Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
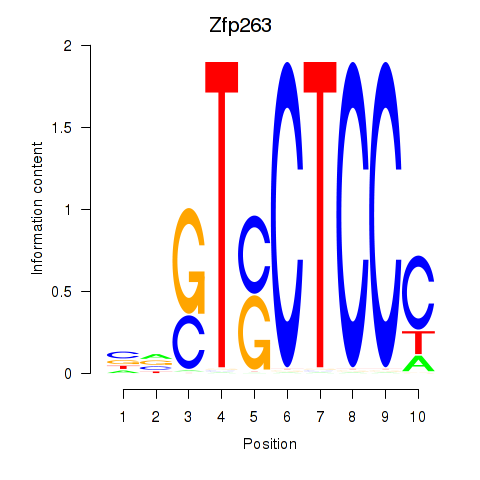
Results for Zfp263
Z-value: 0.93
Transcription factors associated with Zfp263
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp263
|
ENSRNOG00000007678 | zinc finger protein 263 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp263 | rn6_v1_chr10_-_12030416_12030416 | 0.18 | 1.3e-03 | Click! |
Activity profile of Zfp263 motif
Sorted Z-values of Zfp263 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_93520132 | 34.71 |
ENSRNOT00000055137
|
Mrc2
|
mannose receptor, C type 2 |
| chr5_-_75319189 | 25.60 |
ENSRNOT00000047200
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr5_-_75319765 | 25.21 |
ENSRNOT00000085698
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr14_+_3506339 | 21.62 |
ENSRNOT00000002867
|
Tgfbr3
|
transforming growth factor beta receptor 3 |
| chr10_+_89251370 | 20.10 |
ENSRNOT00000076820
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr1_+_261389804 | 19.93 |
ENSRNOT00000064160
|
Marveld1
|
MARVEL domain containing 1 |
| chr3_-_107760550 | 19.89 |
ENSRNOT00000077091
ENSRNOT00000051638 |
Meis2
|
Meis homeobox 2 |
| chr16_+_83522162 | 16.82 |
ENSRNOT00000057386
|
Col4a1
|
collagen type IV alpha 1 chain |
| chr8_+_48443767 | 16.52 |
ENSRNOT00000010127
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr14_+_75852060 | 16.52 |
ENSRNOT00000075975
|
Hs3st1
|
heparan sulfate-glucosamine 3-sulfotransferase 1 |
| chr1_-_246785360 | 16.36 |
ENSRNOT00000019816
|
Glis3
|
GLIS family zinc finger 3 |
| chrX_-_74706214 | 16.21 |
ENSRNOT00000040637
|
Slc16a2
|
solute carrier family 16 member 2 |
| chr15_+_2374582 | 15.92 |
ENSRNOT00000019644
|
Zfp503
|
zinc finger protein 503 |
| chr10_+_84147836 | 15.36 |
ENSRNOT00000010527
|
Hoxb6
|
homeo box B6 |
| chr2_+_182723854 | 15.17 |
ENSRNOT00000012658
|
Sfrp2
|
secreted frizzled-related protein 2 |
| chr17_+_4846789 | 14.53 |
ENSRNOT00000073271
|
Gas1
|
growth arrest-specific 1 |
| chr6_-_99843245 | 13.72 |
ENSRNOT00000080270
|
Gpx2
|
glutathione peroxidase 2 |
| chr4_+_6931495 | 12.95 |
ENSRNOT00000079770
|
Wdr86
|
WD repeat domain 86 |
| chr13_-_86671515 | 12.75 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr20_+_13670066 | 12.70 |
ENSRNOT00000031400
|
LOC103694874
|
stromelysin-3 |
| chr1_-_56683731 | 12.63 |
ENSRNOT00000014552
|
Thbs2
|
thrombospondin 2 |
| chr7_-_11648322 | 12.60 |
ENSRNOT00000026871
|
Gadd45b
|
growth arrest and DNA-damage-inducible, beta |
| chr10_-_62009582 | 12.59 |
ENSRNOT00000004143
|
Hic1
|
HIC ZBTB transcriptional repressor 1 |
| chr9_-_70787913 | 12.56 |
ENSRNOT00000072007
ENSRNOT00000017901 |
Klf7
Klf7
|
Kruppel like factor 7 Kruppel like factor 7 |
| chr6_+_3657325 | 12.47 |
ENSRNOT00000010927
|
Tmem178a
|
transmembrane protein 178A |
| chr7_-_117151256 | 12.26 |
ENSRNOT00000078846
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr4_+_26470864 | 12.03 |
ENSRNOT00000021979
|
Fzd1
|
frizzled class receptor 1 |
| chr2_+_125752130 | 11.72 |
ENSRNOT00000038703
|
Fat4
|
FAT atypical cadherin 4 |
| chr4_-_66002444 | 11.50 |
ENSRNOT00000018645
|
Zc3hav1l
|
zinc finger CCCH-type containing, antiviral 1 like |
| chr14_+_19866408 | 11.34 |
ENSRNOT00000060465
|
Adamts3
|
ADAM metallopeptidase with thrombospondin type 1, motif 3 |
| chr18_-_31430973 | 11.32 |
ENSRNOT00000026065
|
Pcdh12
|
protocadherin 12 |
| chr18_+_57286322 | 11.25 |
ENSRNOT00000026174
|
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr11_-_25078740 | 11.07 |
ENSRNOT00000002109
|
Cyyr1
|
cysteine and tyrosine rich 1 |
| chr13_-_90676629 | 10.92 |
ENSRNOT00000058143
|
Atp1a2
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr1_-_222468896 | 10.77 |
ENSRNOT00000028754
|
Cox8a
|
cytochrome c oxidase subunit 8A |
| chr4_+_157836912 | 10.71 |
ENSRNOT00000067271
|
Scnn1a
|
sodium channel epithelial 1 alpha subunit |
| chr1_-_131454689 | 10.27 |
ENSRNOT00000014152
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr11_-_65845418 | 10.16 |
ENSRNOT00000091057
|
Fstl1
|
follistatin-like 1 |
| chr5_-_173611202 | 10.01 |
ENSRNOT00000047854
|
Agrn
|
agrin |
| chr10_+_72272248 | 9.65 |
ENSRNOT00000003908
|
Car4
|
carbonic anhydrase 4 |
| chr10_+_86303727 | 9.63 |
ENSRNOT00000037752
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr8_+_22750336 | 9.54 |
ENSRNOT00000013496
|
Ldlr
|
low density lipoprotein receptor |
| chr7_+_143707237 | 9.53 |
ENSRNOT00000074212
|
Tns2
|
tensin 2 |
| chr14_+_79538911 | 9.47 |
ENSRNOT00000009960
|
Sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr12_+_37490584 | 9.30 |
ENSRNOT00000001401
ENSRNOT00000090831 |
Rilpl1
|
Rab interacting lysosomal protein-like 1 |
| chr2_-_61949926 | 9.09 |
ENSRNOT00000025966
|
Npr3
|
natriuretic peptide receptor 3 |
| chrX_+_112020646 | 9.03 |
ENSRNOT00000079841
|
Mid2
|
midline 2 |
| chr6_-_92527711 | 8.83 |
ENSRNOT00000007529
|
Nin
|
ninein |
| chr3_+_72540538 | 8.66 |
ENSRNOT00000012379
|
Aplnr
|
apelin receptor |
| chr14_+_100217289 | 8.65 |
ENSRNOT00000077171
ENSRNOT00000007104 |
Cnrip1
|
cannabinoid receptor interacting protein 1 |
| chr9_-_82699551 | 8.63 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chr4_-_64981384 | 8.62 |
ENSRNOT00000017338
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chrX_+_40460047 | 8.56 |
ENSRNOT00000010970
|
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr1_+_81328183 | 8.44 |
ENSRNOT00000074985
ENSRNOT00000075167 |
Plaur
|
plasminogen activator, urokinase receptor |
| chr18_+_2416871 | 8.42 |
ENSRNOT00000081399
|
Gata6
|
GATA binding protein 6 |
| chr2_+_189922996 | 8.41 |
ENSRNOT00000016499
|
S100a16
|
S100 calcium binding protein A16 |
| chrX_-_68562873 | 8.38 |
ENSRNOT00000076193
|
Ophn1
|
oligophrenin 1 |
| chrX_-_68563137 | 8.33 |
ENSRNOT00000034772
|
Ophn1
|
oligophrenin 1 |
| chr18_+_2416552 | 7.95 |
ENSRNOT00000030726
|
Gata6
|
GATA binding protein 6 |
| chr13_-_97173469 | 7.79 |
ENSRNOT00000017784
|
Kif26b
|
kinesin family member 26B |
| chr15_-_48445588 | 7.69 |
ENSRNOT00000077197
ENSRNOT00000018583 |
Extl3
|
exostosin-like glycosyltransferase 3 |
| chr16_-_81434038 | 7.68 |
ENSRNOT00000067508
|
Rasa3
|
RAS p21 protein activator 3 |
| chr8_+_72405748 | 7.67 |
ENSRNOT00000023952
|
Car12
|
carbonic anhydrase 12 |
| chr1_+_225129097 | 7.65 |
ENSRNOT00000026938
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr7_-_47586137 | 7.64 |
ENSRNOT00000006086
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr4_+_155653718 | 7.60 |
ENSRNOT00000065419
|
Foxj2
|
forkhead box J2 |
| chr6_-_95934296 | 7.57 |
ENSRNOT00000034338
|
Six1
|
SIX homeobox 1 |
| chr6_-_110904288 | 7.50 |
ENSRNOT00000014645
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr1_-_221370322 | 7.24 |
ENSRNOT00000028431
|
Capn1
|
calpain 1 |
| chr12_-_19592000 | 7.17 |
ENSRNOT00000001855
|
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr8_-_47393503 | 6.98 |
ENSRNOT00000059868
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr2_-_172459165 | 6.88 |
ENSRNOT00000057473
|
Schip1
|
schwannomin interacting protein 1 |
| chr7_-_62972084 | 6.87 |
ENSRNOT00000064251
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chr6_-_51356383 | 6.85 |
ENSRNOT00000012415
|
Prkar2b
|
protein kinase cAMP-dependent type 2 regulatory subunit beta |
| chrX_-_15693473 | 6.78 |
ENSRNOT00000013758
|
Prickle3
|
prickle planar cell polarity protein 3 |
| chr7_-_142210738 | 6.76 |
ENSRNOT00000006095
|
Pou6f1
|
POU class 6 homeobox 1 |
| chr7_+_140742418 | 6.62 |
ENSRNOT00000089211
|
Prph
|
peripherin |
| chr17_-_70045865 | 6.37 |
ENSRNOT00000031504
|
Calml5
|
calmodulin-like 5 |
| chr16_-_40555576 | 6.16 |
ENSRNOT00000015529
|
Vegfc
|
vascular endothelial growth factor C |
| chr14_-_85484275 | 6.15 |
ENSRNOT00000083770
|
Kremen1
|
kringle containing transmembrane protein 1 |
| chr15_-_2966576 | 6.06 |
ENSRNOT00000070893
ENSRNOT00000017383 |
Kat6b
|
lysine acetyltransferase 6B |
| chr7_+_41475163 | 5.78 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr18_+_30885789 | 5.69 |
ENSRNOT00000044434
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr2_-_207541645 | 5.66 |
ENSRNOT00000019680
|
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr1_+_165724451 | 5.65 |
ENSRNOT00000025827
|
Fam168a
|
family with sequence similarity 168, member A |
| chr5_-_159662754 | 5.57 |
ENSRNOT00000045226
|
Szrd1
|
SUZ RNA binding domain containing 1 |
| chr18_+_30869628 | 5.56 |
ENSRNOT00000060470
|
Pcdhgb4
|
protocadherin gamma subfamily B, 4 |
| chrX_+_77076106 | 5.56 |
ENSRNOT00000091527
ENSRNOT00000089381 |
Atp7a
|
ATPase copper transporting alpha |
| chr1_-_214317466 | 5.52 |
ENSRNOT00000048817
|
Deaf1
|
DEAF1 transcription factor |
| chr15_-_20783063 | 5.52 |
ENSRNOT00000083268
|
Bmp4
|
bone morphogenetic protein 4 |
| chr18_-_5314511 | 5.52 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr1_+_115975324 | 5.49 |
ENSRNOT00000080907
|
Atp10a
|
ATPase phospholipid transporting 10A (putative) |
| chr12_+_29266602 | 5.45 |
ENSRNOT00000076194
|
Wbscr17
|
Williams-Beuren syndrome chromosome region 17 |
| chr3_+_126335863 | 5.39 |
ENSRNOT00000028904
|
Bmp2
|
bone morphogenetic protein 2 |
| chr5_+_1417478 | 5.37 |
ENSRNOT00000008153
ENSRNOT00000085564 |
Jph1
|
junctophilin 1 |
| chr15_-_43542939 | 5.32 |
ENSRNOT00000012996
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr4_-_49439867 | 5.32 |
ENSRNOT00000090583
|
Fam3c
|
family with sequence similarity 3, member C |
| chr9_-_15214596 | 5.26 |
ENSRNOT00000039102
|
Tfeb
|
transcription factor EB |
| chr9_+_54212767 | 5.25 |
ENSRNOT00000078073
ENSRNOT00000090026 |
Gls
|
glutaminase |
| chr18_-_6474990 | 5.20 |
ENSRNOT00000061504
|
Kctd1
|
potassium channel tetramerization domain containing 1 |
| chr3_+_160908769 | 5.19 |
ENSRNOT00000030054
|
Sys1
|
Sys1 golgi trafficking protein |
| chr1_+_177764445 | 5.18 |
ENSRNOT00000037185
|
Rassf10
|
Ras association domain family member 10 |
| chr15_+_34493138 | 5.15 |
ENSRNOT00000089584
ENSRNOT00000027789 |
Nfatc4
|
nuclear factor of activated T-cells 4 |
| chr15_+_57340579 | 5.14 |
ENSRNOT00000015467
|
Zc3h13
|
zinc finger CCCH type containing 13 |
| chr1_-_142183884 | 5.12 |
ENSRNOT00000016032
|
Fes
|
FES proto-oncogene, tyrosine kinase |
| chr13_+_49195325 | 5.09 |
ENSRNOT00000004810
|
Dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr4_+_22859622 | 5.04 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr7_+_145117951 | 5.03 |
ENSRNOT00000055272
|
Pde1b
|
phosphodiesterase 1B |
| chr5_-_144779212 | 4.91 |
ENSRNOT00000016230
|
Ncdn
|
neurochondrin |
| chr2_+_188748359 | 4.85 |
ENSRNOT00000028038
|
Shc1
|
SHC adaptor protein 1 |
| chr1_+_226091774 | 4.78 |
ENSRNOT00000027693
|
Fads3
|
fatty acid desaturase 3 |
| chr13_+_52413241 | 4.77 |
ENSRNOT00000035157
|
AABR07020999.1
|
|
| chr17_+_28504623 | 4.76 |
ENSRNOT00000021568
|
F13a1
|
coagulation factor XIII A1 chain |
| chr6_-_107678156 | 4.70 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr5_-_137104993 | 4.66 |
ENSRNOT00000027271
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F |
| chr6_+_57516713 | 4.59 |
ENSRNOT00000063874
|
Dgkb
|
diacylglycerol kinase, beta |
| chr1_+_84293102 | 4.56 |
ENSRNOT00000028468
|
Sertad1
|
SERTA domain containing 1 |
| chr4_-_77706994 | 4.51 |
ENSRNOT00000038517
|
Zfp777
|
zinc finger protein 777 |
| chr2_+_95320283 | 4.38 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr1_+_101859346 | 4.36 |
ENSRNOT00000028627
|
Kdelr1
|
KDEL endoplasmic reticulum protein retention receptor 1 |
| chr20_+_6018374 | 4.31 |
ENSRNOT00000000621
|
Mapk13
|
mitogen activated protein kinase 13 |
| chr18_+_53915807 | 4.28 |
ENSRNOT00000026543
|
Adamts19
|
ADAM metallopeptidase with thrombospondin type 1 motif, 19 |
| chr7_-_143852119 | 4.25 |
ENSRNOT00000016801
|
Rarg
|
retinoic acid receptor, gamma |
| chr4_-_82160240 | 4.19 |
ENSRNOT00000038775
ENSRNOT00000058985 |
Hoxa4
|
homeo box A4 |
| chr4_+_7076759 | 4.18 |
ENSRNOT00000066598
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chrX_-_34794589 | 4.17 |
ENSRNOT00000008703
|
Rai2
|
retinoic acid induced 2 |
| chr3_+_148386189 | 4.15 |
ENSRNOT00000011255
|
Mylk2
|
myosin light chain kinase 2 |
| chr1_-_207171760 | 4.12 |
ENSRNOT00000047069
|
Fam196a
|
family with sequence similarity 196, member A |
| chr8_+_36625733 | 4.09 |
ENSRNOT00000016248
|
Cdon
|
cell adhesion associated, oncogene regulated |
| chr3_+_72134731 | 4.07 |
ENSRNOT00000083592
|
Ypel4
|
yippee-like 4 |
| chr15_-_33589522 | 4.01 |
ENSRNOT00000022514
|
Efs
|
embryonal Fyn-associated substrate |
| chr10_+_110893457 | 4.00 |
ENSRNOT00000088495
|
Metrnl
|
meteorin-like, glial cell differentiation regulator |
| chr4_+_66405166 | 3.99 |
ENSRNOT00000077486
ENSRNOT00000067097 |
Clec2l
|
C-type lectin domain family 2, member L |
| chr19_-_26094756 | 3.98 |
ENSRNOT00000067780
|
Junb
|
JunB proto-oncogene, AP-1 transcription factor subunit |
| chr3_+_176093640 | 3.97 |
ENSRNOT00000086541
|
Ogfr
|
opioid growth factor receptor |
| chr18_+_30375798 | 3.97 |
ENSRNOT00000060497
|
Pcdhb2
|
protocadherin beta 2 |
| chr6_+_53401109 | 3.94 |
ENSRNOT00000014763
|
Twist1
|
twist family bHLH transcription factor 1 |
| chr10_+_46511271 | 3.93 |
ENSRNOT00000080828
|
Rai1
|
retinoic acid induced 1 |
| chr4_-_148845267 | 3.89 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr4_+_57050214 | 3.87 |
ENSRNOT00000025165
|
Ahcyl2
|
adenosylhomocysteinase-like 2 |
| chr1_+_262987957 | 3.87 |
ENSRNOT00000023111
|
Entpd7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr9_+_16427589 | 3.85 |
ENSRNOT00000091809
|
Gltscr1l
|
GLTSCR1-like |
| chr20_-_2211995 | 3.83 |
ENSRNOT00000077201
|
Trim26
|
tripartite motif-containing 26 |
| chr7_-_144993652 | 3.82 |
ENSRNOT00000087748
|
Itga5
|
integrin subunit alpha 5 |
| chr10_+_85744568 | 3.81 |
ENSRNOT00000005522
|
Lasp1
|
LIM and SH3 protein 1 |
| chrX_+_123751293 | 3.79 |
ENSRNOT00000089883
|
Nkap
|
NFKB activating protein |
| chr1_+_114258719 | 3.77 |
ENSRNOT00000088459
ENSRNOT00000016376 |
Cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr15_-_4022223 | 3.77 |
ENSRNOT00000081997
|
Zswim8
|
zinc finger, SWIM-type containing 8 |
| chr4_+_83713666 | 3.77 |
ENSRNOT00000086473
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr19_+_39067363 | 3.64 |
ENSRNOT00000083515
|
Has3
|
hyaluronan synthase 3 |
| chr3_+_117938500 | 3.61 |
ENSRNOT00000042411
|
Eid1
|
EP300 interacting inhibitor of differentiation 1 |
| chr13_+_98615287 | 3.61 |
ENSRNOT00000004032
|
Itpkb
|
inositol-trisphosphate 3-kinase B |
| chr1_-_64147251 | 3.60 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr12_-_17972737 | 3.60 |
ENSRNOT00000001783
|
Fam20c
|
FAM20C, golgi associated secretory pathway kinase |
| chr10_+_103934797 | 3.59 |
ENSRNOT00000035865
|
Cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr7_-_51353068 | 3.50 |
ENSRNOT00000008222
|
Pawr
|
pro-apoptotic WT1 regulator |
| chr3_+_155160481 | 3.45 |
ENSRNOT00000021133
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr11_+_30550141 | 3.44 |
ENSRNOT00000002866
|
Hunk
|
hormonally upregulated Neu-associated kinase |
| chr14_+_36687134 | 3.44 |
ENSRNOT00000002879
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr6_+_123361864 | 3.44 |
ENSRNOT00000057577
|
AABR07065353.1
|
|
| chr2_-_192960294 | 3.42 |
ENSRNOT00000012420
|
LOC102552326
|
late cornified envelope protein 5A-like |
| chr14_+_18231860 | 3.41 |
ENSRNOT00000003686
|
Btc
|
betacellulin |
| chr1_+_274766283 | 3.34 |
ENSRNOT00000071541
|
Adra2a
|
adrenoceptor alpha 2A |
| chr6_+_76675418 | 3.29 |
ENSRNOT00000076169
ENSRNOT00000010948 ENSRNOT00000082286 |
Brms1l
Brms1l
|
breast cancer metastasis-suppressor 1-like breast cancer metastasis-suppressor 1-like |
| chr5_+_58855773 | 3.27 |
ENSRNOT00000072869
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr17_+_56109549 | 3.14 |
ENSRNOT00000022190
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chrX_-_72370044 | 3.12 |
ENSRNOT00000004224
|
Hdac8
|
histone deacetylase 8 |
| chr1_+_220322940 | 3.05 |
ENSRNOT00000074972
|
LOC108348085
|
beta-1,4-glucuronyltransferase 1 |
| chr4_+_140703619 | 3.01 |
ENSRNOT00000009563
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr15_+_15275541 | 3.01 |
ENSRNOT00000012153
|
Cadps
|
calcium dependent secretion activator |
| chr4_-_28953067 | 2.98 |
ENSRNOT00000013989
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr12_-_22021851 | 2.98 |
ENSRNOT00000039280
|
Tsc22d4
|
TSC22 domain family, member 4 |
| chr11_+_74834050 | 2.98 |
ENSRNOT00000002333
|
Atp13a4
|
ATPase 13A4 |
| chr1_-_57327379 | 2.91 |
ENSRNOT00000080429
|
Dll1
|
delta like canonical Notch ligand 1 |
| chr8_+_63036976 | 2.90 |
ENSRNOT00000011383
|
Stoml1
|
stomatin like 1 |
| chr10_+_105156632 | 2.89 |
ENSRNOT00000086288
|
Galr2
|
galanin receptor 2 |
| chr8_+_71822129 | 2.88 |
ENSRNOT00000089147
|
Dapk2
|
death-associated protein kinase 2 |
| chr1_-_170318935 | 2.87 |
ENSRNOT00000024119
|
Prkcdbp
|
protein kinase C, delta binding protein |
| chr7_-_139907640 | 2.85 |
ENSRNOT00000045473
|
Zfp641
|
zinc finger protein 641 |
| chr2_+_54191538 | 2.82 |
ENSRNOT00000019524
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr4_-_157439507 | 2.81 |
ENSRNOT00000081802
|
Ptms
|
parathymosin |
| chr16_-_48692476 | 2.79 |
ENSRNOT00000013118
|
Irf2
|
interferon regulatory factor 2 |
| chr12_-_22726982 | 2.76 |
ENSRNOT00000001921
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr1_+_266358728 | 2.75 |
ENSRNOT00000027118
|
Wbp1l
|
WW domain binding protein 1-like |
| chr11_-_90406797 | 2.74 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chr9_-_93377643 | 2.71 |
ENSRNOT00000024712
|
Ncl
|
nucleolin |
| chr10_-_72142533 | 2.69 |
ENSRNOT00000030885
|
Mrm1
|
mitochondrial rRNA methyltransferase 1 |
| chr7_-_92882068 | 2.68 |
ENSRNOT00000037809
|
Ext1
|
exostosin glycosyltransferase 1 |
| chr18_-_37776453 | 2.66 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr17_+_72429618 | 2.66 |
ENSRNOT00000026187
|
Gata3
|
GATA binding protein 3 |
| chrX_-_111325186 | 2.57 |
ENSRNOT00000086785
|
Rbm41
|
RNA binding motif protein 41 |
| chr3_+_95133713 | 2.55 |
ENSRNOT00000067940
|
Wt1
|
Wilms tumor 1 |
| chr7_-_143863186 | 2.54 |
ENSRNOT00000017096
|
Rarg
|
retinoic acid receptor, gamma |
| chr10_-_107386072 | 2.50 |
ENSRNOT00000004290
|
Timp2
|
TIMP metallopeptidase inhibitor 2 |
| chr7_+_77763512 | 2.49 |
ENSRNOT00000006411
|
Baalc
|
brain and acute leukemia, cytoplasmic |
| chrX_+_128493614 | 2.48 |
ENSRNOT00000044240
|
Stag2
|
stromal antigen 2 |
| chr12_+_12227010 | 2.42 |
ENSRNOT00000060843
ENSRNOT00000092610 |
Baiap2l1
|
BAI1-associated protein 2-like 1 |
| chr18_+_76559811 | 2.37 |
ENSRNOT00000084621
|
Pard6g
|
par-6 family cell polarity regulator gamma |
| chr2_-_204032023 | 2.37 |
ENSRNOT00000040430
|
Atp1a1
|
ATPase Na+/K+ transporting subunit alpha 1 |
| chr12_-_46989876 | 2.36 |
ENSRNOT00000079043
ENSRNOT00000001534 |
NEWGENE_1586233
|
TP53 regulated inhibitor of apoptosis 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp263
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.6 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 5.5 | 16.4 | GO:0007493 | endodermal cell fate determination(GO:0007493) negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 5.1 | 15.2 | GO:2000041 | negative regulation of mesodermal cell fate specification(GO:0042662) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 4.6 | 13.7 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 4.0 | 12.0 | GO:0035425 | autocrine signaling(GO:0035425) |
| 4.0 | 15.9 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 3.9 | 11.7 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 3.8 | 11.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 3.4 | 10.3 | GO:0060849 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 3.3 | 10.0 | GO:2000539 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 3.1 | 9.3 | GO:1903445 | protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 3.1 | 6.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 2.9 | 8.6 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 2.8 | 16.8 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 2.7 | 10.9 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 2.7 | 10.9 | GO:0072138 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 2.7 | 2.7 | GO:0061209 | cell proliferation involved in mesonephros development(GO:0061209) |
| 2.5 | 7.6 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 2.4 | 9.5 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 2.3 | 6.9 | GO:0030091 | protein repair(GO:0030091) |
| 2.3 | 6.8 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 2.0 | 47.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 1.9 | 7.8 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 1.7 | 5.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 1.7 | 5.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.5 | 16.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.5 | 4.4 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 1.5 | 2.9 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 1.5 | 14.5 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 1.4 | 5.8 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 1.4 | 8.4 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 1.4 | 5.6 | GO:0034760 | negative regulation of iron ion transmembrane transport(GO:0034760) elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 1.4 | 4.2 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 1.4 | 9.6 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 1.4 | 6.9 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 1.4 | 2.7 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 1.3 | 3.9 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 1.3 | 5.2 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 1.3 | 2.5 | GO:0072125 | negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) |
| 1.2 | 3.6 | GO:0097187 | dentinogenesis(GO:0097187) |
| 1.2 | 3.5 | GO:1904457 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) positive regulation of neuronal action potential(GO:1904457) |
| 1.1 | 3.4 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 1.1 | 9.1 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 1.1 | 11.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 1.1 | 20.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 1.0 | 4.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.0 | 3.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.0 | 11.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 1.0 | 2.9 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.9 | 4.7 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.9 | 2.8 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.9 | 3.6 | GO:0046379 | hyaluranon cable assembly(GO:0036118) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.9 | 7.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.9 | 10.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.9 | 2.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.9 | 8.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.9 | 7.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.8 | 9.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.8 | 4.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.8 | 9.6 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.7 | 12.6 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.7 | 5.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.7 | 2.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.7 | 3.3 | GO:0050955 | thermoception(GO:0050955) |
| 0.7 | 2.0 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.6 | 4.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.6 | 11.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.6 | 13.5 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.6 | 1.7 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.6 | 5.0 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.6 | 3.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.6 | 2.2 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.5 | 3.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.5 | 2.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.5 | 3.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.5 | 12.5 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.5 | 2.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.5 | 2.4 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.5 | 2.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.5 | 1.8 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.4 | 3.6 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.4 | 3.0 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.4 | 3.4 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.4 | 2.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.4 | 1.6 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.4 | 4.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.4 | 2.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.4 | 2.7 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.4 | 7.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.4 | 1.4 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.4 | 1.4 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.4 | 2.5 | GO:1905049 | negative regulation of metallopeptidase activity(GO:1905049) |
| 0.3 | 6.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.3 | 3.8 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.3 | 5.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.3 | 3.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.3 | 2.4 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.3 | 5.0 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.3 | 16.7 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.3 | 5.3 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.3 | 10.7 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.3 | 20.4 | GO:0008542 | visual learning(GO:0008542) |
| 0.3 | 10.7 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.3 | 1.3 | GO:0010045 | response to nickel cation(GO:0010045) |
| 0.3 | 5.5 | GO:1900424 | regulation of defense response to bacterium(GO:1900424) |
| 0.3 | 9.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.3 | 2.9 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.3 | 1.4 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.3 | 3.9 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.3 | 2.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.3 | 17.2 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.3 | 3.8 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.3 | 1.8 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 0.7 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.2 | 2.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 5.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 2.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.2 | 1.4 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.2 | 1.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 7.7 | GO:0006024 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.2 | 0.5 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.2 | 1.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 | 1.6 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.2 | 1.9 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.2 | 3.8 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.4 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 13.0 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.1 | 7.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 3.0 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 2.1 | GO:0060746 | parental behavior(GO:0060746) |
| 0.1 | 1.9 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.5 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.1 | 0.3 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.1 | 1.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 8.2 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 5.6 | GO:0045739 | base-excision repair(GO:0006284) positive regulation of DNA repair(GO:0045739) |
| 0.1 | 7.7 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.1 | 1.8 | GO:1903830 | magnesium ion transport(GO:0015693) magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 3.0 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 15.4 | GO:0034101 | erythrocyte homeostasis(GO:0034101) |
| 0.1 | 13.0 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.1 | 0.9 | GO:0045630 | negative regulation of granulocyte differentiation(GO:0030853) positive regulation of T-helper 2 cell differentiation(GO:0045630) chondroblast differentiation(GO:0060591) |
| 0.1 | 3.6 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.1 | 1.7 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 6.4 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 5.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 3.9 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 2.2 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 5.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 1.2 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.1 | 4.4 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.1 | 3.8 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 7.3 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.1 | 1.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 3.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 4.8 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 0.8 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 4.1 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.1 | 1.1 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 9.0 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.1 | 1.7 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 3.4 | GO:0007569 | cell aging(GO:0007569) |
| 0.1 | 2.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 6.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.8 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.0 | GO:0009176 | pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) dUMP metabolic process(GO:0046078) |
| 0.0 | 3.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.6 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 1.9 | GO:1903363 | negative regulation of cellular protein catabolic process(GO:1903363) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.4 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 1.0 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 1.0 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 1.9 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 2.7 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.9 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 2.1 | 16.8 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 1.8 | 5.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.7 | 6.6 | GO:0044299 | C-fiber(GO:0044299) |
| 1.4 | 8.6 | GO:1990393 | 3M complex(GO:1990393) |
| 1.4 | 16.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 1.3 | 8.8 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.2 | 3.6 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 1.2 | 9.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 1.1 | 13.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.8 | 4.0 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.8 | 6.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.7 | 3.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.7 | 7.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.7 | 2.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.7 | 10.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.7 | 10.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 9.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.5 | 12.6 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.5 | 14.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.5 | 7.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.4 | 3.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 2.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.4 | 2.7 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.4 | 2.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 7.1 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.3 | 27.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 2.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 4.4 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.2 | 26.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 3.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 11.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 88.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.2 | 1.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.2 | 2.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 3.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 3.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.4 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 2.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 7.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.5 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 2.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 5.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.4 | GO:0043159 | acrosomal matrix(GO:0043159) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 29.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 4.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 70.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 2.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 5.5 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 14.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 4.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 4.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 4.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.7 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.0 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 6.9 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 4.3 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 12.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 12.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 3.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 3.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 7.6 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 3.0 | 9.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 2.4 | 23.7 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 2.2 | 15.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 2.0 | 16.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.9 | 9.5 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 1.8 | 20.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.8 | 7.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 1.7 | 5.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 1.7 | 6.9 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 1.6 | 12.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.6 | 4.7 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.5 | 4.4 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.3 | 17.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 1.3 | 13.3 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 1.3 | 5.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 1.2 | 4.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 1.2 | 16.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.2 | 10.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 1.2 | 11.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 1.1 | 6.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.1 | 5.6 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 1.0 | 4.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.0 | 10.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 1.0 | 6.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.0 | 3.9 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.9 | 2.8 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.9 | 4.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.9 | 2.6 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.8 | 3.3 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.7 | 3.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.7 | 12.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.7 | 2.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.7 | 2.7 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.7 | 8.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.6 | 5.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.6 | 7.7 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.6 | 1.9 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.6 | 13.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.6 | 2.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.5 | 8.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.5 | 18.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.5 | 6.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.5 | 2.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.5 | 2.0 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.5 | 4.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 2.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.4 | 3.0 | GO:0043426 | MRF binding(GO:0043426) |
| 0.4 | 5.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 34.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.4 | 3.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.4 | 5.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.4 | 5.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.4 | 2.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.4 | 10.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 40.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.3 | 1.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.3 | 2.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 4.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 16.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.3 | 2.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.3 | 7.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 3.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 3.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 3.0 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.3 | 1.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 5.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 3.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 4.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 5.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 4.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 3.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 2.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 2.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 5.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 1.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 2.0 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 3.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 5.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 4.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 114.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.2 | 6.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 3.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 2.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 2.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.4 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 2.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 5.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.7 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 6.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 28.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 7.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 2.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 7.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 3.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 15.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 2.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.4 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.0 | 2.7 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 4.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.6 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 2.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 3.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 18.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.6 | 14.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.6 | 14.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.6 | 42.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.5 | 11.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.5 | 3.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.5 | 14.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.4 | 13.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.4 | 12.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.4 | 4.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 22.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.4 | 23.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.3 | 17.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 50.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 3.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 7.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.3 | 5.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 10.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 4.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 4.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 6.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 3.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 5.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 8.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 2.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 5.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 32.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 2.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 4.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 3.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 3.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 19.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 13.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.7 | 19.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.6 | 5.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.6 | 6.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.6 | 9.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.6 | 17.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.6 | 28.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.5 | 31.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.5 | 24.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.5 | 12.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 3.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.3 | 4.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 8.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.3 | 4.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 5.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 4.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.3 | 3.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 2.9 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.2 | 3.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 5.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.2 | 2.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 7.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 3.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 5.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 7.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 16.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 13.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.2 | 10.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 1.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 12.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 3.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 14.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 16.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 6.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.5 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 1.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.0 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 1.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.4 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 3.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 8.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |