
| 0.1 | 1.0 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.1 | 0.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.1 | GO:1901352 | regulation of phosphatidylglycerol biosynthetic process(GO:1901351) negative regulation of phosphatidylglycerol biosynthetic process(GO:1901352) |
| 0.1 | 0.4 | GO:0071450 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.6 | GO:0006797 | polyphosphate metabolic process(GO:0006797) |
| 0.1 | 0.1 | GO:0042990 | regulation of transcription factor import into nucleus(GO:0042990) negative regulation of transcription factor import into nucleus(GO:0042992) |
| 0.1 | 0.5 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.1 | 0.6 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 0.4 | GO:0034503 | protein localization to nucleolar rDNA repeats(GO:0034503) |
| 0.1 | 0.8 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 1.2 | GO:0001100 | negative regulation of exit from mitosis(GO:0001100) |
| 0.1 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:0060188 | regulation of protein desumoylation(GO:0060188) |
| 0.1 | 0.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.2 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin structure organization(GO:0031032) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.5 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 1.3 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.1 | 0.4 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.1 | 0.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.5 | GO:0000372 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.1 | 2.0 | GO:0016126 | steroid biosynthetic process(GO:0006694) sterol biosynthetic process(GO:0016126) |
| 0.1 | 1.8 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.4 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 1.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.3 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) |
| 0.1 | 1.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 0.3 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) barbed-end actin filament capping(GO:0051016) actin filament capping(GO:0051693) |
| 0.1 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.1 | 0.3 | GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity(GO:0051090) |
| 0.1 | 0.5 | GO:0007323 | peptide pheromone maturation(GO:0007323) |
| 0.1 | 0.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 0.2 | GO:0070637 | nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) |
| 0.1 | 0.1 | GO:0016114 | terpenoid metabolic process(GO:0006721) terpenoid biosynthetic process(GO:0016114) farnesyl diphosphate biosynthetic process(GO:0045337) farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 0.2 | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) |
| 0.1 | 0.4 | GO:0001402 | signal transduction involved in filamentous growth(GO:0001402) |
| 0.1 | 0.3 | GO:0009263 | deoxyribonucleotide metabolic process(GO:0009262) deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.5 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.5 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.2 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.1 | 0.2 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.1 | 0.2 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 0.2 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 0.6 | GO:0070651 | nonfunctional rRNA decay(GO:0070651) |
| 0.1 | 0.2 | GO:1902223 | L-phenylalanine biosynthetic process(GO:0009094) erythrose 4-phosphate/phosphoenolpyruvate family amino acid biosynthetic process(GO:1902223) |
| 0.1 | 0.2 | GO:0090110 | cargo loading into vesicle(GO:0035459) cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 1.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0019935 | cAMP-mediated signaling(GO:0019933) cyclic-nucleotide-mediated signaling(GO:0019935) |
| 0.1 | 0.2 | GO:0048279 | vesicle fusion with endoplasmic reticulum(GO:0048279) |
| 0.1 | 0.8 | GO:0043094 | cellular metabolic compound salvage(GO:0043094) |
| 0.1 | 0.6 | GO:0000917 | barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.1 | 0.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 0.2 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.5 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 2.2 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.1 | 0.2 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.3 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:0055070 | cellular copper ion homeostasis(GO:0006878) copper ion homeostasis(GO:0055070) |
| 0.0 | 0.1 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.0 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.0 | 0.2 | GO:0030847 | termination of RNA polymerase II transcription, exosome-dependent(GO:0030847) |
| 0.0 | 0.1 | GO:0045141 | meiotic telomere clustering(GO:0045141) establishment of chromosome localization(GO:0051303) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 | 0.2 | GO:0007234 | osmosensory signaling via phosphorelay pathway(GO:0007234) |
| 0.0 | 0.3 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 3.1 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 1.0 | GO:0006885 | regulation of pH(GO:0006885) |
| 0.0 | 0.0 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.2 | GO:0010907 | positive regulation of glucose metabolic process(GO:0010907) positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.2 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.2 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.6 | GO:0018208 | protein peptidyl-prolyl isomerization(GO:0000413) peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.2 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.7 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.0 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.1 | GO:0015848 | spermidine transport(GO:0015848) |
| 0.0 | 0.9 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.1 | GO:0009090 | homoserine biosynthetic process(GO:0009090) |
| 0.0 | 0.0 | GO:0051596 | methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.9 | GO:1903509 | liposaccharide metabolic process(GO:1903509) |
| 0.0 | 0.1 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.2 | GO:0043489 | RNA stabilization(GO:0043489) mRNA stabilization(GO:0048255) |
| 0.0 | 0.6 | GO:0046474 | glycerophospholipid biosynthetic process(GO:0046474) |
| 0.0 | 0.5 | GO:0042797 | tRNA transcription(GO:0009304) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) |
| 0.0 | 0.4 | GO:0033673 | negative regulation of protein kinase activity(GO:0006469) negative regulation of kinase activity(GO:0033673) |
| 0.0 | 0.5 | GO:0006915 | apoptotic process(GO:0006915) cell death(GO:0008219) programmed cell death(GO:0012501) |
| 0.0 | 0.2 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) calcium ion homeostasis(GO:0055074) |
| 0.0 | 0.2 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.4 | GO:0051668 | localization within membrane(GO:0051668) |
| 0.0 | 0.1 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.3 | GO:0031070 | intronic snoRNA processing(GO:0031070) intronic box C/D snoRNA processing(GO:0034965) |
| 0.0 | 0.2 | GO:0006903 | vesicle targeting(GO:0006903) vesicle tethering involved in exocytosis(GO:0090522) |
| 0.0 | 0.3 | GO:0006038 | cell wall chitin biosynthetic process(GO:0006038) |
| 0.0 | 0.1 | GO:0071406 | response to methylmercury(GO:0051597) cellular response to methylmercury(GO:0071406) |
| 0.0 | 0.1 | GO:0042401 | tryptophan biosynthetic process(GO:0000162) amine biosynthetic process(GO:0009309) cellular biogenic amine biosynthetic process(GO:0042401) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.0 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.0 | 0.1 | GO:0001015 | snoRNA transcription from an RNA polymerase II promoter(GO:0001015) snoRNA transcription(GO:0009302) |
| 0.0 | 0.1 | GO:0000967 | rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.8 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.4 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.0 | GO:0045017 | glycerolipid biosynthetic process(GO:0045017) |
| 0.0 | 0.0 | GO:0030846 | termination of RNA polymerase II transcription, poly(A)-coupled(GO:0030846) |
| 0.0 | 0.1 | GO:0031134 | sister chromatid biorientation(GO:0031134) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) ketone biosynthetic process(GO:0042181) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.1 | GO:0035753 | maintenance of DNA trinucleotide repeats(GO:0035753) |
| 0.0 | 0.1 | GO:0043488 | regulation of RNA stability(GO:0043487) regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.2 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0010674 | negative regulation of transcription from RNA polymerase II promoter involved in meiotic cell cycle(GO:0010674) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:0040031 | snRNA pseudouridine synthesis(GO:0031120) snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:0009088 | threonine biosynthetic process(GO:0009088) |
| 0.0 | 0.0 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0018202 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.0 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0006771 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.0 | 0.2 | GO:0034249 | negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 | 0.1 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.0 | GO:0001109 | promoter clearance during DNA-templated transcription(GO:0001109) promoter clearance from RNA polymerase II promoter(GO:0001111) |
| 0.0 | 0.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.0 | GO:1901985 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.0 | GO:0006037 | cell wall chitin metabolic process(GO:0006037) |
| 0.0 | 0.0 | GO:0045926 | negative regulation of growth(GO:0045926) negative regulation of filamentous growth(GO:0060258) |
| 0.0 | 0.2 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.1 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.2 | GO:0071900 | regulation of protein serine/threonine kinase activity(GO:0071900) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0010526 | negative regulation of transposition, RNA-mediated(GO:0010526) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0030287 | cell wall-bounded periplasmic space(GO:0030287) |
| 0.4 | 1.7 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.4 | 1.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 0.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 0.7 | GO:0005827 | polar microtubule(GO:0005827) tubulin complex(GO:0045298) |
| 0.2 | 18.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 0.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.2 | 0.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 11.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 0.6 | GO:0030428 | cell septum(GO:0030428) |
| 0.2 | 1.0 | GO:0034456 | UTP-C complex(GO:0034456) |
| 0.2 | 0.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 0.6 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 0.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.4 | GO:0071261 | Ssh1 translocon complex(GO:0071261) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.5 | GO:0044697 | HICS complex(GO:0044697) |
| 0.1 | 0.5 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.1 | 0.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.5 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.1 | 1.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 1.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.1 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 0.8 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 0.7 | GO:0031933 | nuclear heterochromatin(GO:0005720) nuclear telomeric heterochromatin(GO:0005724) telomeric heterochromatin(GO:0031933) |
| 0.1 | 1.5 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 0.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.6 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 1.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.6 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.4 | GO:0033254 | vacuolar transporter chaperone complex(GO:0033254) |
| 0.1 | 0.3 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.1 | 1.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.4 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.3 | GO:0030874 | nucleolar chromatin(GO:0030874) |
| 0.1 | 0.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 3.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) Sec62/Sec63 complex(GO:0031207) |
| 0.1 | 0.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.2 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.2 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 0.2 | GO:0000837 | Doa10p ubiquitin ligase complex(GO:0000837) |
| 0.1 | 1.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.6 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.3 | GO:0035859 | Seh1-associated complex(GO:0035859) |
| 0.1 | 0.3 | GO:0070860 | RNA polymerase I core factor complex(GO:0070860) |
| 0.1 | 2.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 0.3 | GO:0030892 | mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) |
| 0.1 | 1.8 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.2 | GO:0097344 | Rix1 complex(GO:0097344) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.4 | GO:0033180 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.2 | GO:0043529 | GET complex(GO:0043529) |
| 0.1 | 0.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.6 | GO:0005940 | septin ring(GO:0005940) |
| 0.1 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.2 | GO:0044462 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 0.1 | 0.1 | GO:0044453 | nuclear membrane part(GO:0044453) |
| 0.1 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.2 | GO:0000417 | HIR complex(GO:0000417) |
| 0.1 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 1.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 7.7 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.2 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.0 | GO:0031391 | Elg1 RFC-like complex(GO:0031391) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 6.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 1.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.2 | GO:0035097 | histone methyltransferase complex(GO:0035097) Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 3.3 | GO:0005933 | cellular bud(GO:0005933) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.0 | GO:0000131 | incipient cellular bud site(GO:0000131) |
| 0.0 | 0.4 | GO:0097526 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 0.1 | GO:0033597 | mitotic checkpoint complex(GO:0033597) |
| 0.0 | 0.0 | GO:0044284 | mitochondrial crista junction(GO:0044284) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 0.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.0 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0044445 | cytosolic part(GO:0044445) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.9 | 2.8 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.7 | 2.2 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.6 | 1.9 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.6 | 3.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.5 | 1.6 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.5 | 1.6 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.5 | 2.6 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.4 | 1.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 1.7 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.4 | 1.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.4 | 1.6 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 0.3 | 2.7 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.3 | 1.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 0.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.3 | 1.1 | GO:0042973 | glucan endo-1,3-beta-D-glucosidase activity(GO:0042973) |
| 0.3 | 0.8 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.3 | 1.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 0.3 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) |
| 0.3 | 1.0 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.2 | 0.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.2 | 1.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 0.9 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.2 | 0.7 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.2 | 0.6 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.2 | 0.6 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.2 | 0.8 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 1.0 | GO:0004026 | alcohol O-acetyltransferase activity(GO:0004026) alcohol O-acyltransferase activity(GO:0034318) |
| 0.2 | 1.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 0.8 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 0.7 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.2 | 1.1 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.2 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 0.5 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.2 | 1.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.2 | 0.5 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.2 | 0.5 | GO:0016872 | intramolecular lyase activity(GO:0016872) |
| 0.2 | 0.8 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.2 | 0.5 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 0.2 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 2.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.3 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.1 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.4 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 26.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 0.4 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.1 | 0.6 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 2.7 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 1.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 2.0 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 1.1 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 0.8 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 1.0 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.1 | 1.8 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 1.3 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.1 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 1.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.6 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 0.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 1.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.8 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 0.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.6 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.1 | 0.5 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.3 | GO:0004805 | trehalose-phosphatase activity(GO:0004805) |
| 0.1 | 0.5 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.3 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.2 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 1.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.1 | 0.3 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.4 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.6 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.3 | GO:0001186 | core promoter sequence-specific DNA binding(GO:0001046) transcription factor activity, RNA polymerase I transcription factor recruiting(GO:0001186) |
| 0.1 | 0.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.1 | 0.2 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.8 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 1.7 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.3 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.0 | 0.3 | GO:0019205 | nucleobase-containing compound kinase activity(GO:0019205) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.3 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.3 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.2 | GO:0005034 | osmosensor activity(GO:0005034) |
| 0.0 | 0.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 1.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0015606 | spermidine transmembrane transporter activity(GO:0015606) |
| 0.0 | 0.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.1 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0008251 | adenosine deaminase activity(GO:0004000) tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.0 | 0.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.6 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.4 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.1 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.6 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.1 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.1 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.0 | 0.1 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.0 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.0 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.7 | GO:0016791 | phosphatase activity(GO:0016791) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 1.3 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.3 | 1.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 0.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.3 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 1.7 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.2 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 3.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 1.2 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.2 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 0.3 | REACTOME TCA CYCLE AND RESPIRATORY ELECTRON TRANSPORT | Genes involved in The citric acid (TCA) cycle and respiratory electron transport |
| 0.1 | 0.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.7 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |
| 0.1 | 0.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 0.2 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 0.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL PROTEIN MODIFICATION | Genes involved in Post-translational protein modification |
| 0.0 | 0.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME APOPTOSIS | Genes involved in Apoptosis |
| 0.0 | 0.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.0 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |