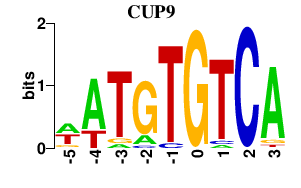
Results for CUP9
Z-value: 0.98
Transcription factors associated with CUP9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUP9
|
S000006098 | Homeodomain-containing transcriptional repressor |
Activity-expression correlation:
Activity profile of CUP9 motif
Sorted Z-values of CUP9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YMR017W | 4.61 |
SPO20
|
Meiosis-specific subunit of the t-SNARE complex, required for prospore membrane formation during sporulation; similar to but not functionally redundant with Sec9p; SNAP-25 homolog |
|
| YGR087C | 4.20 |
PDC6
|
Minor isoform of pyruvate decarboxylase, decarboxylates pyruvate to acetaldehyde, involved in amino acid catabolism; transcription is glucose- and ethanol-dependent, and is strongly induced during sulfur limitation |
|
| YHR139C | 3.32 |
SPS100
|
Protein required for spore wall maturation; expressed during sporulation; may be a component of the spore wall |
|
| YOR100C | 3.10 |
CRC1
|
Mitochondrial inner membrane carnitine transporter, required for carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria during fatty acid beta-oxidation |
|
| YMR107W | 2.93 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YGR059W | 2.86 |
SPR3
|
Sporulation-specific homolog of the yeast CDC3/10/11/12 family of bud neck microfilament genes; septin protein involved in sporulation; regulated by ABFI |
|
| YOR255W | 2.62 |
OSW1
|
Protein involved in sporulation; required for the construction of the outer spore wall layers; required for proper localization of Spo14p |
|
| YIL057C | 2.55 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YFL011W | 2.51 |
HXT10
|
Putative hexose transporter, expressed at low levels and expression is repressed by glucose |
|
| YER065C | 2.50 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YLR307W | 2.39 |
CDA1
|
Chitin deacetylase, together with Cda2p involved in the biosynthesis ascospore wall component, chitosan; required for proper rigidity of the ascospore wall |
|
| YFL012W | 2.39 |
Putative protein of unknown function; transcribed during sporulation; null mutant exhibits increased resistance to rapamycin |
||
| YKL109W | 2.31 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YDL114W | 2.31 |
Putative protein of unknown function with similarity to acyl-carrier-protein reductases; YDL114W is not an essential gene |
||
| YAL062W | 2.25 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YMR118C | 2.23 |
Protein of unknown function with similarity to succinate dehydrogenase cytochrome b subunit; YMR118C is not an essential gene |
||
| YLR307C-A | 2.22 |
Putative protein of unknown function |
||
| YCL048W | 2.18 |
SPS22
|
Protein of unknown function, redundant with Sps2p for the organization of the beta-glucan layer of the spore wall |
|
| YER106W | 2.16 |
MAM1
|
Monopolin, kinetochore associated protein involved in chromosome attachment to meiotic spindle |
|
| YML089C | 2.14 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression induced by calcium shortage |
||
| YGL205W | 2.07 |
POX1
|
Fatty-acyl coenzyme A oxidase, involved in the fatty acid beta-oxidation pathway; localized to the peroxisomal matrix |
|
| YPL021W | 2.06 |
ECM23
|
Non-essential protein of unconfirmed function; affects pre-rRNA processing, may act as a negative regulator of the transcription of genes involved in pseudohyphal growth; homologous to Srd1p |
|
| YNL318C | 1.84 |
HXT14
|
Protein with similarity to hexose transporter family members, expression is induced in low glucose and repressed in high glucose; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YFR023W | 1.84 |
PES4
|
Poly(A) binding protein, suppressor of DNA polymerase epsilon mutation, similar to Mip6p |
|
| YBR180W | 1.80 |
DTR1
|
Putative dityrosine transporter, required for spore wall synthesis; expressed during sporulation; member of the major facilitator superfamily (DHA1 family) of multidrug resistance transporters |
|
| YKL177W | 1.72 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene STE3 |
||
| YBR117C | 1.64 |
TKL2
|
Transketolase, similar to Tkl1p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YPR078C | 1.63 |
Putative protein of unknown function; possible role in DNA metabolism and/or in genome stability; expression is heat-inducible |
||
| YLR136C | 1.62 |
TIS11
|
mRNA-binding protein expressed during iron starvation; binds to a sequence element in the 3'-untranslated regions of specific mRNAs to mediate their degradation; involved in iron homeostasis |
|
| YNL117W | 1.62 |
MLS1
|
Malate synthase, enzyme of the glyoxylate cycle, involved in utilization of non-fermentable carbon sources; expression is subject to carbon catabolite repression; localizes in peroxisomes during growth in oleic acid medium |
|
| YAL067C | 1.56 |
SEO1
|
Putative permease, member of the allantoate transporter subfamily of the major facilitator superfamily; mutation confers resistance to ethionine sulfoxide |
|
| YFL051C | 1.54 |
Putative protein of unknown function; YFL051C is not an essential gene |
||
| YAR069C | 1.52 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR218C | 1.48 |
SPR28
|
Sporulation-specific homolog of the yeast CDC3/10/11/12 family of bud neck microfilament genes; meiotic septin expressed at high levels during meiotic divisions and ascospore formation |
|
| YJR154W | 1.47 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YML090W | 1.46 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YML089C; exhibits growth defect on a non-fermentable (respiratory) carbon source |
||
| YAL018C | 1.46 |
Putative protein of unknown function |
||
| YAR070C | 1.46 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR346C | 1.44 |
Putative protein of unknown function found in mitochondria; expression is regulated by transcription factors involved in pleiotropic drug resistance, Pdr1p and Yrr1p; YLR346C is not an essential gene |
||
| YML091C | 1.44 |
RPM2
|
Protein subunit of mitochondrial RNase P, has roles in nuclear transcription, cytoplasmic and mitochondrial RNA processing, and mitochondrial translation; distributed to mitochondria, cytoplasmic processing bodies, and the nucleus |
|
| YAR050W | 1.43 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YBR051W | 1.41 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partiallly overlaps the REG2/YBR050C regulatory subunit of the Glc7p type-1 protein phosphatase |
||
| YDR281C | 1.41 |
PHM6
|
Protein of unknown function, expression is regulated by phosphate levels |
|
| YJR095W | 1.36 |
SFC1
|
Mitochondrial succinate-fumarate transporter, transports succinate into and fumarate out of the mitochondrion; required for ethanol and acetate utilization |
|
| YHR015W | 1.36 |
MIP6
|
Putative RNA-binding protein, interacts with Mex67p, which is a component of the nuclear pore involved in nuclear mRNA export |
|
| YAR053W | 1.34 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YCL054W | 1.33 |
SPB1
|
AdoMet-dependent methyltransferase involved in rRNA processing and 60S ribosomal subunit maturation; methylates G2922 in the tRNA docking site of the large subunit rRNA and in the absence of snR52, U2921; suppressor of PAB1 mutants |
|
| YNR062C | 1.32 |
Putative membrane protein of unknown function |
||
| YKR102W | 1.32 |
FLO10
|
Lectin-like protein with similarity to Flo1p, thought to be involved in flocculation |
|
| YNL183C | 1.30 |
NPR1
|
Protein kinase that stabilizes several plasma membrane amino acid transporters by antagonizing their ubiquitin-mediated degradation |
|
| YER024W | 1.30 |
YAT2
|
Carnitine acetyltransferase; has similarity to Yat1p, which is a carnitine acetyltransferase associated with the mitochondrial outer membrane |
|
| YDR446W | 1.29 |
ECM11
|
Non-essential protein apparently involved in meiosis, GFP fusion protein is present in discrete clusters in the nucleus throughout mitosis; may be involved in maintaining chromatin structure |
|
| YPR169W-A | 1.28 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps two other dubious ORFs: YPR170C and YPR170W-B |
||
| YLR308W | 1.28 |
CDA2
|
Chitin deacetylase, together with Cda1p involved in the biosynthesis ascospore wall component, chitosan; required for proper rigidity of the ascospore wall |
|
| YGR243W | 1.27 |
FMP43
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YJR048W | 1.26 |
CYC1
|
Cytochrome c, isoform 1; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YGR088W | 1.25 |
CTT1
|
Cytosolic catalase T, has a role in protection from oxidative damage by hydrogen peroxide |
|
| YNR063W | 1.24 |
Putative zinc-cluster protein of unknown function |
||
| YBR144C | 1.23 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR144C is not an essential gene |
||
| YDL210W | 1.23 |
UGA4
|
Permease that serves as a gamma-aminobutyrate (GABA) transport protein involved in the utilization of GABA as a nitrogen source; catalyzes the transport of putrescine and delta-aminolevulinic acid (ALA); localized to the vacuolar membrane |
|
| YAR047C | 1.22 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR048W | 1.22 |
YHK8
|
Presumed antiporter of the DHA1 family of multidrug resistance transporters; contains 12 predicted transmembrane spans; expression of gene is up-regulated in cells exhibiting reduced susceptibility to azoles |
|
| YDR242W | 1.22 |
AMD2
|
Putative amidase |
|
| YBR050C | 1.20 |
REG2
|
Regulatory subunit of the Glc7p type-1 protein phosphatase; involved with Reg1p, Glc7p, and Snf1p in regulation of glucose-repressible genes, also involved in glucose-induced proteolysis of maltose permease |
|
| YNR064C | 1.20 |
Epoxide hydrolase, member of the alpha/beta hydrolase fold family; may have a role in detoxification of epoxides |
||
| YAL066W | 1.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGL170C | 1.17 |
SPO74
|
Component of the meiotic outer plaque of the spindle pole body, involved in modifying the meiotic outer plaque that is required prior to prospore membrane formation |
|
| YHR145C | 1.16 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YKL178C | 1.15 |
STE3
|
Receptor for a factor receptor, transcribed in alpha cells and required for mating by alpha cells, couples to MAP kinase cascade to mediate pheromone response; ligand bound receptors are endocytosed and recycled to the plasma membrane; GPCR |
|
| YLR327C | 1.12 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YBR296C | 1.11 |
PHO89
|
Na+/Pi cotransporter, active in early growth phase; similar to phosphate transporters of Neurospora crassa; transcription regulated by inorganic phosphate concentrations and Pho4p |
|
| YJL045W | 1.11 |
Minor succinate dehydrogenase isozyme; homologous to Sdh1p, the major isozyme reponsible for the oxidation of succinate and transfer of electrons to ubiquinone; induced during the diauxic shift in a Cat8p-dependent manner |
||
| YPR002W | 1.06 |
PDH1
|
Mitochondrial protein that participates in respiration, induced by diauxic shift; homologous to E. coli PrpD, may take part in the conversion of 2-methylcitrate to 2-methylisocitrate |
|
| YHR139C-A | 1.06 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR194C-B | 1.05 |
Putative protein of unknown function |
||
| YFL012W-A | 1.05 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene IES1/YFL013C |
||
| YHL040C | 1.05 |
ARN1
|
Transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; responsible for uptake of iron bound to ferrirubin, ferrirhodin, and related siderophores |
|
| YOR186W | 1.04 |
Putative protein of unknown function; proper regulation of expression during heat stress is sphingolipid-dependent |
||
| YMR244W | 1.03 |
Putative protein of unknown function |
||
| YDR445C | 1.02 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL117W | 1.00 |
Putative protein of unknown function |
||
| YOL052C-A | 0.98 |
DDR2
|
Multistress response protein, expression is activated by a variety of xenobiotic agents and environmental or physiological stresses |
|
| YFR032C-B | 0.96 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YEL049W | 0.96 |
PAU2
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YKL161C | 0.96 |
Protein kinase implicated in the Slt2p mitogen-activated (MAP) kinase signaling pathway; associates with Rlm1p |
||
| YKR034W | 0.96 |
DAL80
|
Negative regulator of genes in multiple nitrogen degradation pathways; expression is regulated by nitrogen levels and by Gln3p; member of the GATA-binding family, forms homodimers and heterodimers with Deh1p |
|
| YPL092W | 0.95 |
SSU1
|
Plasma membrane sulfite pump involved in sulfite metabolism and required for efficient sulfite efflux; major facilitator superfamily protein |
|
| YGR258C | 0.95 |
RAD2
|
Single-stranded DNA endonuclease, cleaves single-stranded DNA during nucleotide excision repair to excise damaged DNA; subunit of Nucleotide Excision Repair Factor 3 (NEF3); homolog of human XPG protein |
|
| YCL018W | 0.93 |
LEU2
|
Beta-isopropylmalate dehydrogenase (IMDH), catalyzes the third step in the leucine biosynthesis pathway |
|
| YBR250W | 0.93 |
SPO23
|
Protein of unknown function; associates with meiosis-specific protein Spo1p |
|
| YPL222C-A | 0.92 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YBR179C | 0.92 |
FZO1
|
Mitochondrial integral membrane protein involved in mitochondrial fusion and maintenance of the mitochondrial genome; contains N-terminal GTPase domain |
|
| YOR237W | 0.92 |
HES1
|
Protein implicated in the regulation of ergosterol biosynthesis; one of a seven member gene family with a common essential function and non-essential unique functions; similar to human oxysterol binding protein (OSBP) |
|
| YMR280C | 0.91 |
CAT8
|
Zinc cluster transcriptional activator necessary for derepression of a variety of genes under non-fermentative growth conditions, active after diauxic shift, binds carbon source responsive elements |
|
| YIL160C | 0.91 |
POT1
|
3-ketoacyl-CoA thiolase with broad chain length specificity, cleaves 3-ketoacyl-CoA into acyl-CoA and acetyl-CoA during beta-oxidation of fatty acids |
|
| YAR035W | 0.91 |
YAT1
|
Outer mitochondrial carnitine acetyltransferase, minor ethanol-inducible enzyme involved in transport of activated acyl groups from the cytoplasm into the mitochondrial matrix; phosphorylated |
|
| YKR105C | 0.91 |
VBA5
|
Putative transporter of the Major Facilitator Superfamily (MFS); proposed role as a basic amino acid permease based on phylogeny |
|
| YOL082W | 0.90 |
ATG19
|
Receptor protein for the cytoplasm-to-vacuole targeting (Cvt) pathway; recognizes cargo proteins aminopeptidase I (Lap4p) and alpha-mannosidase (Ams1p) and delivers them to the preautophagosomal structure for packaging into Cvt vesicles |
|
| YHR095W | 0.89 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBR094W | 0.89 |
PBY1
|
Putative tubulin tyrosine ligase associated with P-bodies |
|
| YMR040W | 0.87 |
YET2
|
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; homolog of human BAP31 protein |
|
| YDR042C | 0.86 |
Putative protein of unknown function; expression is increased in ssu72-ts69 mutant |
||
| YOL050C | 0.86 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps verified gene GAL11; deletion confers sensitivity to 4-(N-(S-glutathionylacetyl)amino) phenylarsenoxide (GSAO) |
||
| YFR029W | 0.85 |
PTR3
|
Component of the SPS plasma membrane amino acid sensor system (Ssy1p-Ptr3p-Ssy5p), which senses external amino acid concentration and transmits intracellular signals that result in regulation of expression of amino acid permease genes |
|
| YPL147W | 0.85 |
PXA1
|
Subunit of a heterodimeric peroxisomal ATP-binding cassette transporter complex (Pxa1p-Pxa2p), required for import of long-chain fatty acids into peroxisomes; similarity to human adrenoleukodystrophy transporter and ALD-related proteins |
|
| YJR151C | 0.84 |
DAN4
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YFL013C | 0.84 |
IES1
|
Subunit of the INO80 chromatin remodeling complex |
|
| YCL069W | 0.84 |
VBA3
|
Permease of basic amino acids in the vacuolar membrane |
|
| YNL180C | 0.83 |
RHO5
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely involved in protein kinase C (Pkc1p)-dependent signal transduction pathway that controls cell integrity |
|
| YDL085W | 0.83 |
NDE2
|
Mitochondrial external NADH dehydrogenase, catalyzes the oxidation of cytosolic NADH; Nde1p and Nde2p are involved in providing the cytosolic NADH to the mitochondrial respiratory chain |
|
| YOL024W | 0.82 |
Putative protein of unknown function, predicted to have thiol-disulfide oxidoreductase active site |
||
| YDL204W | 0.82 |
RTN2
|
Protein of unknown function; has similarity to mammalian reticulon proteins; member of the RTNLA (reticulon-like A) subfamily |
|
| YBR040W | 0.82 |
FIG1
|
Integral membrane protein required for efficient mating; may participate in or regulate the low affinity Ca2+ influx system, which affects intracellular signaling and cell-cell fusion during mating |
|
| YMR090W | 0.81 |
Putative protein of unknown function with similarity to DTDP-glucose 4,6-dehydratases; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YMR090W is not an essential gene |
||
| YHL035C | 0.81 |
VMR1
|
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; member of the ATP-binding cassette (ABC) family; potential Cdc28p substrate; detected in purified mitochondria in high-throughput studies |
|
| YPR193C | 0.81 |
HPA2
|
Tetrameric histone acetyltransferase with similarity to Gcn5p, Hat1p, Elp3p, and Hpa3p; acetylates histones H3 and H4 in vitro and exhibits autoacetylation activity |
|
| YDR043C | 0.80 |
NRG1
|
Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters; mediates glucose repression and negatively regulates a variety of processes including filamentous growth and alkaline pH response |
|
| YNL179C | 0.80 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YFL030W | 0.79 |
AGX1
|
Alanine:glyoxylate aminotransferase (AGT), catalyzes the synthesis of glycine from glyoxylate, which is one of three pathways for glycine biosynthesis in yeast; has similarity to mammalian and plant alanine:glyoxylate aminotransferases |
|
| YML058W-A | 0.79 |
HUG1
|
Protein involved in the Mec1p-mediated checkpoint pathway that responds to DNA damage or replication arrest, transcription is induced by DNA damage |
|
| YOR211C | 0.78 |
MGM1
|
Mitochondrial GTPase related to dynamin, present in a complex containing Ugo1p and Fzo1p; required for normal morphology of cristae and for stability of Tim11p; homolog of human OPA1 involved in autosomal dominant optic atrophy |
|
| YKL163W | 0.78 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YBR072W | 0.78 |
HSP26
|
Small heat shock protein (sHSP) with chaperone activity; forms hollow, sphere-shaped oligomers that suppress unfolded proteins aggregation; oligomer activation requires a heat-induced conformational change; not expressed in unstressed cells |
|
| YER116C | 0.76 |
SLX8
|
Subunit of the Slx5-Slx8 substrate-specific ubiquitin ligase complex; stimulated by prior attachment of SUMO to the substrate |
|
| YDR310C | 0.76 |
SUM1
|
Transcriptional repressor required for mitotic repression of middle sporulation-specific genes; involved in telomere maintenance, regulated by the pachytene checkpoint |
|
| YGL163C | 0.76 |
RAD54
|
DNA-dependent ATPase, stimulates strand exchange by modifying the topology of double-stranded DNA; involved in the recombinational repair of double-strand breaks in DNA during vegetative growth and meiosis; member of the SWI/SNF family |
|
| YDR223W | 0.75 |
CRF1
|
Transcriptional corepressor involved in the regulation of ribosomal protein gene transcription via the TOR signaling pathway and protein kinase A, phosphorylated by activated Yak1p which promotes accumulation of Crf1p in the nucleus |
|
| YHR150W | 0.75 |
PEX28
|
Peroxisomal integral membrane peroxin, involved in the regulation of peroxisomal size, number and distribution; genetic interactions suggest that Pex28p and Pex29p act at steps upstream of those mediated by Pex30p, Pex31p, and Pex32p |
|
| YJL043W | 0.75 |
Putative protein of unknown function; YJL043W is a non-essential gene |
||
| YOR192C-C | 0.75 |
Putative protein of unknown function; identified by expression profiling and mass spectrometry |
||
| YER097W | 0.75 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR195W | 0.74 |
ICY1
|
Protein of unknown function, required for viability in rich media of cells lacking mitochondrial DNA; mutants have an invasive growth defect with elongated morphology; induced by amino acid starvation |
|
| YDL115C | 0.74 |
IWR1
|
Protein of unknown function, deletion causes hypersensitivity to the K1 killer toxin |
|
| YGL118C | 0.74 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YIL037C | 0.74 |
PRM2
|
Pheromone-regulated protein, predicted to have 4 transmembrane segments and a coiled coil domain; regulated by Ste12p |
|
| YOR192C | 0.74 |
THI72
|
Transporter of thiamine or related compound; shares sequence similarity with Thi7p |
|
| YAR060C | 0.74 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR540C | 0.72 |
IRC4
|
Putative protein of unknown function; null mutant displays increased levels of spontaneous Rad52p foci; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
|
| YNL328C | 0.72 |
MDJ2
|
Constituent of the mitochondrial import motor associated with the presequence translocase; function overlaps with that of Pam18p; stimulates the ATPase activity of Ssc1p to drive mitochondrial import; contains a J domain |
|
| YBR215W | 0.71 |
HPC2
|
Subunit of the HIR complex, a nucleosome assembly complex involved in regulation of histone gene transcription; mutants display synthetic defects with subunits of FACT, a complex that allows passage of RNA Pol II through nucleosomes |
|
| YGR110W | 0.71 |
Putative protein of unknown function; transcription is increased in response to genotoxic stress; plays a role in restricting Ty1 transposition |
||
| YAL063C | 0.70 |
FLO9
|
Lectin-like protein with similarity to Flo1p, thought to be expressed and involved in flocculation |
|
| YHR211W | 0.69 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YGR065C | 0.69 |
VHT1
|
High-affinity plasma membrane H+-biotin (vitamin H) symporter; mutation results in fatty acid auxotrophy; 12 transmembrane domain containing major facilitator subfamily member; mRNA levels negatively regulated by iron deprivation and biotin |
|
| YOR289W | 0.69 |
Putative protein of unknown function; transcription induced by the unfolded protein response; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and the nucleus |
||
| YJR047C | 0.68 |
ANB1
|
Translation initiation factor eIF-5A, promotes formation of the first peptide bond; similar to and functionally redundant with Hyp2p; undergoes an essential hypusination modification; expressed under anaerobic conditions |
|
| YMR317W | 0.68 |
Putative protein of unknown function with some similarity to sialidase from Trypanosoma; YMR317W is not an essential gene |
||
| YDR528W | 0.67 |
HLR1
|
Protein involved in regulation of cell wall composition and integrity and response to osmotic stress; overproduction suppresses a lysis sensitive PKC mutation; similar to Lre1p, which functions antagonistically to protein kinase A |
|
| YKL217W | 0.67 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YDR259C | 0.66 |
YAP6
|
Putative basic leucine zipper (bZIP) transcription factor; overexpression increases sodium and lithium tolerance |
|
| YER014C-A | 0.66 |
BUD25
|
Protein involved in bud-site selection; diploid mutants display a random budding pattern instead of the wild-type bipolar pattern |
|
| YLL056C | 0.65 |
Putative protein of unknown function, transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in pleiotropic drug resistance (PDR) phenomenon; YLL056C is not an essential gene |
||
| YMR316C-B | 0.65 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR054C | 0.65 |
OSW2
|
Protein of unknown function proposed to be involved in the assembly of the spore wall |
|
| YOR365C | 0.65 |
Putative protein of unknown function; YOR365C is not an essential protein |
||
| YDR119W-A | 0.64 |
Putative protein of unknown function |
||
| YPL271W | 0.64 |
ATP15
|
Epsilon subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; phosphorylated |
|
| YMR014W | 0.64 |
BUD22
|
Protein involved in bud-site selection; diploid mutants display a random budding pattern instead of the wild-type bipolar pattern |
|
| YIR014W | 0.63 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; expression directly regulated by the metabolic and meiotic transcriptional regulator Ume6p; YIR014W is a non-essential gene |
||
| YOR345C | 0.62 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene REV1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YBR018C | 0.62 |
GAL7
|
Galactose-1-phosphate uridyl transferase, synthesizes glucose-1-phosphate and UDP-galactose from UDP-D-glucose and alpha-D-galactose-1-phosphate in the second step of galactose catabolism |
|
| YCR091W | 0.62 |
KIN82
|
Putative serine/threonine protein kinase, most similar to cyclic nucleotide-dependent protein kinase subfamily and the protein kinase C subfamily |
|
| YER098W | 0.62 |
UBP9
|
Ubiquitin carboxyl-terminal hydrolase, ubiquitin-specific protease that cleaves ubiquitin-protein fusions |
|
| YKL202W | 0.61 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR347C | 0.61 |
PYK2
|
Pyruvate kinase that appears to be modulated by phosphorylation; PYK2 transcription is repressed by glucose, and Pyk2p may be active under low glycolytic flux |
|
| YMR081C | 0.61 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YIR029W | 0.61 |
DAL2
|
Allantoicase, converts allantoate to urea and ureidoglycolate in the second step of allantoin degradation; expression sensitive to nitrogen catabolite repression and induced by allophanate, an intermediate in allantoin degradation |
|
| YOR306C | 0.60 |
MCH5
|
Plasma membrane riboflavin transporter; facilitates the uptake of vitamin B2; required for FAD-dependent processes; sequence similarity to mammalian monocarboxylate permeases, however mutants are not deficient in monocarboxylate transport |
|
| YGR182C | 0.60 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF TIM13/YGR181W |
||
| YPR065W | 0.60 |
ROX1
|
Heme-dependent repressor of hypoxic genes; contains an HMG domain that is responsible for DNA bending activity |
|
| YMR013C | 0.60 |
SEC59
|
Dolichol kinase, catalyzes the terminal step in dolichyl monophosphate (Dol-P) biosynthesis; required for viability and for normal rates of lipid intermediate synthesis and protein N-glycosylation |
|
| YOR339C | 0.60 |
UBC11
|
Ubiquitin-conjugating enzyme most similar in sequence to Xenopus ubiquitin-conjugating enzyme E2-C, but not a true functional homolog of this E2; unlike E2-C, not required for the degradation of mitotic cyclin Clb2 |
|
| YOL100W | 0.59 |
PKH2
|
Serine/threonine protein kinase involved in sphingolipid-mediated signaling pathway that controls endocytosis; activates Ypk1p and Ykr2p, components of signaling cascade required for maintenance of cell wall integrity; redundant with Pkh1p |
|
| YHR093W | 0.59 |
AHT1
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; multicopy suppressor of glucose transport defects, likely due to the presence of an HXT4 regulatory element in the region |
|
| YER061C | 0.59 |
CEM1
|
Mitochondrial beta-keto-acyl synthase with possible role in fatty acid synthesis; required for mitochondrial respiration |
|
| YBR269C | 0.58 |
FMP21
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YPL018W | 0.58 |
CTF19
|
Outer kinetochore protein, required for accurate mitotic chromosome segregation; component of the kinetochore sub-complex COMA (Ctf19p, Okp1p, Mcm21p, Ame1p) that functions as a platform for kinetochore assembly |
|
| YHR033W | 0.57 |
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm |
||
| YMR016C | 0.57 |
SOK2
|
Nuclear protein that plays a regulatory role in the cyclic AMP (cAMP)-dependent protein kinase (PKA) signal transduction pathway; negatively regulates pseudohyphal differentiation; homologous to several transcription factors |
|
| YFL003C | 0.57 |
MSH4
|
Protein involved in meiotic recombination, required for normal levels of crossing over, colocalizes with Zip2p to discrete foci on meiotic chromosomes, has homology to bacterial MutS protein |
|
| YML047C | 0.57 |
PRM6
|
Pheromone-regulated protein, predicted to have 2 transmembrane segments; regulated by Ste12p during mating |
|
| YOR346W | 0.56 |
REV1
|
Deoxycytidyl transferase, forms a complex with the subunits of DNA polymerase zeta, Rev3p and Rev7p; involved in repair of abasic sites in damaged DNA |
|
| YPL258C | 0.55 |
THI21
|
Hydroxymethylpyrimidine phosphate kinase, involved in the last steps in thiamine biosynthesis; member of a gene family with THI20 and THI22; functionally redundant with Thi20p |
|
| YLR369W | 0.54 |
SSQ1
|
Mitochondrial hsp70-type molecular chaperone, required for assembly of iron/sulfur clusters into proteins at a step after cluster synthesis, and for maturation of Yfh1p, which is a homolog of human frataxin implicated in Friedreich's ataxia |
|
| YGR039W | 0.54 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the Autonomously Replicating Sequence ARS722 |
||
| YDR542W | 0.54 |
PAU10
|
Hypothetical protein |
|
| YFR012W | 0.54 |
Putative protein of unknown function |
||
| YIL077C | 0.54 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; deletion confers sensitivity to 4-(N-(S-glutathionylacetyl)amino) phenylarsenoxide (GSAO) |
||
| YDL187C | 0.53 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLL047W | 0.53 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified gene RNP1 |
||
| YIL023C | 0.53 |
YKE4
|
Zinc transporter; localizes to the ER; null mutant is sensitive to calcofluor white, leads to zinc accumulation in cytosol; ortholog of the mouse KE4 and member of the ZIP (ZRT, IRT-like Protein) family |
|
| YJR049C | 0.53 |
UTR1
|
ATP-NADH kinase; phosphorylates both NAD and NADH; active as a hexamer; enhances the activity of ferric reductase (Fre1p) |
|
| YHR212C | 0.53 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YKR015C | 0.53 |
Putative protein of unknown function |
||
| YHR023W | 0.52 |
MYO1
|
Type II myosin heavy chain, required for wild-type cytokinesis and cell separation; localizes to the actomyosin ring; binds to myosin light chains Mlc1p and Mlc2p through its IQ1 and IQ2 motifs respectively |
|
| YBR147W | 0.52 |
RTC2
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays fluconazole resistance and suppresses cdc13-1 temperature sensitivity |
|
| YMR018W | 0.52 |
Putative protein of unknown function with similarity to human PEX5Rp (peroxin protein 5 related protein); transcription increases during colony development similar to genes involved in peroxisome biogenesis; YMR018W is not an essential gene |
||
| YMR141C | 0.52 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNL052W | 0.51 |
COX5A
|
Subunit Va of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; predominantly expressed during aerobic growth while its isoform Vb (Cox5Bp) is expressed during anaerobic growth |
|
| YOR382W | 0.51 |
FIT2
|
Mannoprotein that is incorporated into the cell wall via a glycosylphosphatidylinositol (GPI) anchor, involved in the retention of siderophore-iron in the cell wall |
Network of associatons between targets according to the STRING database.
First level regulatory network of CUP9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.2 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.7 | 2.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.7 | 4.5 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.7 | 2.2 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.5 | 4.8 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.5 | 1.9 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.5 | 2.4 | GO:0006032 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.4 | 1.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.4 | 1.1 | GO:0019543 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 0.4 | 1.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.3 | 0.7 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.3 | 2.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.3 | 1.0 | GO:0090295 | nitrogen catabolite repression of transcription(GO:0090295) |
| 0.3 | 2.5 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.3 | 0.9 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.3 | 11.6 | GO:0070591 | ascospore wall assembly(GO:0030476) spore wall assembly(GO:0042244) spore wall biogenesis(GO:0070590) ascospore wall biogenesis(GO:0070591) fungal-type cell wall assembly(GO:0071940) |
| 0.3 | 2.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 1.1 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.3 | 0.9 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.3 | 3.8 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 0.3 | 0.8 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.3 | 0.8 | GO:0042710 | biofilm formation(GO:0042710) |
| 0.3 | 1.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.2 | 1.0 | GO:0015809 | arginine transport(GO:0015809) |
| 0.2 | 0.5 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.2 | 1.9 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.2 | 0.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 1.2 | GO:0019660 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.2 | 0.9 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.2 | 0.9 | GO:0043200 | response to amino acid(GO:0043200) |
| 0.2 | 1.9 | GO:0000755 | cytogamy(GO:0000755) |
| 0.2 | 0.6 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.2 | 2.2 | GO:0015893 | drug transport(GO:0015893) |
| 0.2 | 0.8 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 0.9 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.2 | 0.8 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) |
| 0.2 | 0.8 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.2 | 0.5 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of metal ion(GO:0051238) sequestering of iron ion(GO:0097577) |
| 0.2 | 1.2 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.2 | 0.5 | GO:0000916 | actomyosin contractile ring contraction(GO:0000916) contractile ring contraction(GO:0036213) |
| 0.2 | 1.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.2 | 0.8 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) |
| 0.2 | 0.8 | GO:0043954 | cellular component maintenance(GO:0043954) |
| 0.2 | 0.5 | GO:0000304 | response to singlet oxygen(GO:0000304) |
| 0.2 | 0.2 | GO:0001666 | response to hypoxia(GO:0001666) cellular response to hypoxia(GO:0071456) |
| 0.2 | 0.8 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.2 | 1.1 | GO:0034762 | regulation of transmembrane transport(GO:0034762) |
| 0.2 | 0.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.6 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 0.6 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.1 | 1.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.6 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 0.7 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.7 | GO:0045764 | positive regulation of cellular amino acid metabolic process(GO:0045764) |
| 0.1 | 0.5 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 0.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 1.3 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 5.6 | GO:0034293 | sexual sporulation(GO:0034293) sexual sporulation resulting in formation of a cellular spore(GO:0043935) |
| 0.1 | 0.6 | GO:0034486 | amino acid export(GO:0032973) amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 0.1 | 0.5 | GO:1903137 | inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.1 | 0.1 | GO:1900544 | positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.1 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.4 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.1 | 1.6 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.1 | 1.3 | GO:0000436 | carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.1 | 1.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.4 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 | 1.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.4 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.1 | 0.7 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.2 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.1 | 0.5 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.2 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.3 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 0.4 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.1 | 0.3 | GO:0098610 | agglutination involved in conjugation with cellular fusion(GO:0000752) agglutination involved in conjugation(GO:0000771) heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) cell-cell adhesion(GO:0098609) adhesion between unicellular organisms(GO:0098610) multi organism cell adhesion(GO:0098740) cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.1 | 0.3 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.1 | 0.4 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 1.6 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 0.5 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.4 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.3 | GO:0034517 | ribophagy(GO:0034517) |
| 0.1 | 1.5 | GO:0031503 | protein complex localization(GO:0031503) |
| 0.1 | 0.2 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.1 | 0.3 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 0.2 | GO:0043007 | maintenance of rDNA(GO:0043007) |
| 0.1 | 1.0 | GO:0042724 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.1 | 0.7 | GO:0045910 | negative regulation of DNA recombination(GO:0045910) |
| 0.1 | 0.4 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 0.3 | GO:0007534 | gene conversion at mating-type locus(GO:0007534) |
| 0.1 | 0.6 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.1 | GO:0010942 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.1 | 0.7 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.7 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.3 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.1 | 0.3 | GO:0006101 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.1 | 0.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.1 | GO:0001198 | regulation of mating-type specific transcription from RNA polymerase II promoter(GO:0001196) negative regulation of mating-type specific transcription from RNA polymerase II promoter(GO:0001198) negative regulation of mating-type specific transcription, DNA-templated(GO:0045894) |
| 0.1 | 0.8 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 0.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.1 | GO:0000078 | obsolete cytokinesis after mitosis checkpoint(GO:0000078) |
| 0.1 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.1 | 0.3 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.1 | 0.1 | GO:0006827 | high-affinity iron ion transmembrane transport(GO:0006827) |
| 0.1 | 0.2 | GO:0090294 | nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) nitrogen catabolite activation of transcription(GO:0090294) |
| 0.1 | 0.1 | GO:0071248 | cellular response to metal ion(GO:0071248) |
| 0.1 | 0.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.2 | GO:0000196 | MAPK cascade involved in cell wall organization or biogenesis(GO:0000196) |
| 0.1 | 0.1 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.1 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) |
| 0.1 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.1 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 0.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.2 | GO:0071041 | antisense RNA transcript catabolic process(GO:0071041) |
| 0.0 | 0.1 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) copper ion homeostasis(GO:0055070) |
| 0.0 | 0.3 | GO:0043629 | ncRNA polyadenylation(GO:0043629) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 0.2 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.0 | 0.2 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 0.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.7 | GO:0043934 | sporulation(GO:0043934) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.6 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.1 | GO:0034395 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0045895 | positive regulation of mating-type specific transcription, DNA-templated(GO:0045895) |
| 0.0 | 0.1 | GO:0032048 | cardiolipin metabolic process(GO:0032048) |
| 0.0 | 0.1 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.0 | 0.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.0 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) |
| 0.0 | 0.2 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.5 | GO:0015918 | sterol transport(GO:0015918) |
| 0.0 | 0.8 | GO:0008645 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0000753 | cell morphogenesis involved in conjugation with cellular fusion(GO:0000753) |
| 0.0 | 0.1 | GO:0072530 | cytosine transport(GO:0015856) purine-containing compound transmembrane transport(GO:0072530) |
| 0.0 | 0.0 | GO:0006119 | oxidative phosphorylation(GO:0006119) respiratory electron transport chain(GO:0022904) ATP synthesis coupled electron transport(GO:0042773) mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) |
| 0.0 | 0.0 | GO:1902931 | negative regulation of alcohol biosynthetic process(GO:1902931) |
| 0.0 | 0.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.1 | GO:0051126 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.0 | 0.0 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.1 | GO:0000409 | regulation of transcription by galactose(GO:0000409) positive regulation of transcription by galactose(GO:0000411) |
| 0.0 | 0.1 | GO:0070096 | mitochondrial outer membrane translocase complex assembly(GO:0070096) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.1 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 0.3 | GO:0034965 | intronic snoRNA processing(GO:0031070) intronic box C/D snoRNA processing(GO:0034965) |
| 0.0 | 0.4 | GO:0000742 | karyogamy involved in conjugation with cellular fusion(GO:0000742) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0016241 | regulation of macroautophagy(GO:0016241) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 1.0 | GO:0007124 | pseudohyphal growth(GO:0007124) |
| 0.0 | 0.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.0 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 0.1 | GO:0046036 | CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) |
| 0.0 | 0.0 | GO:0006641 | neutral lipid metabolic process(GO:0006638) acylglycerol metabolic process(GO:0006639) triglyceride metabolic process(GO:0006641) |
| 0.0 | 0.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0000114 | obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.0 | 0.1 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.1 | GO:0051469 | vesicle fusion with vacuole(GO:0051469) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.1 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.3 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.0 | GO:1903313 | positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.2 | GO:0042327 | positive regulation of phosphorylation(GO:0042327) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.1 | GO:0016562 | receptor recycling(GO:0001881) protein import into peroxisome matrix, receptor recycling(GO:0016562) receptor metabolic process(GO:0043112) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.0 | GO:0001308 | negative regulation of chromatin silencing involved in replicative cell aging(GO:0001308) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.4 | GO:0030466 | chromatin silencing at silent mating-type cassette(GO:0030466) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.0 | 0.1 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.2 | GO:0033617 | respiratory chain complex IV assembly(GO:0008535) mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.2 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.0 | GO:0051259 | protein oligomerization(GO:0051259) |
| 0.0 | 0.0 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 13.9 | GO:0031160 | spore wall(GO:0031160) |
| 0.6 | 2.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.5 | 2.2 | GO:0033551 | monopolin complex(GO:0033551) |
| 0.4 | 4.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 1.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 7.5 | GO:0005628 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.3 | 0.8 | GO:0005756 | mitochondrial proton-transporting ATP synthase, central stalk(GO:0005756) proton-transporting ATP synthase, central stalk(GO:0045269) |
| 0.3 | 1.1 | GO:0097002 | mitochondrial inner boundary membrane(GO:0097002) |
| 0.3 | 0.8 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.2 | 0.7 | GO:0000417 | HIR complex(GO:0000417) |
| 0.2 | 0.6 | GO:0045265 | mitochondrial proton-transporting ATP synthase, stator stalk(GO:0000274) proton-transporting ATP synthase, stator stalk(GO:0045265) |
| 0.2 | 0.6 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.2 | 0.5 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.2 | 1.6 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.2 | 1.7 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.2 | 0.7 | GO:0030892 | mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) |
| 0.2 | 1.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.1 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 0.2 | 0.5 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 0.7 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.6 | GO:0000817 | COMA complex(GO:0000817) |
| 0.1 | 0.8 | GO:0032126 | eisosome(GO:0032126) |
| 0.1 | 0.8 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 1.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 1.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.2 | GO:0009353 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial oxoglutarate dehydrogenase complex(GO:0009353) dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.6 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.1 | GO:0044453 | nuclear membrane part(GO:0044453) |
| 0.1 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.3 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.1 | 0.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.0 | GO:0031307 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.5 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 0.2 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0000446 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) nucleoplasmic THO complex(GO:0000446) |
| 0.0 | 0.5 | GO:0000142 | cellular bud neck contractile ring(GO:0000142) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.5 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle(GO:0030139) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0031499 | TRAMP complex(GO:0031499) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 4.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0016587 | Isw1 complex(GO:0016587) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0044233 | ERMES complex(GO:0032865) ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0005825 | half bridge of spindle pole body(GO:0005825) |
| 0.0 | 0.0 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 1.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 1.0 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.1 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.0 | 0.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.0 | 0.0 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.0 | GO:0008275 | gamma-tubulin small complex, spindle pole body(GO:0000928) gamma-tubulin complex(GO:0000930) gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0090544 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.2 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 1.0 | 4.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.7 | 2.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.6 | 3.6 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.6 | 2.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.4 | 1.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.4 | 1.2 | GO:0015489 | putrescine transmembrane transporter activity(GO:0015489) |
| 0.3 | 1.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.3 | 1.3 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.3 | 1.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 1.0 | GO:0008137 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 1.3 | GO:0015343 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.3 | 1.8 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.3 | 2.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 4.9 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.2 | 0.7 | GO:0008972 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.2 | 0.7 | GO:0042736 | NAD+ kinase activity(GO:0003951) NADH kinase activity(GO:0042736) |
| 0.2 | 0.7 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.2 | 1.3 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.2 | 0.6 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.2 | 0.4 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.2 | 0.6 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.2 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 0.5 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.2 | 4.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 1.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 3.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.2 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.2 | 1.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.6 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.1 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.2 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 6.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.5 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.1 | 1.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.3 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 1.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.1 | GO:0042947 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.1 | 0.9 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.1 | 0.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.5 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.1 | 1.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 1.5 | GO:0020037 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.1 | 0.5 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.4 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.5 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.3 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.1 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.1 | 0.3 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) |
| 0.1 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.3 | GO:0004375 | glycine dehydrogenase (decarboxylating) activity(GO:0004375) oxidoreductase activity, acting on the CH-NH2 group of donors, disulfide as acceptor(GO:0016642) |
| 0.1 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 0.2 | GO:0001128 | RNA polymerase II transcription coactivator activity involved in preinitiation complex assembly(GO:0001128) |
| 0.1 | 0.2 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.3 | GO:0015079 | potassium ion transmembrane transporter activity(GO:0015079) |
| 0.1 | 0.3 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 0.1 | 0.2 | GO:0001097 | RNA polymerase II basal transcription factor binding(GO:0001091) TFIIH-class transcription factor binding(GO:0001097) |
| 0.1 | 0.4 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.1 | 0.5 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 2.2 | GO:0022804 | active transmembrane transporter activity(GO:0022804) |
| 0.0 | 0.1 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.0 | 0.2 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.0 | 0.1 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.7 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.4 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.2 | GO:0038023 | transmembrane signaling receptor activity(GO:0004888) signaling receptor activity(GO:0038023) transmembrane receptor activity(GO:0099600) |
| 0.0 | 0.1 | GO:0015658 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.1 | GO:0015077 | monovalent inorganic cation transmembrane transporter activity(GO:0015077) |
| 0.0 | 0.1 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 1.4 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.1 | GO:0042124 | glucanosyltransferase activity(GO:0042123) 1,3-beta-glucanosyltransferase activity(GO:0042124) |
| 0.0 | 0.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.3 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.0 | 2.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.1 | GO:0000772 | mating pheromone activity(GO:0000772) pheromone activity(GO:0005186) |
| 0.0 | 0.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.0 | GO:0004458 | lactate dehydrogenase activity(GO:0004457) D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.1 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 0.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.5 | GO:0016773 | phosphotransferase activity, alcohol group as acceptor(GO:0016773) |
| 0.0 | 0.0 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.0 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 0.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 49.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.4 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 1.0 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.6 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 0.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.2 | REACTOME HEMOSTASIS | Genes involved in Hemostasis |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 48.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.1 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |