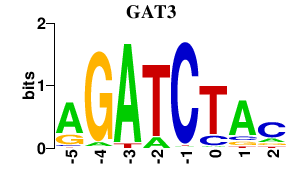
Results for GAT3
Z-value: 0.33
Transcription factors associated with GAT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GAT3
|
S000004003 | Protein containing GATA family zinc finger motifs |
Activity-expression correlation:
Activity profile of GAT3 motif
Sorted Z-values of GAT3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YKL217W | 1.28 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YPL223C | 1.11 |
GRE1
|
Hydrophilin of unknown function; stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YFR053C | 1.07 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YHR139C | 0.99 |
SPS100
|
Protein required for spore wall maturation; expressed during sporulation; may be a component of the spore wall |
|
| YER103W | 0.98 |
SSA4
|
Heat shock protein that is highly induced upon stress; plays a role in SRP-dependent cotranslational protein-membrane targeting and translocation; member of the HSP70 family; cytoplasmic protein that concentrates in nuclei upon starvation |
|
| YOR343C | 0.97 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YKR097W | 0.95 |
PCK1
|
Phosphoenolpyruvate carboxykinase, key enzyme in gluconeogenesis, catalyzes early reaction in carbohydrate biosynthesis, glucose represses transcription and accelerates mRNA degradation, regulated by Mcm1p and Cat8p, located in the cytosol |
|
| YFL052W | 0.93 |
Putative zinc cluster protein that contains a DNA binding domain; null mutant sensitive to calcofluor white, low osmolarity and heat, suggesting a role for YFL052Wp in cell wall integrity |
||
| YPL250C | 0.92 |
ICY2
|
Protein of unknown function; mobilized into polysomes upon a shift from a fermentable to nonfermentable carbon source; potential Cdc28p substrate |
|
| YJR146W | 0.89 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene HMS2 |
||
| YGL255W | 0.87 |
ZRT1
|
High-affinity zinc transporter of the plasma membrane, responsible for the majority of zinc uptake; transcription is induced under low-zinc conditions by the Zap1p transcription factor |
|
| YEL039C | 0.85 |
CYC7
|
Cytochrome c isoform 2, expressed under hypoxic conditions; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YML128C | 0.84 |
MSC1
|
Protein of unknown function; mutant is defective in directing meiotic recombination events to homologous chromatids; the authentic, non-tagged protein is detected in highly purified mitochondria and is phosphorylated |
|
| YEL070W | 0.80 |
DSF1
|
Deletion suppressor of mpt5 mutation |
|
| YPL054W | 0.77 |
LEE1
|
Zinc-finger protein of unknown function |
|
| YFR017C | 0.76 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and is induced in response to the DNA-damaging agent MMS; YFR017C is not an essential gene |
||
| YNR034W-A | 0.75 |
Putative protein of unknown function; expression is regulated by Msn2p/Msn4p |
||
| YJR078W | 0.70 |
BNA2
|
Putative tryptophan 2,3-dioxygenase or indoleamine 2,3-dioxygenase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p and Aft2p |
|
| YBR072W | 0.68 |
HSP26
|
Small heat shock protein (sHSP) with chaperone activity; forms hollow, sphere-shaped oligomers that suppress unfolded proteins aggregation; oligomer activation requires a heat-induced conformational change; not expressed in unstressed cells |
|
| YHR096C | 0.68 |
HXT5
|
Hexose transporter with moderate affinity for glucose, induced in the presence of non-fermentable carbon sources, induced by a decrease in growth rate, contains an extended N-terminal domain relative to other HXTs |
|
| YIL057C | 0.67 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YEL008W | 0.65 |
Hypothetical protein predicted to be involved in metabolism |
||
| YAR060C | 0.64 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR248W | 0.64 |
SOL4
|
6-phosphogluconolactonase with similarity to Sol3p |
|
| YDR453C | 0.63 |
TSA2
|
Stress inducible cytoplasmic thioredoxin peroxidase; cooperates with Tsa1p in the removal of reactive oxygen, nitrogen and sulfur species using thioredoxin as hydrogen donor; deletion enhances the mutator phenotype of tsa1 mutants |
|
| YML054C | 0.61 |
CYB2
|
Cytochrome b2 (L-lactate cytochrome-c oxidoreductase), component of the mitochondrial intermembrane space, required for lactate utilization; expression is repressed by glucose and anaerobic conditions |
|
| YAL054C | 0.61 |
ACS1
|
Acetyl-coA synthetase isoform which, along with Acs2p, is the nuclear source of acetyl-coA for histone acetlyation; expressed during growth on nonfermentable carbon sources and under aerobic conditions |
|
| YIL012W | 0.60 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR139C-A | 0.58 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR316C-B | 0.58 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR023W | 0.57 |
MTL1
|
Protein with both structural and functional similarity to Mid2p, which is a plasma membrane sensor required for cell integrity signaling during pheromone-induced morphogenesis; suppresses rgd1 null mutations |
|
| YJL217W | 0.57 |
Cytoplasmic protein of unknown function; expression induced by calcium shortage and via the copper sensing transciption factor Mac1p during conditons of copper deficiency; mRNA is cell cycle regulated, peaking in G1 phase |
||
| YMR317W | 0.57 |
Putative protein of unknown function with some similarity to sialidase from Trypanosoma; YMR317W is not an essential gene |
||
| YHL021C | 0.56 |
AIM17
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) |
|
| YDR536W | 0.56 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YJR077C | 0.55 |
MIR1
|
Mitochondrial phosphate carrier, imports inorganic phosphate into mitochondria; functionally redundant with Pic2p but more abundant than Pic2p under normal conditions; phosphorylated |
|
| YNL052W | 0.53 |
COX5A
|
Subunit Va of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; predominantly expressed during aerobic growth while its isoform Vb (Cox5Bp) is expressed during anaerobic growth |
|
| YNL074C | 0.52 |
MLF3
|
Serine-rich protein of unknown function; overproduction suppresses the growth inhibition caused by exposure to the immunosuppressant leflunomide |
|
| YLR296W | 0.50 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR312C | 0.49 |
QNQ1
|
Protein of unknown function; null mutant quiescent and non-quiescent cells exhibit reduced reproductive capacity |
|
| YGR191W | 0.49 |
HIP1
|
High-affinity histidine permease, also involved in the transport of manganese ions |
|
| YAR053W | 0.48 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR375W | 0.48 |
STP3
|
Zinc-finger protein of unknown function, possibly involved in pre-tRNA splicing and in uptake of branched-chain amino acids |
|
| YNL042W-B | 0.48 |
Putative protein of unknown function |
||
| YJR048W | 0.48 |
CYC1
|
Cytochrome c, isoform 1; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YOR178C | 0.48 |
GAC1
|
Regulatory subunit for Glc7p type-1 protein phosphatase (PP1), tethers Glc7p to Gsy2p glycogen synthase, binds Hsf1p heat shock transcription factor, required for induction of some HSF-regulated genes under heat shock |
|
| YOL154W | 0.47 |
ZPS1
|
Putative GPI-anchored protein; transcription is induced under low-zinc conditions, as mediated by the Zap1p transcription factor, and at alkaline pH |
|
| YDR530C | 0.47 |
APA2
|
Diadenosine 5',5''-P1,P4-tetraphosphate phosphorylase II (AP4A phosphorylase), involved in catabolism of bis(5'-nucleosidyl) tetraphosphates; has similarity to Apa1p |
|
| YPL156C | 0.46 |
PRM4
|
Pheromone-regulated protein, predicted to have 1 transmembrane segment; transcriptionally regulated by Ste12p during mating and by Cat8p during the diauxic shift |
|
| YGR022C | 0.46 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF MTL1/YGR023W |
||
| YLL026W | 0.46 |
HSP104
|
Heat shock protein that cooperates with Ydj1p (Hsp40) and Ssa1p (Hsp70) to refold and reactivate previously denatured, aggregated proteins; responsive to stresses including: heat, ethanol, and sodium arsenite; involved in [PSI+] propagation |
|
| YHR092C | 0.45 |
HXT4
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YJL067W | 0.45 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPR030W | 0.45 |
CSR2
|
Nuclear protein with a potential regulatory role in utilization of galactose and nonfermentable carbon sources; overproduction suppresses the lethality at high temperature of a chs5 spa2 double null mutation; potential Cdc28p substrate |
|
| YIR027C | 0.44 |
DAL1
|
Allantoinase, converts allantoin to allantoate in the first step of allantoin degradation; expression sensitive to nitrogen catabolite repression |
|
| YML100W | 0.44 |
TSL1
|
Large subunit of trehalose 6-phosphate synthase (Tps1p)/phosphatase (Tps2p) complex, which converts uridine-5'-diphosphoglucose and glucose 6-phosphate to trehalose, homologous to Tps3p and may share function |
|
| YGR043C | 0.44 |
NQM1
|
Protein of unknown function; transcription is repressed by Mot1p and induced by alpha-factor and during diauxic shift; null mutant non-quiescent cells exhibit reduced reproductive capacity |
|
| YBR214W | 0.44 |
SDS24
|
One of two S. cerevisiae homologs (Sds23p and Sds24p) of the Schizosaccharomyces pombe Sds23 protein, which genetic studies have implicated in APC/cyclosome regulation; may play an indirect role in fluid-phase endocytosis |
|
| YJL045W | 0.44 |
Minor succinate dehydrogenase isozyme; homologous to Sdh1p, the major isozyme reponsible for the oxidation of succinate and transfer of electrons to ubiquinone; induced during the diauxic shift in a Cat8p-dependent manner |
||
| YHR212C | 0.43 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR332W | 0.43 |
MID2
|
O-glycosylated plasma membrane protein that acts as a sensor for cell wall integrity signaling and activates the pathway; interacts with Rom2p, a guanine nucleotide exchange factor for Rho1p, and with cell integrity pathway protein Zeo1p |
|
| YCR021C | 0.43 |
HSP30
|
Hydrophobic plasma membrane localized, stress-responsive protein that negatively regulates the H(+)-ATPase Pma1p; induced by heat shock, ethanol treatment, weak organic acid, glucose limitation, and entry into stationary phase |
|
| YBR132C | 0.42 |
AGP2
|
High affinity polyamine permease, preferentially uses spermidine over putrescine; expression is down-regulated by osmotic stress; plasma membrane carnitine transporter, also functions as a low-affinity amino acid permease |
|
| YPL222W | 0.42 |
FMP40
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YOR235W | 0.42 |
IRC13
|
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YNL036W | 0.42 |
NCE103
|
Carbonic anhydrase; poorly transcribed under aerobic conditions and at an undetectable level under anaerobic conditions; involved in non-classical protein export pathway |
|
| YAR019C | 0.41 |
CDC15
|
Protein kinase of the Mitotic Exit Network that is localized to the spindle pole bodies at late anaphase; promotes mitotic exit by directly switching on the kinase activity of Dbf2p |
|
| YLR331C | 0.40 |
JIP3
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 98% of ORF overlaps the verified gene MID2 |
|
| YBR077C | 0.40 |
SLM4
|
Component of the EGO complex, which is involved in the regulation of microautophagy, and of the GSE complex, which is required for proper sorting of amino acid permease Gap1p; gene exhibits synthetic genetic interaction with MSS4 |
|
| YDR171W | 0.39 |
HSP42
|
Small heat shock protein (sHSP) with chaperone activity; forms barrel-shaped oligomers that suppress unfolded protein aggregation; involved in cytoskeleton reorganization after heat shock |
|
| YGL162W | 0.39 |
SUT1
|
Transcription factor of the Zn[II]2Cys6 family involved in sterol uptake; involved in induction of hypoxic gene expression |
|
| YDL085W | 0.38 |
NDE2
|
Mitochondrial external NADH dehydrogenase, catalyzes the oxidation of cytosolic NADH; Nde1p and Nde2p are involved in providing the cytosolic NADH to the mitochondrial respiratory chain |
|
| YBR085C-A | 0.38 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and to the nucleus |
||
| YNL142W | 0.38 |
MEP2
|
Ammonium permease involved in regulation of pseudohyphal growth; belongs to a ubiquitous family of cytoplasmic membrane proteins that transport only ammonium (NH4+); expression is under the nitrogen catabolite repression regulation |
|
| YNL055C | 0.38 |
POR1
|
Mitochondrial porin (voltage-dependent anion channel), outer membrane protein required for the maintenance of mitochondrial osmotic stability and mitochondrial membrane permeability; phosphorylated |
|
| YOR348C | 0.38 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YEL009C | 0.37 |
GCN4
|
Transcriptional activator of amino acid biosynthetic genes in response to amino acid starvation; expression is tightly regulated at both the transcriptional and translational levels |
|
| YIL037C | 0.37 |
PRM2
|
Pheromone-regulated protein, predicted to have 4 transmembrane segments and a coiled coil domain; regulated by Ste12p |
|
| YOL032W | 0.37 |
OPI10
|
Protein with a possible role in phospholipid biosynthesis, based on inositol-excreting phenotype of the null mutant and its suppression by exogenous choline |
|
| YNL125C | 0.36 |
ESBP6
|
Protein with similarity to monocarboxylate permeases, appears not to be involved in transport of monocarboxylates such as lactate, pyruvate or acetate across the plasma membrane |
|
| YNL144C | 0.36 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YNL144C is not an essential gene |
||
| YDL181W | 0.36 |
INH1
|
Protein that inhibits ATP hydrolysis by the F1F0-ATP synthase; inhibitory function is enhanced by stabilizing proteins Stf1p and Stf2p; has similarity to Stf1p; has a calmodulin-binding motif and binds calmodulin in vitro |
|
| YPL262W | 0.36 |
FUM1
|
Fumarase, converts fumaric acid to L-malic acid in the TCA cycle; cytosolic and mitochondrial localization determined by the N-terminal mitochondrial targeting sequence and protein conformation; phosphorylated in mitochondria |
|
| YLR374C | 0.36 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF STP3/YLR375W |
||
| YLR294C | 0.36 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene ATP14 |
||
| YBR145W | 0.35 |
ADH5
|
Alcohol dehydrogenase isoenzyme V; involved in ethanol production |
|
| YGR213C | 0.35 |
RTA1
|
Protein involved in 7-aminocholesterol resistance; has seven potential membrane-spanning regions; expression is induced under both low-heme and low-oxygen conditions |
|
| YHR212W-A | 0.35 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YJL144W | 0.35 |
Cytoplasmic hydrophilin of unknown function, possibly involved in the dessication response; expression induced by osmotic stress, starvation and during stationary phase; GFP-fusion protein is induced by the DNA-damaging agent MMS |
||
| YKL163W | 0.35 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YPR160W | 0.35 |
GPH1
|
Non-essential glycogen phosphorylase required for the mobilization of glycogen, activity is regulated by cyclic AMP-mediated phosphorylation, expression is regulated by stress-response elements and by the HOG MAP kinase pathway |
|
| YKR011C | 0.34 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus |
||
| YKL100C | 0.34 |
Putative protein of unknown function with similarity to a human minor histocompatibility antigen; YKL100C is not an essential gene |
||
| YGR069W | 0.34 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR268C | 0.34 |
HUA1
|
Cytoplasmic protein containing a zinc finger domain with sequence similarity to that of Type I J-proteins; computational analysis of large-scale protein-protein interaction data suggests a possible role in actin patch assembly |
|
| YOL119C | 0.33 |
MCH4
|
Protein with similarity to mammalian monocarboxylate permeases, which are involved in transport of monocarboxylic acids across the plasma membrane; mutant is not deficient in monocarboxylate transport |
|
| YBL099W | 0.33 |
ATP1
|
Alpha subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; phosphorylated |
|
| YMR191W | 0.33 |
SPG5
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YHR086W | 0.33 |
NAM8
|
RNA binding protein, component of the U1 snRNP protein; mutants are defective in meiotic recombination and in formation of viable spores, involved in the formation of DSBs through meiosis-specific splicing of MER2 pre-mRNA |
|
| YOL118C | 0.33 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YIL059C | 0.32 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YIL060W |
||
| YER147C | 0.32 |
SCC4
|
Subunit of cohesin loading factor (Scc2p-Scc4p), a complex required for the loading of cohesin complexes onto chromosomes; involved in establishing sister chromatid cohesion during double-strand break repair via phosphorylated histone H2AX |
|
| YNL093W | 0.32 |
YPT53
|
GTPase, similar to Ypt51p and Ypt52p and to mammalian rab5; required for vacuolar protein sorting and endocytosis |
|
| YJR150C | 0.32 |
DAN1
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YPR027C | 0.32 |
Putative protein of unknown function |
||
| YAR050W | 0.31 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YFL051C | 0.31 |
Putative protein of unknown function; YFL051C is not an essential gene |
||
| YOR298C-A | 0.31 |
MBF1
|
Transcriptional coactivator that bridges the DNA-binding region of Gcn4p and TATA-binding protein Spt15p; suppressor of frameshift mutations |
|
| YOR152C | 0.31 |
Putative protein of unknown function; has no similarity to any known protein; YOR152C is not an essential gene |
||
| YGR070W | 0.31 |
ROM1
|
GDP/GTP exchange protein (GEP) for Rho1p; mutations are synthetically lethal with mutations in rom2, which also encodes a GEP |
|
| YJR147W | 0.31 |
HMS2
|
Protein with similarity to heat shock transcription factors; overexpression suppresses the pseudohyphal filamentation defect of a diploid mep1 mep2 homozygous null mutant |
|
| YIL047C | 0.31 |
SYG1
|
Plasma membrane protein of unknown function; truncation and overexpression suppresses lethality of G-alpha protein deficiency |
|
| YAR047C | 0.30 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR190C | 0.30 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene HIP1/YGR191W |
||
| YDR034C | 0.30 |
LYS14
|
Transcriptional activator involved in regulation of genes of the lysine biosynthesis pathway; requires 2-aminoadipate semialdehyde as co-inducer |
|
| YGL140C | 0.30 |
Putative protein of unknown function; non-essential gene; contains multiple predicted transmembrane domains |
||
| YJL093C | 0.30 |
TOK1
|
Outward-rectifier potassium channel of the plasma membrane with two pore domains in tandem, each of which forms a functional channel permeable to potassium; carboxy tail functions to prevent inner gate closures; target of K1 toxin |
|
| YLL019C | 0.30 |
KNS1
|
Nonessential putative protein kinase of unknown cellular role; member of the LAMMER family of protein kinases, which are serine/threonine kinases also capable of phosphorylating tyrosine residues |
|
| YBL100C | 0.29 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; almost completely overlaps the 5' end of ATP1 |
||
| YLR311C | 0.29 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR295C | 0.28 |
ATP14
|
Subunit h of the F0 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YIL046W-A | 0.28 |
Putative protein of unknown function; identified by expression profiling and mass spectrometry |
||
| YGR149W | 0.28 |
Putative protein of unknown function; predicted to be an integal membrane protein |
||
| YMR280C | 0.28 |
CAT8
|
Zinc cluster transcriptional activator necessary for derepression of a variety of genes under non-fermentative growth conditions, active after diauxic shift, binds carbon source responsive elements |
|
| YCL048W-A | 0.28 |
Putative protein of unknown function |
||
| YGR279C | 0.27 |
SCW4
|
Cell wall protein with similarity to glucanases; scw4 scw10 double mutants exhibit defects in mating |
|
| YFL039C | 0.27 |
ACT1
|
Actin, structural protein involved in cell polarization, endocytosis, and other cytoskeletal functions |
|
| YBL015W | 0.27 |
ACH1
|
Acetyl-coA hydrolase, primarily localized to mitochondria; phosphorylated; required for acetate utilization and for diploid pseudohyphal growth |
|
| YPR061C | 0.27 |
JID1
|
Probable Hsp40p co-chaperone, has a DnaJ-like domain and appears to be involved in ER-associated degradation of misfolded proteins containing a tightly folded cytoplasmic domain; inhibits replication of Brome mosaic virus in S. cerevisiae |
|
| YNR055C | 0.27 |
HOL1
|
Putative transporter in the major facilitator superfamily (DHA1 family) of multidrug resistance transporters; mutations in membrane-spanning domains permit cation and histidinol uptake |
|
| YKL051W | 0.27 |
SFK1
|
Plasma membrane protein that may act together with or upstream of Stt4p to generate normal levels of the essential phospholipid PI4P, at least partially mediates proper localization of Stt4p to the plasma membrane |
|
| YGR197C | 0.27 |
SNG1
|
Protein involved in nitrosoguanidine (MNNG) resistance; expression is regulated by transcription factors involved in multidrug resistance |
|
| YPR106W | 0.27 |
ISR1
|
Predicted protein kinase, overexpression causes sensitivity to staurosporine, which is a potent inhibitor of protein kinase C |
|
| YGL262W | 0.27 |
Putative protein of unknown function; null mutant displays elevated sensitivity to expression of a mutant huntingtin fragment or of alpha-synuclein; YGL262W is not an essential gene |
||
| YDR059C | 0.26 |
UBC5
|
Ubiquitin-conjugating enzyme that mediates selective degradation of short-lived and abnormal proteins, central component of the cellular stress response; expression is heat inducible |
|
| YDR540C | 0.26 |
IRC4
|
Putative protein of unknown function; null mutant displays increased levels of spontaneous Rad52p foci; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
|
| YKR042W | 0.26 |
UTH1
|
Mitochondrial outer membrane and cell wall localized SUN family member required for mitochondrial autophagy; involved in the oxidative stress response, life span during starvation, mitochondrial biogenesis, and cell death |
|
| YPR191W | 0.26 |
QCR2
|
Subunit 2 of the ubiquinol cytochrome-c reductase complex, which is a component of the mitochondrial inner membrane electron transport chain; phosphorylated; transcription is regulated by Hap1p, Hap2p/Hap3p, and heme |
|
| YGL114W | 0.26 |
Putative protein of unknown function; predicted member of the oligopeptide transporter (OPT) family of membrane transporters |
||
| YBR108W | 0.26 |
AIM3
|
Protein interacting with Rsv167p; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) and severe growth defect in minimal glycerol media |
|
| YNL037C | 0.26 |
IDH1
|
Subunit of mitochondrial NAD(+)-dependent isocitrate dehydrogenase, which catalyzes the oxidation of isocitrate to alpha-ketoglutarate in the TCA cycle |
|
| YIL060W | 0.25 |
Putative protein of unknown function; mutant accumulates less glycogen than does wild type; YIL060W is not an essential gene |
||
| YML087C | 0.25 |
AIM33
|
Putative protein of unknown function; highly conserved across species and orthologous to human CYB5R4; null mutant shows increased frequency of mitochondrial genome loss (petite formation) and severe growth defect in minimal glycerol media |
|
| YLR297W | 0.25 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YLR297W is not an essential gene; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YAR019W-A | 0.25 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR020W | 0.25 |
ATP20
|
Subunit g of the mitochondrial F1F0 ATP synthase; reversibly phosphorylated on two residues; unphosphorylated form is required for dimerization of the ATP synthase complex |
|
| YJR151C | 0.25 |
DAN4
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YCL001W-B | 0.24 |
Putative protein of unknown function; identified by homology |
||
| YIL147C | 0.24 |
SLN1
|
Histidine kinase osmosensor that regulates a MAP kinase cascade; transmembrane protein with an intracellular kinase domain that signals to Ypd1p and Ssk1p, thereby forming a phosphorelay system similar to bacterial two-component regulators |
|
| YML131W | 0.24 |
Putative protein of unknown function with similarity to oxidoreductases; HOG1 and SKO1-dependent mRNA expression is induced after osmotic shock; GFP-fusion protein localizes to the cytoplasm and is induced by the DNA-damaging agent MMS |
||
| YER163C | 0.24 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YHR145C | 0.24 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR412C-A | 0.24 |
Putative protein of unknown function |
||
| YGR250C | 0.24 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YLR315W | 0.24 |
NKP2
|
Non-essential kinetochore protein, subunit of the Ctf19 central kinetochore complex (Ctf19p-Mcm21p-Okp1p-Mcm22p-Mcm16p-Ctf3p-Chl4p-Mcm19p-Nkp1p-Nkp2p-Ame1p-Mtw1p) |
|
| YDR342C | 0.24 |
HXT7
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels |
|
| YJR152W | 0.23 |
DAL5
|
Allantoin permease; ureidosuccinate permease; expression is constitutive but sensitive to nitrogen catabolite repression |
|
| YBR056W-A | 0.23 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR056C-B |
||
| YDR322C-A | 0.23 |
TIM11
|
Subunit e of mitochondrial F1F0-ATPase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; essential for the dimeric and oligomeric state of ATP synthase |
|
| YDR445C | 0.23 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR142W | 0.23 |
BTN2
|
v-SNARE binding protein that facilitates specific protein retrieval from a late endosome to the Golgi; modulates arginine uptake, possible role in mediating pH homeostasis between the vacuole and plasma membrane H(+)-ATPase |
|
| YKL044W | 0.23 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDL224C | 0.23 |
WHI4
|
Putative RNA binding protein and partially redundant Whi3p homolog that regulates the cell size requirement for passage through Start and commitment to cell division |
|
| YOR100C | 0.22 |
CRC1
|
Mitochondrial inner membrane carnitine transporter, required for carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria during fatty acid beta-oxidation |
|
| YPR028W | 0.22 |
YOP1
|
Membrane protein that interacts with Yip1p to mediate membrane traffic; overexpression results in cell death and accumulation of internal cell membranes; regulates vesicular traffic in stressed cells |
|
| YPL271W | 0.22 |
ATP15
|
Epsilon subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; phosphorylated |
|
| YKL148C | 0.22 |
SDH1
|
Flavoprotein subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
|
| YDR446W | 0.22 |
ECM11
|
Non-essential protein apparently involved in meiosis, GFP fusion protein is present in discrete clusters in the nucleus throughout mitosis; may be involved in maintaining chromatin structure |
|
| YEL011W | 0.22 |
GLC3
|
Glycogen branching enzyme, involved in glycogen accumulation; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YIL162W | 0.22 |
SUC2
|
Invertase, sucrose hydrolyzing enzyme; a secreted, glycosylated form is regulated by glucose repression, and an intracellular, nonglycosylated enzyme is produced constitutively |
|
| YDR250C | 0.22 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YOR379C | 0.22 |
Hypothetical protein |
||
| YMR020W | 0.22 |
FMS1
|
Polyamine oxidase, converts spermine to spermidine, which is required for the essential hypusination modification of translation factor eIF-5A; also involved in pantothenic acid biosynthesis |
|
| YKL103C | 0.22 |
LAP4
|
Vacuolar aminopeptidase, often used as a marker protein in studies of autophagy and cytosol to vacuole targeting (CVT) pathway |
|
| YKL220C | 0.21 |
FRE2
|
Ferric reductase and cupric reductase, reduces siderophore-bound iron and oxidized copper prior to uptake by transporters; expression induced by low iron levels but not by low copper levels |
|
| YPL018W | 0.21 |
CTF19
|
Outer kinetochore protein, required for accurate mitotic chromosome segregation; component of the kinetochore sub-complex COMA (Ctf19p, Okp1p, Mcm21p, Ame1p) that functions as a platform for kinetochore assembly |
|
| YJR148W | 0.21 |
BAT2
|
Cytosolic branched-chain amino acid aminotransferase, homolog of murine ECA39; highly expressed during stationary phase and repressed during logarithmic phase |
|
| YPL224C | 0.21 |
MMT2
|
Putative metal transporter involved in mitochondrial iron accumulation; closely related to Mmt1p |
|
| YPR015C | 0.21 |
Putative protein of unknown function |
||
| YBR076C-A | 0.21 |
Dubious open reading frame unlikely to encode a protein; partially overlaps verified gene ECM8; identified by fungal homology and RT-PCR |
||
| YHR002W | 0.21 |
LEU5
|
Mitochondrial carrier protein involved in the accumulation of CoA in the mitochondrial matrix; homolog of human Graves disease protein; does not encode an isozyme of Leu4p, as first hypothesized |
|
| YDR533C | 0.21 |
HSP31
|
Possible chaperone and cysteine protease with similarity to E. coli Hsp31; member of the DJ-1/ThiJ/PfpI superfamily, which includes human DJ-1 involved in Parkinson's disease; exists as a dimer and contains a putative metal-binding site |
|
| YBL074C | 0.20 |
AAR2
|
Component of the U5 snRNP, required for splicing of U3 precursors; originally described as a splicing factor specifically required for splicing pre-mRNA of the MATa1 cistron |
|
| YER065C | 0.20 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YGR067C | 0.20 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YNL276C | 0.20 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene MET2/YNL277W |
||
| YAR023C | 0.20 |
Putative integral membrane protein, member of DUP240 gene family |
||
| YBR060C | 0.20 |
ORC2
|
Subunit of the origin recognition complex, which directs DNA replication by binding to replication origins and is also involved in transcriptional silencing; phosphorylated by Cdc28p |
|
| YMR189W | 0.20 |
GCV2
|
P subunit of the mitochondrial glycine decarboxylase complex, required for the catabolism of glycine to 5,10-methylene-THF; expression is regulated by levels of 5,10-methylene-THF in the cytoplasm |
|
| YLR037C | 0.20 |
DAN2
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YOR378W | 0.20 |
Putative protein of unknown function; belongs to the DHA2 family of drug:H+ antiporters; YOR378W is not an essential gene |
||
| YBL073W | 0.20 |
Hypothetical protein; open reading frame overlaps 5' end of essential AAR2 gene encoding a component of the U5 snRNP required for splicing of U3 precursors |
||
| YGR236C | 0.20 |
SPG1
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YLR203C | 0.20 |
MSS51
|
Nuclear encoded protein required for translation of COX1 mRNA; binds to Cox1 protein |
|
| YER004W | 0.20 |
FMP52
|
Protein of unknown function, localized to the mitochondrial outer membrane; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YMR272C | 0.20 |
SCS7
|
Sphingolipid alpha-hydroxylase, functions in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids, has both cytochrome b5-like and hydroxylase/desaturase domains, not essential for growth |
|
| YOR231W | 0.19 |
MKK1
|
Mitogen-activated kinase kinase involved in protein kinase C signaling pathway that controls cell integrity; upon activation by Bck1p phosphorylates downstream target, Slt2p; functionally redundant with Mkk2p |
|
| YPL058C | 0.19 |
PDR12
|
Plasma membrane ATP-binding cassette (ABC) transporter, weak-acid-inducible multidrug transporter required for weak organic acid resistance; induced by sorbate and benzoate and regulated by War1p; mutants exhibit sorbate hypersensitivity |
|
| YBL049W | 0.19 |
MOH1
|
Protein of unknown function, has homology to kinase Snf7p; not required for growth on nonfermentable carbon sources; essential for viability in stationary phase |
|
| YKL177W | 0.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene STE3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GAT3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0015755 | fructose transport(GO:0015755) |
| 0.3 | 1.3 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.3 | 0.8 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 0.7 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.2 | 2.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.6 | GO:0035786 | protein complex oligomerization(GO:0035786) |
| 0.2 | 0.2 | GO:0034225 | cellular response to zinc ion starvation(GO:0034224) regulation of transcription from RNA polymerase II promoter in response to zinc ion starvation(GO:0034225) |
| 0.2 | 0.8 | GO:0034354 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.1 | 0.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.5 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.1 | 0.6 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 1.1 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.4 | GO:0050685 | positive regulation of mRNA processing(GO:0050685) |
| 0.1 | 0.8 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.7 | GO:0045117 | azole transport(GO:0045117) |
| 0.1 | 0.4 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.1 | 0.8 | GO:0019655 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.1 | 0.4 | GO:0032371 | positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) positive regulation of sterol transport(GO:0032373) positive regulation of transmembrane transport(GO:0034764) regulation of sterol import by regulation of transcription from RNA polymerase II promoter(GO:0035968) positive regulation of sterol import by positive regulation of transcription from RNA polymerase II promoter(GO:0035969) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.1 | 0.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.6 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.3 | GO:0035948 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 0.1 | 0.3 | GO:0034762 | regulation of transmembrane transport(GO:0034762) |
| 0.1 | 0.7 | GO:0000256 | allantoin metabolic process(GO:0000255) allantoin catabolic process(GO:0000256) |
| 0.1 | 0.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.6 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.1 | 0.4 | GO:0046351 | trehalose biosynthetic process(GO:0005992) oligosaccharide biosynthetic process(GO:0009312) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.3 | GO:0030048 | actin filament-based movement(GO:0030048) actin filament-based transport(GO:0099515) |
| 0.1 | 1.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.3 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.7 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.5 | GO:0009164 | nucleoside catabolic process(GO:0009164) glycosyl compound catabolic process(GO:1901658) |
| 0.1 | 0.3 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.1 | 0.3 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.1 | 0.3 | GO:0006074 | (1->3)-beta-D-glucan metabolic process(GO:0006074) |
| 0.1 | 0.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.3 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.2 | GO:0007234 | osmosensory signaling via phosphorelay pathway(GO:0007234) |
| 0.1 | 0.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.1 | 0.6 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.1 | 0.3 | GO:0001308 | negative regulation of chromatin silencing involved in replicative cell aging(GO:0001308) |
| 0.1 | 0.5 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of metal ion(GO:0051238) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.1 | GO:0015749 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.1 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.0 | 0.3 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.1 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.0 | 0.2 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.0 | 0.5 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.4 | GO:0048017 | inositol lipid-mediated signaling(GO:0048017) |
| 0.0 | 0.1 | GO:0072529 | pyrimidine-containing compound catabolic process(GO:0072529) |
| 0.0 | 0.1 | GO:0052547 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 0.0 | 0.2 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.3 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.2 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 1.0 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.2 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.0 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.0 | 1.0 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.1 | GO:0006784 | heme a biosynthetic process(GO:0006784) heme a metabolic process(GO:0046160) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.4 | GO:0006101 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.0 | 0.1 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.0 | 0.2 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:0000435 | regulation of transcription from RNA polymerase II promoter by galactose(GO:0000431) positive regulation of transcription from RNA polymerase II promoter by galactose(GO:0000435) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0036257 | multivesicular body organization(GO:0036257) |
| 0.0 | 0.2 | GO:0005987 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.0 | 0.1 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.3 | GO:0000436 | carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 | 0.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.2 | GO:0006113 | fermentation(GO:0006113) |
| 0.0 | 0.1 | GO:0010994 | free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.5 | GO:0000767 | cell morphogenesis involved in conjugation(GO:0000767) |
| 0.0 | 0.1 | GO:0019627 | urea metabolic process(GO:0019627) |
| 0.0 | 0.1 | GO:0000709 | meiotic joint molecule formation(GO:0000709) |
| 0.0 | 0.2 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.2 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.2 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.2 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.1 | GO:0043388 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.0 | 0.1 | GO:0006343 | establishment of chromatin silencing(GO:0006343) |
| 0.0 | 0.1 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.1 | GO:0000388 | spliceosome conformational change to release U4 (or U4atac) and U1 (or U11)(GO:0000388) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.3 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0000736 | removal of nonhomologous ends(GO:0000735) double-strand break repair via single-strand annealing, removal of nonhomologous ends(GO:0000736) |
| 0.0 | 0.1 | GO:0070814 | hydrogen sulfide metabolic process(GO:0070813) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 | 0.1 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.3 | GO:0044396 | actin cortical patch assembly(GO:0000147) actin cortical patch organization(GO:0044396) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) |
| 0.0 | 0.4 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.2 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.0 | GO:0006032 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 1.1 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0000755 | cytogamy(GO:0000755) |
| 0.0 | 0.0 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.2 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:1900461 | positive regulation of cell growth(GO:0030307) positive regulation of pseudohyphal growth by positive regulation of transcription from RNA polymerase II promoter(GO:1900461) positive regulation of pseudohyphal growth(GO:2000222) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.7 | GO:0070726 | cell wall assembly(GO:0070726) |
| 0.0 | 0.1 | GO:1903138 | inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.0 | 0.0 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.1 | GO:0032973 | amino acid export(GO:0032973) amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:0042744 | hydrogen peroxide metabolic process(GO:0042743) hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.1 | GO:0031135 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.0 | 0.2 | GO:0034087 | establishment of sister chromatid cohesion(GO:0034085) establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.0 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.3 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.3 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.0 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.1 | GO:0009228 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.0 | 0.0 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.2 | GO:0000742 | karyogamy involved in conjugation with cellular fusion(GO:0000742) |
| 0.0 | 0.0 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.1 | GO:0045996 | negative regulation of transcription by pheromones(GO:0045996) negative regulation of transcription from RNA polymerase II promoter by pheromones(GO:0046020) |
| 0.0 | 0.1 | GO:0032120 | ascospore-type prospore membrane assembly(GO:0032120) membrane biogenesis(GO:0044091) membrane assembly(GO:0071709) |
| 0.0 | 0.0 | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) |
| 0.0 | 0.1 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.0 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.0 | 0.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0006276 | plasmid maintenance(GO:0006276) |
| 0.0 | 0.0 | GO:0006183 | GTP biosynthetic process(GO:0006183) GTP metabolic process(GO:0046039) |
| 0.0 | 0.0 | GO:0034204 | lipid translocation(GO:0034204) regulation of membrane lipid distribution(GO:0097035) |
| 0.0 | 0.0 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 0.1 | GO:0019400 | glycerol metabolic process(GO:0006071) alditol metabolic process(GO:0019400) |
| 0.0 | 0.1 | GO:0006672 | ceramide metabolic process(GO:0006672) ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0045010 | positive regulation of actin filament polymerization(GO:0030838) actin nucleation(GO:0045010) |
| 0.0 | 0.1 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 0.0 | 0.0 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.0 | GO:0072350 | tricarboxylic acid metabolic process(GO:0072350) |
| 0.0 | 0.0 | GO:0006108 | malate metabolic process(GO:0006108) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.6 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 2.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.6 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.5 | GO:0072379 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 0.1 | 0.4 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.1 | 0.3 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.1 | 0.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.3 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.1 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.2 | GO:0000817 | COMA complex(GO:0000817) |
| 0.0 | 0.1 | GO:0034099 | luminal surveillance complex(GO:0034099) |
| 0.0 | 0.1 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.0 | 0.6 | GO:0031160 | spore wall(GO:0031160) |
| 0.0 | 0.1 | GO:0071561 | nucleus-vacuole junction(GO:0071561) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.0 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.1 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.0 | 0.4 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.2 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.0 | 0.1 | GO:0030139 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle(GO:0030139) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0044232 | ERMES complex(GO:0032865) organelle membrane contact site(GO:0044232) ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 1.0 | GO:0009277 | fungal-type cell wall(GO:0009277) |
| 0.0 | 0.0 | GO:0030312 | cell wall(GO:0005618) external encapsulating structure(GO:0030312) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.3 | 1.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.2 | 0.8 | GO:0016878 | CoA-ligase activity(GO:0016405) acid-thiol ligase activity(GO:0016878) |
| 0.2 | 0.7 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 2.8 | GO:0020037 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.2 | 0.5 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.2 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.4 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) |
| 0.1 | 0.4 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) wide pore channel activity(GO:0022829) |
| 0.1 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 0.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.7 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.1 | 0.3 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.1 | 1.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.2 | GO:0022838 | substrate-specific channel activity(GO:0022838) |
| 0.1 | 0.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.4 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 0.2 | GO:0004575 | beta-fructofuranosidase activity(GO:0004564) sucrose alpha-glucosidase activity(GO:0004575) |
| 0.1 | 0.1 | GO:0042947 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.1 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.1 | 0.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.3 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.1 | 0.9 | GO:0015578 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.1 | 0.1 | GO:0015294 | solute:cation symporter activity(GO:0015294) |
| 0.1 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.1 | GO:0019212 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 0.2 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.1 | 0.2 | GO:0004375 | glycine dehydrogenase (decarboxylating) activity(GO:0004375) oxidoreductase activity, acting on the CH-NH2 group of donors, disulfide as acceptor(GO:0016642) |
| 0.1 | 0.5 | GO:0016723 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 0.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 0.3 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.1 | 0.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.6 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.6 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0008972 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.0 | 0.3 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 0.0 | 0.4 | GO:0099600 | transmembrane signaling receptor activity(GO:0004888) signaling receptor activity(GO:0038023) transmembrane receptor activity(GO:0099600) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.2 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.0 | 0.1 | GO:0042973 | glucan endo-1,3-beta-D-glucosidase activity(GO:0042973) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0042927 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.0 | 0.2 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 1.1 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.3 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0030414 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.0 | 0.2 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.2 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.3 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.1 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0052742 | phosphatidylinositol kinase activity(GO:0052742) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0050253 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 1.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0097372 | NAD-dependent histone deacetylase activity (H3-K18 specific)(GO:0097372) |
| 0.0 | 0.1 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0042123 | glucanosyltransferase activity(GO:0042123) 1,3-beta-glucanosyltransferase activity(GO:0042124) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.1 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.7 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.1 | GO:0034318 | alcohol O-acetyltransferase activity(GO:0004026) alcohol O-acyltransferase activity(GO:0034318) |
| 0.0 | 0.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.0 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.0 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.0 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.0 | 0.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 0.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.1 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.1 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.0 | REACTOME MEIOSIS | Genes involved in Meiosis |