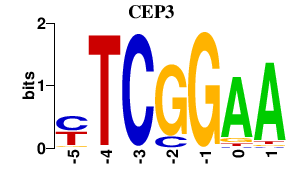
Results for CEP3
Z-value: 0.74
Transcription factors associated with CEP3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEP3
|
S000004778 | Essential kinetochore protein |
Activity-expression correlation:
Activity profile of CEP3 motif
Sorted Z-values of CEP3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YHR092C | 2.39 |
HXT4
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YBR054W | 1.39 |
YRO2
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in a phosphorylated state in highly purified mitochondria in high-throughput studies; transcriptionally regulated by Haa1p |
|
| YLL053C | 1.25 |
Putative protein; in the Sigma 1278B strain background YLL053C is contiguous with AQY2 which encodes an aquaporin |
||
| YDR278C | 1.21 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YAR060C | 1.13 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YJL216C | 1.12 |
Protein of unknown function, similar to alpha-D-glucosidases; transcriptionally activated by both Pdr8p and Yrm1p, along with transporters and other genes involved in the pleiotropic drug resistance (PDR) phenomenon |
||
| YLL052C | 1.12 |
AQY2
|
Water channel that mediates the transport of water across cell membranes, only expressed in proliferating cells, controlled by osmotic signals, may be involved in freeze tolerance; disrupted by a stop codon in many S. cerevisiae strains |
|
| YER067C-A | 1.11 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YER067W |
||
| YCR102W-A | 1.08 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YMR082C | 1.06 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNL160W | 1.04 |
YGP1
|
Cell wall-related secretory glycoprotein; induced by nutrient deprivation-associated growth arrest and upon entry into stationary phase; may be involved in adaptation prior to stationary phase entry; has similarity to Sps100p |
|
| YNR001W-A | 1.01 |
Dubious open reading frame unlikely to encode a functional protein; identified by homology |
||
| YGR139W | 1.01 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL033W | 1.00 |
HOP2
|
Meiosis-specific protein that localizes to chromosomes, preventing synapsis between nonhomologous chromosomes and ensuring synapsis between homologs; complexes with Mnd1p to promote homolog pairing and meiotic double-strand break repair |
|
| YPR157W | 0.91 |
Putative protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YLL055W | 0.91 |
YCT1
|
High-affinity cysteine-specific transporter with similarity to the Dal5p family of transporters; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; YCT1 is not an essential gene |
|
| YEL008W | 0.90 |
Hypothetical protein predicted to be involved in metabolism |
||
| YOL136C | 0.89 |
PFK27
|
6-phosphofructo-2-kinase, catalyzes synthesis of fructose-2,6-bisphosphate; inhibited by phosphoenolpyruvate and sn-glycerol 3-phosphate, expression induced by glucose and sucrose, transcriptional regulation involves protein kinase A |
|
| YBR072W | 0.88 |
HSP26
|
Small heat shock protein (sHSP) with chaperone activity; forms hollow, sphere-shaped oligomers that suppress unfolded proteins aggregation; oligomer activation requires a heat-induced conformational change; not expressed in unstressed cells |
|
| YCR104W | 0.87 |
PAU3
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YHR095W | 0.76 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR023W | 0.73 |
MTL1
|
Protein with both structural and functional similarity to Mid2p, which is a plasma membrane sensor required for cell integrity signaling during pheromone-induced morphogenesis; suppresses rgd1 null mutations |
|
| YBR067C | 0.72 |
TIP1
|
Major cell wall mannoprotein with possible lipase activity; transcription is induced by heat- and cold-shock; member of the Srp1p/Tip1p family of serine-alanine-rich proteins |
|
| YER066W | 0.70 |
Putative protein of unknown function; YER066W is not an essential gene |
||
| YHR212W-A | 0.70 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YPL014W | 0.66 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and to the nucleus |
||
| YAR047C | 0.65 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR036C | 0.63 |
EHD3
|
3-hydroxyisobutyryl-CoA hydrolase, member of a family of enoyl-CoA hydratase/isomerases; non-tagged protein is detected in highly purified mitochondria in high-throughput studies; phosphorylated; mutation affects fluid-phase endocytosis |
|
| YPL025C | 0.62 |
Hypothetical protein |
||
| YFR022W | 0.61 |
ROG3
|
Protein that binds to Rsp5p, which is a hect-type ubiquitin ligase, via its 2 PY motifs; has similarity to Rod1p; mutation suppresses the temperature sensitivity of an mck1 rim11 double mutant |
|
| YIL166C | 0.61 |
Putative protein with similarity to the allantoate permease (Dal5p) subfamily of the major facilitator superfamily; mRNA expression is elevated by sulfur limitation; YIL166C is a non-essential gene |
||
| YKL163W | 0.61 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YGR138C | 0.60 |
TPO2
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; transcription of TPO2 is regulated by Haa1p; member of the major facilitator superfamily |
|
| YPR149W | 0.60 |
NCE102
|
Protein of unknown function; contains transmembrane domains; involved in secretion of proteins that lack classical secretory signal sequences; component of the detergent-insoluble glycolipid-enriched complexes (DIGs) |
|
| YBR222C | 0.59 |
PCS60
|
Peroxisomal AMP-binding protein, localizes to both the peroxisomal peripheral membrane and matrix, expression is highly inducible by oleic acid, similar to E. coli long chain acyl-CoA synthetase |
|
| YMR034C | 0.58 |
Putative transporter, member of the SLC10 carrier family; identified in a transposon mutagenesis screen as a gene involved in azole resistance; YMR034C is not an essential gene |
||
| YDR276C | 0.56 |
PMP3
|
Small plasma membrane protein related to a family of plant polypeptides that are overexpressed under high salt concentration or low temperature, not essential for viability, deletion causes hyperpolarization of the plasma membrane potential |
|
| YKL217W | 0.55 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YBR177C | 0.55 |
EHT1
|
Acyl-coenzymeA:ethanol O-acyltransferase that plays a minor role in medium-chain fatty acid ethyl ester biosynthesis; possesses short-chain esterase activity; localizes to lipid particles and the mitochondrial outer membrane |
|
| YDL037C | 0.54 |
BSC1
|
Protein of unconfirmed function, similar to cell surface flocculin Muc1p; ORF exhibits genomic organization compatible with a translational readthrough-dependent mode of expression |
|
| YKR040C | 0.54 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YKR041W |
||
| YGL179C | 0.54 |
TOS3
|
Protein kinase, related to and functionally redundant with Elm1p and Sak1p for the phosphorylation and activation of Snf1p; functionally orthologous to LKB1, a mammalian kinase associated with Peutz-Jeghers cancer-susceptibility syndrome |
|
| YCR021C | 0.53 |
HSP30
|
Hydrophobic plasma membrane localized, stress-responsive protein that negatively regulates the H(+)-ATPase Pma1p; induced by heat shock, ethanol treatment, weak organic acid, glucose limitation, and entry into stationary phase |
|
| YER067W | 0.53 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YER067W is not an essential gene |
||
| YBR183W | 0.52 |
YPC1
|
Alkaline ceramidase that also has reverse (CoA-independent) ceramide synthase activity, catalyzes both breakdown and synthesis of phytoceramide; overexpression confers fumonisin B1 resistance |
|
| YHR212C | 0.52 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER130C | 0.50 |
Hypothetical protein |
||
| YDR222W | 0.49 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
||
| YNR067C | 0.49 |
DSE4
|
Daughter cell-specific secreted protein with similarity to glucanases, degrades cell wall from the daughter side causing daughter to separate from mother |
|
| YPR196W | 0.49 |
Putative maltose activator |
||
| YDR269C | 0.48 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR382W | 0.47 |
FIT2
|
Mannoprotein that is incorporated into the cell wall via a glycosylphosphatidylinositol (GPI) anchor, involved in the retention of siderophore-iron in the cell wall |
|
| YKL162C | 0.47 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the mitochondrion |
||
| YJL142C | 0.47 |
IRC9
|
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified gene YJL141C; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YGR250C | 0.46 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YDR129C | 0.46 |
SAC6
|
Fimbrin, actin-bundling protein; cooperates with Scp1p (calponin/transgelin) in the organization and maintenance of the actin cytoskeleton |
|
| YOL097W-A | 0.46 |
Putative protein of unknown function; identified by expression profiling and mass spectrometry |
||
| YPL111W | 0.45 |
CAR1
|
Arginase, responsible for arginine degradation, expression responds to both induction by arginine and nitrogen catabolite repression; disruption enhances freeze tolerance |
|
| YKR102W | 0.45 |
FLO10
|
Lectin-like protein with similarity to Flo1p, thought to be involved in flocculation |
|
| YDR343C | 0.43 |
HXT6
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt7p, expressed at high basal levels relative to other HXTs, repression of expression by high glucose requires SNF3 |
|
| YGR249W | 0.43 |
MGA1
|
Protein similar to heat shock transcription factor; multicopy suppressor of pseudohyphal growth defects of ammonium permease mutants |
|
| YJL222W-A | 0.43 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YJL107C | 0.42 |
Putative protein of unknown function; expression is induced by activation of the HOG1 mitogen-activated signaling pathway and this induction is Hog1p/Pbs2p dependent; YJL107C and adjacent ORF, YJL108C are merged in related fungi |
||
| YNL327W | 0.42 |
EGT2
|
Glycosylphosphatidylinositol (GPI)-anchored cell wall endoglucanase required for proper cell separation after cytokinesis, expression is activated by Swi5p and tightly regulated in a cell cycle-dependent manner |
|
| YOL128C | 0.42 |
YGK3
|
Protein kinase related to mammalian glycogen synthase kinases of the GSK-3 family; GSK-3 homologs (Mck1p, Rim11p, Mrk1p, Ygk3p) are involved in control of Msn2p-dependent transcription of stress responsive genes and in protein degradation |
|
| YDR541C | 0.42 |
Putative dihydrokaempferol 4-reductase |
||
| YPL103C | 0.42 |
FMP30
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YGR067C | 0.42 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YGL158W | 0.42 |
RCK1
|
Protein kinase involved in the response to oxidative stress; identified as suppressor of S. pombe cell cycle checkpoint mutations |
|
| YLL058W | 0.41 |
Putative protein of unknown function with similarity to Str2p, which is a cystathionine gamma-synthase important in sulfur metabolism; YLL058W is not an essential gene |
||
| YDR533C | 0.41 |
HSP31
|
Possible chaperone and cysteine protease with similarity to E. coli Hsp31; member of the DJ-1/ThiJ/PfpI superfamily, which includes human DJ-1 involved in Parkinson's disease; exists as a dimer and contains a putative metal-binding site |
|
| YOR178C | 0.41 |
GAC1
|
Regulatory subunit for Glc7p type-1 protein phosphatase (PP1), tethers Glc7p to Gsy2p glycogen synthase, binds Hsf1p heat shock transcription factor, required for induction of some HSF-regulated genes under heat shock |
|
| YBR182C-A | 0.40 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YGR254W | 0.40 |
ENO1
|
Enolase I, a phosphopyruvate hydratase that catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate during glycolysis and the reverse reaction during gluconeogenesis; expression is repressed in response to glucose |
|
| YLR350W | 0.40 |
ORM2
|
Evolutionarily conserved protein with similarity to Orm1p, required for resistance to agents that induce the unfolded protein response; human ortholog is located in the endoplasmic reticulum |
|
| YDR270W | 0.40 |
CCC2
|
Cu(+2)-transporting P-type ATPase, required for export of copper from the cytosol into an extracytosolic compartment; has similarity to human proteins involved in Menkes and Wilsons diseases |
|
| YOL059W | 0.39 |
GPD2
|
NAD-dependent glycerol 3-phosphate dehydrogenase, homolog of Gpd1p, expression is controlled by an oxygen-independent signaling pathway required to regulate metabolism under anoxic conditions; located in cytosol and mitochondria |
|
| YLR251W | 0.39 |
SYM1
|
Protein required for ethanol metabolism; induced by heat shock and localized to the inner mitochondrial membrane; homologous to mammalian peroxisomal membrane protein Mpv17 |
|
| YHL021C | 0.39 |
AIM17
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) |
|
| YLR120C | 0.39 |
YPS1
|
Aspartic protease, attached to the plasma membrane via a glycosylphosphatidylinositol (GPI) anchor |
|
| YGR022C | 0.38 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF MTL1/YGR023W |
||
| YOR376W | 0.38 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YOR376W is not an essential gene. |
||
| YJR155W | 0.38 |
AAD10
|
Putative aryl-alcohol dehydrogenase with similarity to P. chrysosporium aryl-alcohol dehydrogenase; mutational analysis has not yet revealed a physiological role |
|
| YPR145C-A | 0.37 |
Putative protein of unknown function |
||
| YOR003W | 0.37 |
YSP3
|
Putative precursor to the subtilisin-like protease III |
|
| YBR158W | 0.37 |
AMN1
|
Protein required for daughter cell separation, multiple mitotic checkpoints, and chromosome stability; contains 12 degenerate leucine-rich repeat motifs; expression is induced by the Mitotic Exit Network (MEN) |
|
| YGL256W | 0.37 |
ADH4
|
Alcohol dehydrogenase isoenzyme type IV, dimeric enzyme demonstrated to be zinc-dependent despite sequence similarity to iron-activated alcohol dehydrogenases; transcription is induced in response to zinc deficiency |
|
| YDL215C | 0.37 |
GDH2
|
NAD(+)-dependent glutamate dehydrogenase, degrades glutamate to ammonia and alpha-ketoglutarate; expression sensitive to nitrogen catabolite repression and intracellular ammonia levels |
|
| YFR056C | 0.37 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YLR142W | 0.36 |
PUT1
|
Proline oxidase, nuclear-encoded mitochondrial protein involved in utilization of proline as sole nitrogen source; PUT1 transcription is induced by Put3p in the presence of proline and the absence of a preferred nitrogen source |
|
| YKL216W | 0.35 |
URA1
|
Dihydroorotate dehydrogenase, catalyzes the fourth enzymatic step in the de novo biosynthesis of pyrimidines, converting dihydroorotic acid into orotic acid |
|
| YPR156C | 0.35 |
TPO3
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; member of the major facilitator superfamily |
|
| YEL063C | 0.35 |
CAN1
|
Plasma membrane arginine permease, requires phosphatidyl ethanolamine (PE) for localization, exclusively associated with lipid rafts; mutation confers canavanine resistance |
|
| YDL238C | 0.34 |
GUD1
|
Guanine deaminase, a catabolic enzyme of the guanine salvage pathway producing xanthine and ammonia from guanine; activity is low in exponentially-growing cultures but expression is increased in post-diauxic and stationary-phase cultures |
|
| YDR106W | 0.34 |
ARP10
|
Component of the dynactin complex, localized to the pointed end of the Arp1p filament; may regulate membrane association of the complex |
|
| YKL039W | 0.34 |
PTM1
|
Protein of unknown function, copurifies with late Golgi vesicles containing the v-SNARE Tlg2p |
|
| YIL119C | 0.34 |
RPI1
|
Putative transcriptional regulator; overexpression suppresses the heat shock sensitivity of wild-type RAS2 overexpression and also suppresses the cell lysis defect of an mpk1 mutation |
|
| YOL161C | 0.34 |
PAU20
|
Hypothetical protein |
|
| YOR120W | 0.33 |
GCY1
|
Putative NADP(+) coupled glycerol dehydrogenase, proposed to be involved in an alternative pathway for glycerol catabolism; member of the aldo-keto reductase (AKR) family |
|
| YBR047W | 0.33 |
FMP23
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YJL214W | 0.33 |
HXT8
|
Protein of unknown function with similarity to hexose transporter family members, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YAL067W-A | 0.33 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YDR442W | 0.33 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL196W | 0.33 |
DSD1
|
D-serine dehydratase (aka D-serine ammonia-lyase); converts D-serine to pyruvate and ammonia; specifc for D-serine, unlike the bacterial enzyme which recognizes both D-serine and L-serine as substrates |
|
| YNR034W-A | 0.32 |
Putative protein of unknown function; expression is regulated by Msn2p/Msn4p |
||
| YGL229C | 0.32 |
SAP4
|
Protein required for function of the Sit4p protein phosphatase, member of a family of similar proteins that form complexes with Sit4p, including Sap155p, Sap185p, and Sap190p |
|
| YLR342W | 0.32 |
FKS1
|
Catalytic subunit of 1,3-beta-D-glucan synthase, functionally redundant with alternate catalytic subunit Gsc2p; binds to regulatory subunit Rho1p; involved in cell wall synthesis and maintenance; localizes to sites of cell wall remodeling |
|
| YGR142W | 0.32 |
BTN2
|
v-SNARE binding protein that facilitates specific protein retrieval from a late endosome to the Golgi; modulates arginine uptake, possible role in mediating pH homeostasis between the vacuole and plasma membrane H(+)-ATPase |
|
| YCR106W | 0.31 |
RDS1
|
Zinc cluster transcription factor involved in conferring resistance to cycloheximide |
|
| YHL024W | 0.31 |
RIM4
|
Putative RNA-binding protein required for the expression of early and middle sporulation genes |
|
| YBL043W | 0.31 |
ECM13
|
Non-essential protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YOL015W | 0.31 |
IRC10
|
Putative protein of unknown function; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YER001W | 0.31 |
MNN1
|
Alpha-1,3-mannosyltransferase, integral membrane glycoprotein of the Golgi complex, required for addition of alpha1,3-mannose linkages to N-linked and O-linked oligosaccharides, one of five S. cerevisiae proteins of the MNN1 family |
|
| YLR296W | 0.31 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YEL039C | 0.31 |
CYC7
|
Cytochrome c isoform 2, expressed under hypoxic conditions; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YER150W | 0.31 |
SPI1
|
GPI-anchored cell wall protein involved in weak acid resistance; basal expression requires Msn2p/Msn4p; expression is induced under conditions of stress and during the diauxic shift; similar to Sed1p |
|
| YOL143C | 0.30 |
RIB4
|
Lumazine synthase (6,7-dimethyl-8-ribityllumazine synthase, also known as DMRL synthase); catalyzes synthesis of immediate precursor to riboflavin |
|
| YDR133C | 0.30 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR134C |
||
| YBR003W | 0.30 |
COQ1
|
Hexaprenyl pyrophosphate synthetase, catalyzes the first step in ubiquinone (coenzyme Q) biosynthesis |
|
| YJL082W | 0.30 |
IML2
|
Protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YAL066W | 0.30 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR287C | 0.29 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; has similarity to alpha-D-glucosidase (maltase); authentic, non-tagged protein detected in purified mitochondria in high-throughput studies |
||
| YHR001W | 0.29 |
OSH7
|
Member of an oxysterol-binding protein family with seven members in S. cerevisiae; family members have overlapping, redundant functions in sterol metabolism and collectively perform a function essential for viability |
|
| YLR420W | 0.29 |
URA4
|
Dihydroorotase, catalyzes the third enzymatic step in the de novo biosynthesis of pyrimidines, converting carbamoyl-L-aspartate into dihydroorotate |
|
| YKL062W | 0.28 |
MSN4
|
Transcriptional activator related to Msn2p; activated in stress conditions, which results in translocation from the cytoplasm to the nucleus; binds DNA at stress response elements of responsive genes, inducing gene expression |
|
| YJL213W | 0.28 |
Protein of unknown function that may interact with ribosomes; periodically expressed during the yeast metabolic cycle; phosphorylated in vitro by the mitotic exit network (MEN) kinase complex, Dbf2p/Mob1p |
||
| YPR108W-A | 0.28 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YEL009C | 0.28 |
GCN4
|
Transcriptional activator of amino acid biosynthetic genes in response to amino acid starvation; expression is tightly regulated at both the transcriptional and translational levels |
|
| YPL262W | 0.27 |
FUM1
|
Fumarase, converts fumaric acid to L-malic acid in the TCA cycle; cytosolic and mitochondrial localization determined by the N-terminal mitochondrial targeting sequence and protein conformation; phosphorylated in mitochondria |
|
| YLR329W | 0.27 |
REC102
|
Protein involved in early stages of meiotic recombination; required for chromosome synapsis; forms a complex with Rec104p and Spo11p necessary during the initiation of recombination |
|
| YFR053C | 0.27 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YER066C-A | 0.27 |
Dubious open reading frame unlikely to encode a protein, partially overlaps uncharacterized ORF YER067W |
||
| YKR103W | 0.27 |
NFT1
|
Putative transporter of the multidrug resistance-associated protein (MRP) subfamily; adjacent ORFs YKR103W and YKR104W are merged in different strain backgrounds. |
|
| YGL257C | 0.26 |
MNT2
|
Mannosyltransferase involved in adding the 4th and 5th mannose residues of O-linked glycans |
|
| YBR027C | 0.26 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR379C-A | 0.26 |
Hypothetical protein identified by homology. See FEBS Letters [2000] 487:31-36. |
||
| YIL050W | 0.26 |
PCL7
|
Pho85p cyclin of the Pho80p subfamily, forms a functional kinase complex with Pho85p which phosphorylates Mmr1p and is regulated by Pho81p; involved in glycogen metabolism, expression is cell-cycle regulated |
|
| YEL017W | 0.26 |
GTT3
|
Protein of unknown function with a possible role in glutathione metabolism, as suggested by computational analysis of large-scale protein-protein interaction data; GFP-fusion protein localizes to the nuclear periphery |
|
| YPL054W | 0.26 |
LEE1
|
Zinc-finger protein of unknown function |
|
| YDL129W | 0.26 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and the nucleus; YDL129W is not an essential gene |
||
| YIL046W-A | 0.26 |
Putative protein of unknown function; identified by expression profiling and mass spectrometry |
||
| YEL007W | 0.25 |
Putative protein with sequence similarity to S. pombe gti1+ (gluconate transport inducer 1) |
||
| YLR252W | 0.25 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the verified gene SYM1, a mitochondrial protein involved in ethanol metabolism |
||
| YCR023C | 0.25 |
Vacuolar membrane protein of unknown function; member of the multidrug resistance family; YCR023C is not an essential gene |
||
| YCR022C | 0.25 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YCR022C is not an essential gene |
||
| YKL142W | 0.24 |
MRP8
|
Putative mitochondrial ribosomal protein, has similarity to E. coli ribosomal protein S2 |
|
| YJR061W | 0.24 |
Putative protein of unknown function; non-essential gene with similarity to Mnn4, a putative membrane protein involved in glycosylation; transcription repressed by Rm101p |
||
| YEL011W | 0.24 |
GLC3
|
Glycogen branching enzyme, involved in glycogen accumulation; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YHR180W-A | 0.24 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps dubious ORF YHR180C-B and long terminal repeat YHRCsigma3 |
||
| YLL056C | 0.24 |
Putative protein of unknown function, transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in pleiotropic drug resistance (PDR) phenomenon; YLL056C is not an essential gene |
||
| YCR009C | 0.24 |
RVS161
|
Amphiphysin-like lipid raft protein; subunit of a complex (Rvs161p-Rvs167p) that regulates polarization of the actin cytoskeleton, endocytosis, cell polarity, cell fusion and viability following starvation or osmotic stress |
|
| YPR026W | 0.23 |
ATH1
|
Acid trehalase required for utilization of extracellular trehalose |
|
| YGL053W | 0.23 |
PRM8
|
Pheromone-regulated protein with 2 predicted transmembrane segments and an FF sequence, a motif involved in COPII binding; forms a complex with Prp9p in the ER; member of DUP240 gene family |
|
| YMR251W-A | 0.23 |
HOR7
|
Protein of unknown function; overexpression suppresses Ca2+ sensitivity of mutants lacking inositol phosphorylceramide mannosyltransferases Csg1p and Csh1p; transcription is induced under hyperosmotic stress and repressed by alpha factor |
|
| YMR105C | 0.23 |
PGM2
|
Phosphoglucomutase, catalyzes the conversion from glucose-1-phosphate to glucose-6-phosphate, which is a key step in hexose metabolism; functions as the acceptor for a Glc-phosphotransferase |
|
| YLR174W | 0.23 |
IDP2
|
Cytosolic NADP-specific isocitrate dehydrogenase, catalyzes oxidation of isocitrate to alpha-ketoglutarate; levels are elevated during growth on non-fermentable carbon sources and reduced during growth on glucose |
|
| YOR029W | 0.23 |
Dubious open reading frame unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YOR135C | 0.23 |
IRC14
|
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YOR136W; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YLR113W | 0.23 |
HOG1
|
Mitogen-activated protein kinase involved in osmoregulation via three independent osmosensors; mediates the recruitment and activation of RNA Pol II at Hot1p-dependent promoters; localization regulated by Ptp2p and Ptp3p |
|
| YGL034C | 0.23 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER103W | 0.23 |
SSA4
|
Heat shock protein that is highly induced upon stress; plays a role in SRP-dependent cotranslational protein-membrane targeting and translocation; member of the HSP70 family; cytoplasmic protein that concentrates in nuclei upon starvation |
|
| YGL001C | 0.23 |
ERG26
|
C-3 sterol dehydrogenase, catalyzes the second of three steps required to remove two C-4 methyl groups from an intermediate in ergosterol biosynthesis |
|
| YOR377W | 0.23 |
ATF1
|
Alcohol acetyltransferase with potential roles in lipid and sterol metabolism; responsible for the major part of volatile acetate ester production during fermentation |
|
| YBL001C | 0.22 |
ECM15
|
Non-essential protein of unknown function, likely exists as tetramer, may be regulated by the binding of small-molecule ligands (possibly sulfate ions), may have a role in yeast cell-wall biogenesis |
|
| YLR297W | 0.22 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YLR297W is not an essential gene; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YDR102C | 0.22 |
Dubious open reading frame; homozygous diploid deletion strain exhibits high budding index |
||
| YEL020C | 0.22 |
Hypothetical protein with low sequence identity to Pdc1p |
||
| YDR342C | 0.22 |
HXT7
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels |
|
| YDR344C | 0.22 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YAL067C | 0.22 |
SEO1
|
Putative permease, member of the allantoate transporter subfamily of the major facilitator superfamily; mutation confers resistance to ethionine sulfoxide |
|
| YAR053W | 0.22 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YMR035W | 0.22 |
IMP2
|
Catalytic subunit of the mitochondrial inner membrane peptidase complex, required for maturation of mitochondrial proteins of the intermembrane space; complex contains Imp1p and Imp2p (both catalytic subunits), and Som1p |
|
| YDR105C | 0.22 |
TMS1
|
Vacuolar membrane protein of unknown function that is conserved in mammals; predicted to contain eleven transmembrane helices; interacts with Pdr5p, a protein involved in multidrug resistance |
|
| YMR194C-A | 0.21 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER158C | 0.21 |
Protein of unknown function, has similarity to Afr1p; potentially phosphorylated by Cdc28p |
||
| YMR145C | 0.21 |
NDE1
|
Mitochondrial external NADH dehydrogenase, a type II NAD(P)H:quinone oxidoreductase that catalyzes the oxidation of cytosolic NADH; Nde1p and Nde2p provide cytosolic NADH to the mitochondrial respiratory chain |
|
| YDR103W | 0.21 |
STE5
|
Pheromone-response scaffold protein; binds kinases Ste11p, Ste7p, and Fus3p to form a MAPK cascade complex that interacts with the plasma membrane, via a PH (pleckstrin homology) and PM/NLS domain, and with Ste4p-Ste18p, during signaling |
|
| YPL024W | 0.21 |
RMI1
|
Involved in response to DNA damage; null mutants have increased rates of recombination and delayed S phase; interacts physically and genetically with Sgs1p (RecQ family member) and Top3p (topoisomerase III) |
|
| YPR016W-A | 0.21 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR151C | 0.21 |
CTH1
|
Member of the CCCH zinc finger family; has similarity to mammalian Tis11 protein, which activates transcription and also has a role in mRNA degradation; may function with Tis11p in iron homeostasis |
|
| YBR178W | 0.20 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YBR177C |
||
| YER078W-A | 0.20 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YLL059C | 0.20 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YBR046C | 0.20 |
ZTA1
|
Zeta-crystallin homolog, found in the cytoplasm and nucleus; has similarity to E. coli quinone oxidoreductase and to human zeta-crystallin, which has quinone oxidoreductase activity |
|
| YLR402W | 0.20 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOL131W | 0.20 |
Putative protein of unknown function |
||
| YLL024C | 0.20 |
SSA2
|
ATP binding protein involved in protein folding and vacuolar import of proteins; member of heat shock protein 70 (HSP70) family; associated with the chaperonin-containing T-complex; present in the cytoplasm, vacuolar membrane and cell wall |
|
| YKL067W | 0.20 |
YNK1
|
Nucleoside diphosphate kinase, catalyzes the transfer of gamma phosphates from nucleoside triphosphates, usually ATP, to nucleoside diphosphates by a mechanism that involves formation of an autophosphorylated enzyme intermediate |
|
| YOR268C | 0.20 |
Putative protein of unknown function; sporulation is abnormal in homozygous diploid; YOR268C is not an essential gene |
||
| YBR203W | 0.20 |
COS111
|
Protein required for resistance to the antifungal drug ciclopirox olamine; not related to the subtelomerically-encoded COS family; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YNL134C | 0.20 |
Putative protein of unknown function with similarity to dehydrogenases from other model organisms; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and nucleus and is induced by the DNA-damaging agent MMS |
||
| YGR136W | 0.20 |
LSB1
|
Protein containing an N-terminal SH3 domain; binds Las17p, which is a homolog of human Wiskott-Aldrich Syndrome protein involved in actin patch assembly and actin polymerization |
|
| YKR098C | 0.19 |
UBP11
|
Ubiquitin-specific protease that cleaves ubiquitin from ubiquitinated proteins |
|
| YMR081C | 0.19 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YBL101C | 0.19 |
ECM21
|
Non-essential protein of unknown function; promoter contains several Gcn4p binding elements |
|
| YJR152W | 0.19 |
DAL5
|
Allantoin permease; ureidosuccinate permease; expression is constitutive but sensitive to nitrogen catabolite repression |
|
| YLR047C | 0.19 |
FRE8
|
Protein with sequence similarity to iron/copper reductases, involved in iron homeostasis; deletion mutant has iron deficiency/accumulation growth defects; expression increased in the absence of copper-responsive transcription factor Mac1p |
|
| YPL185W | 0.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene UIP4/YPL186C |
||
| YDR148C | 0.19 |
KGD2
|
Dihydrolipoyl transsuccinylase, component of the mitochondrial alpha-ketoglutarate dehydrogenase complex, which catalyzes the oxidative decarboxylation of alpha-ketoglutarate to succinyl-CoA in the TCA cycle; phosphorylated |
|
| YGR043C | 0.19 |
NQM1
|
Protein of unknown function; transcription is repressed by Mot1p and induced by alpha-factor and during diauxic shift; null mutant non-quiescent cells exhibit reduced reproductive capacity |
Network of associatons between targets according to the STRING database.
First level regulatory network of CEP3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.3 | 0.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.9 | GO:0000296 | spermine transport(GO:0000296) |
| 0.2 | 0.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 0.5 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.2 | 0.5 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 0.2 | 0.7 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.2 | 0.5 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) regulation of membrane lipid metabolic process(GO:1905038) |
| 0.2 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.6 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.5 | GO:0019627 | urea metabolic process(GO:0019627) |
| 0.1 | 0.3 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.1 | 0.4 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.5 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.1 | 0.7 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.1 | 0.4 | GO:0097306 | positive regulation of transcription from RNA polymerase II promoter in response to ethanol(GO:0061410) cellular response to ethanol(GO:0071361) cellular response to alcohol(GO:0097306) |
| 0.1 | 0.7 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.1 | 0.3 | GO:0042843 | D-xylose catabolic process(GO:0042843) |
| 0.1 | 1.0 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.8 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.1 | 0.1 | GO:0035952 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 0.1 | 0.4 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.1 | 0.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.3 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.1 | 0.3 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.3 | GO:0006827 | high-affinity iron ion transmembrane transport(GO:0006827) |
| 0.1 | 0.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.3 | GO:0019563 | alditol catabolic process(GO:0019405) glycerol catabolic process(GO:0019563) |
| 0.1 | 0.2 | GO:0060988 | lipid tube assembly(GO:0060988) protein-lipid complex assembly(GO:0065005) protein-lipid complex subunit organization(GO:0071825) |
| 0.1 | 0.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 1.3 | GO:0007129 | synapsis(GO:0007129) |
| 0.1 | 0.4 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 0.5 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.1 | 0.3 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.4 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.1 | 0.1 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.1 | GO:0046323 | glucose import(GO:0046323) |
| 0.1 | 0.3 | GO:0034308 | ethanol metabolic process(GO:0006067) primary alcohol metabolic process(GO:0034308) |
| 0.1 | 0.2 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.1 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.3 | GO:0009208 | CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) |
| 0.1 | 0.2 | GO:0043388 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.1 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.1 | GO:0051222 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 0.0 | 0.3 | GO:0044205 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.2 | GO:0051126 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.2 | GO:0046351 | trehalose biosynthetic process(GO:0005992) oligosaccharide biosynthetic process(GO:0009312) disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0072523 | purine-containing compound catabolic process(GO:0072523) |
| 0.0 | 0.2 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.1 | GO:0019379 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.0 | 0.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.3 | GO:0009231 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.0 | 0.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.2 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) |
| 0.0 | 0.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.0 | 0.5 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.1 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.0 | 0.1 | GO:0019357 | nicotinate nucleotide biosynthetic process(GO:0019357) nicotinate nucleotide salvage(GO:0019358) pyridine nucleotide salvage(GO:0019365) nicotinate nucleotide metabolic process(GO:0046497) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) unsaturated fatty acid metabolic process(GO:0033559) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 1.2 | GO:0070726 | cell wall assembly(GO:0070726) |
| 0.0 | 0.8 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.4 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) regulation of vacuole organization(GO:0044088) |
| 0.0 | 0.1 | GO:0045923 | positive regulation of fatty acid metabolic process(GO:0045923) |
| 0.0 | 0.1 | GO:0015786 | pyrimidine nucleotide-sugar transport(GO:0015781) UDP-glucose transport(GO:0015786) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) |
| 0.0 | 0.1 | GO:0032365 | intracellular lipid transport(GO:0032365) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.5 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0006026 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.2 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.4 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.2 | GO:0060238 | regulation of pheromone-dependent signal transduction involved in conjugation with cellular fusion(GO:0010969) regulation of signal transduction involved in conjugation with cellular fusion(GO:0060238) |
| 0.0 | 0.0 | GO:0046160 | heme a biosynthetic process(GO:0006784) heme a metabolic process(GO:0046160) |
| 0.0 | 0.1 | GO:0034090 | maintenance of meiotic sister chromatid cohesion(GO:0034090) |
| 0.0 | 0.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.2 | GO:0090522 | vesicle tethering involved in exocytosis(GO:0090522) |
| 0.0 | 0.3 | GO:0001100 | negative regulation of exit from mitosis(GO:0001100) |
| 0.0 | 0.1 | GO:1903313 | positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.6 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.2 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.1 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.2 | GO:0042886 | amide transport(GO:0042886) |
| 0.0 | 0.2 | GO:0006101 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.0 | 0.1 | GO:2001038 | cellular response to drug(GO:0035690) regulation of response to drug(GO:2001023) positive regulation of response to drug(GO:2001025) regulation of cellular response to drug(GO:2001038) positive regulation of cellular response to drug(GO:2001040) |
| 0.0 | 0.2 | GO:0031333 | negative regulation of protein complex assembly(GO:0031333) |
| 0.0 | 0.5 | GO:0051445 | regulation of meiotic nuclear division(GO:0040020) regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.2 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.0 | GO:0051606 | detection of chemical stimulus(GO:0009593) detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) detection of stimulus(GO:0051606) |
| 0.0 | 0.2 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.1 | GO:0036498 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) IRE1-mediated unfolded protein response(GO:0036498) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.4 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.3 | GO:0043086 | negative regulation of catalytic activity(GO:0043086) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.3 | GO:0009272 | fungal-type cell wall biogenesis(GO:0009272) |
| 0.0 | 0.0 | GO:0052547 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 0.0 | 0.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.0 | 0.2 | GO:0042181 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) ketone biosynthetic process(GO:0042181) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0010526 | negative regulation of transposition, RNA-mediated(GO:0010526) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.5 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.1 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.1 | GO:0043112 | receptor recycling(GO:0001881) protein import into peroxisome matrix, receptor recycling(GO:0016562) receptor metabolic process(GO:0043112) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) allantoin catabolic process(GO:0000256) cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.0 | GO:0071049 | nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.0 | 1.1 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.0 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.0 | GO:0043649 | glutamate catabolic process(GO:0006538) dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.1 | GO:0007009 | plasma membrane organization(GO:0007009) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.0 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0018206 | N-terminal peptidyl-methionine acetylation(GO:0017196) peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0030428 | cell septum(GO:0030428) |
| 0.2 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.8 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.4 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.1 | 0.5 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.1 | 1.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.2 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.1 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 0.2 | GO:0070211 | Snt2C complex(GO:0070211) |
| 0.1 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0030287 | cell wall-bounded periplasmic space(GO:0030287) |
| 0.0 | 0.4 | GO:0045263 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.2 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.0 | 0.1 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.0 | 0.2 | GO:0034044 | exomer complex(GO:0034044) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0098797 | plasma membrane protein complex(GO:0098797) |
| 0.0 | 0.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.3 | GO:0005619 | ascospore wall(GO:0005619) |
| 0.0 | 0.1 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.0 | 0.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.0 | GO:0000928 | gamma-tubulin small complex, spindle pole body(GO:0000928) gamma-tubulin complex(GO:0000930) gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:0001400 | mating projection base(GO:0001400) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 2.9 | GO:0022838 | substrate-specific channel activity(GO:0022838) |
| 0.2 | 0.9 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.2 | 0.6 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.2 | 0.6 | GO:0016208 | AMP binding(GO:0016208) |
| 0.2 | 0.9 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 0.9 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.2 | 0.5 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.2 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 0.8 | GO:0034318 | alcohol O-acetyltransferase activity(GO:0004026) alcohol O-acyltransferase activity(GO:0034318) |
| 0.1 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.1 | 0.5 | GO:0042973 | glucan endo-1,3-beta-D-glucosidase activity(GO:0042973) |
| 0.1 | 0.6 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.3 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 0.4 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.1 | 0.5 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.4 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 0.3 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.1 | 1.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.1 | 0.4 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 0.1 | 0.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.2 | GO:0015603 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.1 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.1 | 0.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 0.4 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 0.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.5 | GO:0016723 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 0.3 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 1.1 | GO:0016298 | lipase activity(GO:0016298) |
| 0.0 | 0.1 | GO:0019203 | trehalose-phosphatase activity(GO:0004805) carbohydrate phosphatase activity(GO:0019203) |
| 0.0 | 0.1 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.6 | GO:0017171 | serine hydrolase activity(GO:0017171) |
| 0.0 | 0.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0015085 | calcium ion transmembrane transporter activity(GO:0015085) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0042947 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0015186 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.1 | GO:0008897 | holo-[acyl-carrier-protein] synthase activity(GO:0008897) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.1 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.0 | 0.6 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.2 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.1 | GO:0034979 | NAD-dependent histone deacetylase activity(GO:0017136) NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 0.1 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 0.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.2 | GO:0046906 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.0 | 0.1 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) |
| 0.0 | 0.0 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0042736 | NAD+ kinase activity(GO:0003951) NADH kinase activity(GO:0042736) |
| 0.0 | 0.0 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.0 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.5 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.2 | GO:0016896 | exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.4 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 0.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.0 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.4 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.0 | GO:0016872 | intramolecular lyase activity(GO:0016872) |
| 0.0 | 0.2 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0015293 | symporter activity(GO:0015293) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 0.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 0.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 0.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 22.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 0.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 0.2 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.1 | 0.2 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.1 | 0.5 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 22.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |