Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
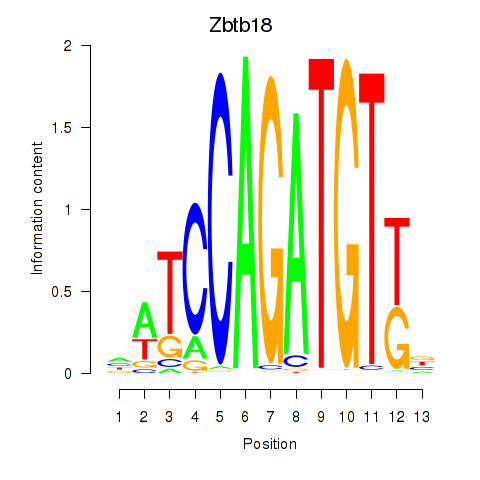
Results for Zbtb18
Z-value: 1.47
Transcription factors associated with Zbtb18
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp238 | rn6_v1_chr13_+_95589668_95589668 | -0.52 | 6.2e-24 | Click! |
Activity profile of Zbtb18 motif
Sorted Z-values of Zbtb18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_128046780 | 130.12 |
ENSRNOT00000078064
|
LOC500712
|
Ab1-233 |
| chr16_+_6970342 | 92.96 |
ENSRNOT00000061294
ENSRNOT00000048344 |
Itih4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr2_+_252452269 | 79.62 |
ENSRNOT00000021970
|
Uox
|
urate oxidase |
| chr16_+_18690246 | 71.25 |
ENSRNOT00000081484
|
Mat1a
|
methionine adenosyltransferase 1A |
| chr13_+_78805347 | 68.61 |
ENSRNOT00000003748
|
Serpinc1
|
serpin family C member 1 |
| chr1_-_170431073 | 66.43 |
ENSRNOT00000024710
|
Hpx
|
hemopexin |
| chr3_+_28416954 | 65.45 |
ENSRNOT00000043533
|
Kynu
|
kynureninase |
| chr16_+_18690649 | 63.27 |
ENSRNOT00000015190
|
Mat1a
|
methionine adenosyltransferase 1A |
| chr4_+_65110746 | 60.11 |
ENSRNOT00000017675
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr1_+_83714347 | 55.88 |
ENSRNOT00000085245
|
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr5_-_134484839 | 54.14 |
ENSRNOT00000045571
ENSRNOT00000012681 |
Cyp4a2
|
cytochrome P450, family 4, subfamily a, polypeptide 2 |
| chr1_+_83163079 | 53.83 |
ENSRNOT00000077725
ENSRNOT00000034845 |
Cyp2b3
|
cytochrome P450, family 2, subfamily b, polypeptide 3 |
| chrX_+_143097525 | 51.48 |
ENSRNOT00000004559
|
F9
|
coagulation factor IX |
| chr17_+_8489266 | 50.59 |
ENSRNOT00000016252
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr2_+_114413410 | 49.76 |
ENSRNOT00000015866
|
Slc2a2
|
solute carrier family 2 member 2 |
| chr4_+_122244711 | 46.97 |
ENSRNOT00000038251
|
Uroc1
|
urocanate hydratase 1 |
| chr16_-_7007287 | 44.57 |
ENSRNOT00000041216
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr20_+_28989491 | 44.50 |
ENSRNOT00000074524
|
Pla2g12b
|
phospholipase A2, group XIIB |
| chr16_-_7007051 | 44.43 |
ENSRNOT00000023984
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr1_+_48273611 | 44.17 |
ENSRNOT00000022254
ENSRNOT00000022068 |
Slc22a1
|
solute carrier family 22 member 1 |
| chr19_-_43911057 | 43.86 |
ENSRNOT00000026017
|
Ctrb1
|
chymotrypsinogen B1 |
| chr6_-_127816055 | 42.37 |
ENSRNOT00000013175
|
Serpina3m
|
serine (or cysteine) proteinase inhibitor, clade A, member 3M |
| chr5_-_19368431 | 41.89 |
ENSRNOT00000012819
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr10_+_90085559 | 41.31 |
ENSRNOT00000028332
|
Nags
|
N-acetylglutamate synthase |
| chr5_+_61425746 | 41.13 |
ENSRNOT00000064113
|
RGD1305807
|
hypothetical LOC298077 |
| chr8_-_85645718 | 41.02 |
ENSRNOT00000032185
|
Gsta2
|
glutathione S-transferase alpha 2 |
| chr14_+_22724399 | 39.25 |
ENSRNOT00000002724
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr14_+_77067503 | 37.98 |
ENSRNOT00000085275
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr2_+_243550627 | 37.45 |
ENSRNOT00000085067
ENSRNOT00000083682 |
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr7_+_93975451 | 37.41 |
ENSRNOT00000011379
|
Colec10
|
collectin subfamily member 10 |
| chr11_-_66034573 | 35.91 |
ENSRNOT00000003645
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr14_+_22142364 | 35.53 |
ENSRNOT00000002699
|
Sult1b1
|
sulfotransferase family 1B member 1 |
| chr3_+_159902441 | 34.66 |
ENSRNOT00000089893
ENSRNOT00000011978 |
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr1_+_189364288 | 34.36 |
ENSRNOT00000080338
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr4_+_65112944 | 31.94 |
ENSRNOT00000083672
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr8_-_50539331 | 31.02 |
ENSRNOT00000088997
|
AABR07073400.1
|
|
| chr1_+_263554453 | 31.00 |
ENSRNOT00000070861
|
Abcc2
|
ATP binding cassette subfamily C member 2 |
| chr10_-_62254287 | 30.92 |
ENSRNOT00000004313
|
Serpinf1
|
serpin family F member 1 |
| chr1_-_163328591 | 29.78 |
ENSRNOT00000034843
|
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr20_-_5806097 | 29.57 |
ENSRNOT00000000611
|
Clps
|
colipase |
| chr13_+_83073544 | 28.77 |
ENSRNOT00000066119
ENSRNOT00000079796 ENSRNOT00000077070 |
Dpt
|
dermatopontin |
| chr13_+_83073866 | 28.61 |
ENSRNOT00000075996
|
Dpt
|
dermatopontin |
| chr13_-_56958549 | 28.58 |
ENSRNOT00000017293
ENSRNOT00000083912 |
RGD1564614
|
similar to complement factor H-related protein |
| chr4_-_176679815 | 28.11 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr17_-_35958077 | 27.78 |
ENSRNOT00000038532
|
Agtr1a
|
angiotensin II receptor, type 1a |
| chr20_-_12820466 | 27.60 |
ENSRNOT00000001699
|
Ftcd
|
formimidoyltransferase cyclodeaminase |
| chr13_-_89433815 | 27.54 |
ENSRNOT00000091541
|
Fcgr2b
|
Fc fragment of IgG receptor IIb |
| chr20_-_5805627 | 27.16 |
ENSRNOT00000085996
|
Clps
|
colipase |
| chr1_+_279896973 | 27.09 |
ENSRNOT00000068119
|
Pnliprp2
|
pancreatic lipase related protein 2 |
| chr17_+_43458553 | 27.06 |
ENSRNOT00000088939
|
Slc17a4
|
solute carrier family 17, member 4 |
| chr9_+_4269160 | 26.80 |
ENSRNOT00000050420
|
Sult1c2
|
sulfotransferase family 1C member 2 |
| chr19_-_57333433 | 25.63 |
ENSRNOT00000024917
|
Agt
|
angiotensinogen |
| chr7_+_95074236 | 24.02 |
ENSRNOT00000067649
|
Col14a1
|
collagen type XIV alpha 1 chain |
| chr6_-_7961207 | 23.96 |
ENSRNOT00000007174
|
Abcg5
|
ATP binding cassette subfamily G member 5 |
| chr1_-_260638816 | 23.51 |
ENSRNOT00000065632
ENSRNOT00000017938 ENSRNOT00000077962 |
Pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr19_-_37907714 | 23.25 |
ENSRNOT00000026361
|
Ctrl
|
chymotrypsin-like |
| chr5_+_60528997 | 22.57 |
ENSRNOT00000051445
|
Grhpr
|
glyoxylate and hydroxypyruvate reductase |
| chr1_+_167937026 | 22.45 |
ENSRNOT00000020655
|
Olr57
|
olfactory receptor 57 |
| chrX_+_15669191 | 22.21 |
ENSRNOT00000013553
|
Magix
|
MAGI family member, X-linked |
| chr14_+_22417206 | 21.52 |
ENSRNOT00000052369
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr20_-_27117663 | 21.30 |
ENSRNOT00000000434
|
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr1_-_252849756 | 21.25 |
ENSRNOT00000025845
ENSRNOT00000078279 |
Lipa
|
lipase A, lysosomal acid type |
| chr19_-_43848937 | 21.05 |
ENSRNOT00000044844
|
Ldhd
|
lactate dehydrogenase D |
| chr2_-_195423787 | 19.98 |
ENSRNOT00000071603
|
LOC103689947
|
selenium-binding protein 1 |
| chr4_-_23135354 | 19.90 |
ENSRNOT00000011432
|
Steap4
|
STEAP4 metalloreductase |
| chr13_-_98478327 | 19.47 |
ENSRNOT00000030135
|
Coq8a
|
coenzyme Q8A |
| chr8_+_112594691 | 19.39 |
ENSRNOT00000038383
ENSRNOT00000081281 |
Acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr1_+_201981357 | 19.37 |
ENSRNOT00000027999
|
Acadsb
|
acyl-CoA dehydrogenase, short/branched chain |
| chr6_-_26828972 | 19.33 |
ENSRNOT00000010932
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr3_-_46361092 | 18.96 |
ENSRNOT00000008987
|
Cd302
|
CD302 molecule |
| chr4_-_22192474 | 18.91 |
ENSRNOT00000043822
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr2_-_196113149 | 17.93 |
ENSRNOT00000088465
|
Selenbp1
|
selenium binding protein 1 |
| chr1_+_252589785 | 17.80 |
ENSRNOT00000025928
|
Fas
|
Fas cell surface death receptor |
| chr10_+_83655460 | 17.58 |
ENSRNOT00000008011
|
Gngt2
|
G protein subunit gamma transducin 2 |
| chr1_-_275882444 | 17.28 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr1_+_21613148 | 17.10 |
ENSRNOT00000018695
|
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr7_-_15225309 | 16.82 |
ENSRNOT00000075589
ENSRNOT00000066095 |
Cyp4f6
|
cytochrome P450, family 4, subfamily f, polypeptide 6 |
| chr10_+_104377748 | 16.73 |
ENSRNOT00000080714
|
Llgl2
|
LLGL2, scribble cell polarity complex component |
| chr5_+_159845774 | 16.58 |
ENSRNOT00000012328
|
Epha2
|
Eph receptor A2 |
| chr3_-_2853272 | 16.46 |
ENSRNOT00000023022
|
Fcna
|
ficolin A |
| chr14_+_2050483 | 15.87 |
ENSRNOT00000000047
|
Slc26a1
|
solute carrier family 26 member 1 |
| chr2_+_200793571 | 15.69 |
ENSRNOT00000091444
|
Hao2
|
hydroxyacid oxidase 2 |
| chr10_+_58776792 | 15.65 |
ENSRNOT00000019574
|
Txndc17
|
thioredoxin domain containing 17 |
| chr1_+_247238798 | 15.65 |
ENSRNOT00000083511
|
Rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chr10_-_98388637 | 15.61 |
ENSRNOT00000005665
|
Abca8a
|
ATP-binding cassette, subfamily A (ABC1), member 8a |
| chr7_+_26522314 | 15.35 |
ENSRNOT00000011572
|
Slc41a2
|
solute carrier family 41 member 2 |
| chr9_+_52023295 | 15.10 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr17_+_81352700 | 15.10 |
ENSRNOT00000024736
|
Mrc1
|
mannose receptor, C type 1 |
| chr5_+_152290084 | 15.08 |
ENSRNOT00000089849
|
Aim1l
|
absent in melanoma 1-like |
| chr8_+_22750336 | 15.02 |
ENSRNOT00000013496
|
Ldlr
|
low density lipoprotein receptor |
| chr13_+_91481461 | 14.96 |
ENSRNOT00000012103
|
Olr1576
|
olfactory receptor 1576 |
| chr1_+_217173199 | 14.91 |
ENSRNOT00000075078
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr17_+_18059382 | 14.64 |
ENSRNOT00000031656
|
Nhlrc1
|
NHL repeat containing E3 ubiquitin protein ligase 1 |
| chr6_+_107517668 | 14.42 |
ENSRNOT00000013753
|
Acot4
|
acyl-CoA thioesterase 4 |
| chr4_-_119188251 | 14.39 |
ENSRNOT00000057350
|
Gkn3
|
gastrokine 3 |
| chr13_-_53870428 | 14.27 |
ENSRNOT00000000812
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr18_+_70739492 | 14.23 |
ENSRNOT00000085601
|
Acaa2
|
acetyl-CoA acyltransferase 2 |
| chr8_+_119261747 | 14.22 |
ENSRNOT00000079648
|
AC132539.1
|
|
| chr8_+_71168097 | 14.12 |
ENSRNOT00000081375
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr6_+_7961413 | 14.08 |
ENSRNOT00000007638
ENSRNOT00000061871 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr1_-_164671577 | 14.01 |
ENSRNOT00000091807
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr14_+_59611434 | 14.00 |
ENSRNOT00000065366
|
Cckar
|
cholecystokinin A receptor |
| chr15_-_49505553 | 13.96 |
ENSRNOT00000028974
|
Adamdec1
|
ADAM-like, decysin 1 |
| chrX_-_6620722 | 13.96 |
ENSRNOT00000066674
|
Maoa
|
monoamine oxidase A |
| chr4_+_156752082 | 13.59 |
ENSRNOT00000084588
ENSRNOT00000068407 |
Cd163
|
CD163 molecule |
| chr9_-_71900044 | 13.37 |
ENSRNOT00000020322
|
Idh1
|
isocitrate dehydrogenase (NADP(+)) 1, cytosolic |
| chr1_-_164659992 | 13.32 |
ENSRNOT00000024281
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chrX_+_110818716 | 13.24 |
ENSRNOT00000086308
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr10_-_104564480 | 13.03 |
ENSRNOT00000008525
|
Galk1
|
galactokinase 1 |
| chr10_-_98018014 | 12.79 |
ENSRNOT00000005367
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr11_-_77593171 | 12.69 |
ENSRNOT00000002645
ENSRNOT00000043498 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr11_+_44192940 | 12.65 |
ENSRNOT00000002255
|
St3gal6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr13_-_108460362 | 12.63 |
ENSRNOT00000048200
|
LOC498308
|
Da2-20 |
| chr4_+_157126935 | 12.43 |
ENSRNOT00000056051
|
C1r
|
complement C1r |
| chr10_+_84718824 | 12.38 |
ENSRNOT00000055464
|
Copz2
|
coatomer protein complex, subunit zeta 2 |
| chr11_-_90406797 | 12.33 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chr19_-_43841795 | 12.24 |
ENSRNOT00000079539
|
Ldhd
|
lactate dehydrogenase D |
| chr11_-_62451149 | 11.99 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr16_+_54332660 | 11.67 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr8_-_78233430 | 11.60 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr1_-_146289465 | 11.56 |
ENSRNOT00000017362
|
Abhd17c
|
abhydrolase domain containing 17C |
| chr3_+_2288667 | 11.54 |
ENSRNOT00000012289
|
Entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr1_-_169856727 | 11.50 |
ENSRNOT00000047978
|
Olr180
|
olfactory receptor 180 |
| chr1_+_184301493 | 11.40 |
ENSRNOT00000078096
|
Insc
|
inscuteable homolog (Drosophila) |
| chr13_+_41883137 | 11.39 |
ENSRNOT00000004581
|
Slc35f5
|
solute carrier family 35, member F5 |
| chr1_+_72810545 | 11.32 |
ENSRNOT00000092117
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr9_+_100156154 | 11.29 |
ENSRNOT00000005935
|
Aqp12a
|
aquaporin 12A |
| chr15_+_4554603 | 11.25 |
ENSRNOT00000075153
|
Kcnk5
|
potassium two pore domain channel subfamily K member 5 |
| chr10_+_4719713 | 11.24 |
ENSRNOT00000003412
|
Litaf
|
lipopolysaccharide-induced TNF factor |
| chr5_+_137356801 | 10.98 |
ENSRNOT00000027432
|
RGD1305347
|
similar to RIKEN cDNA 2610528J11 |
| chr13_+_99221013 | 10.90 |
ENSRNOT00000083608
|
Tmem63a
|
transmembrane protein 63a |
| chr1_-_153740905 | 10.68 |
ENSRNOT00000023239
|
Prss23
|
protease, serine, 23 |
| chr15_+_29073755 | 10.55 |
ENSRNOT00000075409
|
AABR07017595.1
|
|
| chr18_-_25314047 | 10.32 |
ENSRNOT00000079778
|
AABR07031674.4
|
|
| chr1_+_203160323 | 10.31 |
ENSRNOT00000027919
|
AABR07005844.1
|
|
| chr3_+_2642531 | 10.00 |
ENSRNOT00000081798
|
Fut7
|
fucosyltransferase 7 |
| chr13_-_80702315 | 9.95 |
ENSRNOT00000004577
|
Fmo4
|
flavin containing monooxygenase 4 |
| chr1_-_215451156 | 9.59 |
ENSRNOT00000066317
|
AABR07072002.1
|
|
| chr3_+_72460889 | 9.59 |
ENSRNOT00000012209
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr8_+_71167305 | 9.45 |
ENSRNOT00000021337
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chrX_-_111942749 | 9.33 |
ENSRNOT00000087583
|
AABR07040840.1
|
|
| chr17_+_72209373 | 9.18 |
ENSRNOT00000064802
|
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr10_+_14105750 | 9.12 |
ENSRNOT00000090552
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr11_+_61661724 | 9.06 |
ENSRNOT00000079423
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr20_+_6877091 | 8.99 |
ENSRNOT00000093654
|
RGD735065
|
similar to GI:13385412-like protein splice form I |
| chr4_-_64981384 | 8.95 |
ENSRNOT00000017338
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr8_+_70994563 | 8.82 |
ENSRNOT00000051504
ENSRNOT00000077163 |
Spg21
|
spastic paraplegia 21 homolog (human) |
| chr3_+_119805941 | 8.78 |
ENSRNOT00000018584
|
Adra2b
|
adrenoceptor alpha 2B |
| chr11_+_66713888 | 8.71 |
ENSRNOT00000003340
|
Fbxo40
|
F-box protein 40 |
| chr8_-_21995806 | 8.64 |
ENSRNOT00000028034
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr1_-_137359072 | 8.62 |
ENSRNOT00000014553
|
Klhl25
|
kelch-like family member 25 |
| chr11_+_70967439 | 8.58 |
ENSRNOT00000032518
|
Iqcg
|
IQ motif containing G |
| chrX_-_26376467 | 8.55 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr1_+_100199057 | 8.45 |
ENSRNOT00000025831
|
Klk1
|
kallikrein 1 |
| chr11_+_61662270 | 8.42 |
ENSRNOT00000079521
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr10_+_14656812 | 8.41 |
ENSRNOT00000024904
|
Prss34
|
protease, serine, 34 |
| chr18_+_12008759 | 8.29 |
ENSRNOT00000022665
ENSRNOT00000061344 ENSRNOT00000038635 |
Dsg1
|
desmoglein 1 |
| chr3_+_138024508 | 8.28 |
ENSRNOT00000007794
|
Dstn
|
destrin, actin depolymerizing factor |
| chr5_-_155264143 | 8.24 |
ENSRNOT00000017385
|
C1qa
|
complement C1q A chain |
| chr20_+_14101659 | 8.17 |
ENSRNOT00000072696
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr4_-_117631716 | 8.01 |
ENSRNOT00000057475
|
LOC103690006
|
RNA/RNP complex-1-interacting phosphatase |
| chr10_-_40805941 | 7.92 |
ENSRNOT00000017588
|
Atox1
|
antioxidant 1 copper chaperone |
| chr13_-_50514151 | 7.90 |
ENSRNOT00000003951
|
Ren
|
renin |
| chr19_-_23554594 | 7.88 |
ENSRNOT00000004590
|
Il15
|
interleukin 15 |
| chr16_-_68936869 | 7.86 |
ENSRNOT00000058643
|
Tex24
|
testis expressed gene 24 |
| chr8_+_90304148 | 7.81 |
ENSRNOT00000001195
|
AABR07070892.1
|
|
| chr1_+_217018916 | 7.77 |
ENSRNOT00000028195
ENSRNOT00000078979 |
Dhcr7
|
7-dehydrocholesterol reductase |
| chr4_-_78458179 | 7.72 |
ENSRNOT00000078473
ENSRNOT00000011327 |
Tmem176b
|
transmembrane protein 176B |
| chr5_-_139864393 | 7.71 |
ENSRNOT00000030016
|
Zfp69
|
zinc finger protein 69 |
| chr19_-_9765484 | 7.65 |
ENSRNOT00000016626
|
Setd6
|
SET domain containing 6 |
| chr1_-_141188031 | 7.64 |
ENSRNOT00000044567
|
Polg
|
DNA polymerase gamma, catalytic subunit |
| chr20_+_3178250 | 7.59 |
ENSRNOT00000082116
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr7_+_136037230 | 7.43 |
ENSRNOT00000071423
|
AABR07058745.1
|
|
| chr10_+_69737328 | 7.38 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr7_+_120202601 | 7.37 |
ENSRNOT00000082862
|
AC096473.3
|
|
| chr13_+_42008842 | 7.21 |
ENSRNOT00000038811
|
Gpr39
|
G protein-coupled receptor 39 |
| chr1_-_168283863 | 7.17 |
ENSRNOT00000021115
|
Olr80
|
olfactory receptor 80 |
| chr5_-_58917822 | 7.16 |
ENSRNOT00000082554
|
AABR07048059.1
|
|
| chr3_+_11679530 | 7.16 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr18_-_64061472 | 7.15 |
ENSRNOT00000022519
|
Fam210a
|
family with sequence similarity 210, member A |
| chr8_+_14060394 | 7.15 |
ENSRNOT00000014827
|
Smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr3_-_48604097 | 7.12 |
ENSRNOT00000009620
|
Ifih1
|
interferon induced with helicase C domain 1 |
| chr12_+_41155497 | 7.09 |
ENSRNOT00000041741
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr2_-_248484100 | 7.07 |
ENSRNOT00000040856
|
Gbp3
|
guanylate binding protein 3 |
| chr17_+_77176716 | 7.07 |
ENSRNOT00000083342
ENSRNOT00000024162 |
Optn
|
optineurin |
| chr5_+_158042587 | 7.04 |
ENSRNOT00000090593
|
Tas1r2
|
taste 1 receptor member 2 |
| chr10_+_87774552 | 7.03 |
ENSRNOT00000044342
|
Krtap9-1
|
keratin associated protein 9-1 |
| chr1_+_167956337 | 6.98 |
ENSRNOT00000090675
|
AC096030.2
|
|
| chr1_-_252116971 | 6.95 |
ENSRNOT00000035013
|
Lipo1
|
lipase, member O1 |
| chr4_+_7257752 | 6.91 |
ENSRNOT00000088357
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr2_+_112914375 | 6.89 |
ENSRNOT00000092737
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr5_+_154552195 | 6.88 |
ENSRNOT00000072864
|
Asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr1_-_101054851 | 6.85 |
ENSRNOT00000027849
|
Prrg2
|
proline rich and Gla domain 2 |
| chr10_-_35716260 | 6.83 |
ENSRNOT00000004308
ENSRNOT00000059255 |
Sqstm1
|
sequestosome 1 |
| chr4_-_29092753 | 6.80 |
ENSRNOT00000014810
|
Bet1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr1_+_172264473 | 6.75 |
ENSRNOT00000050651
|
Olr246
|
olfactory receptor 246 |
| chr7_+_118692851 | 6.71 |
ENSRNOT00000091911
|
LOC100911562
|
apolipoprotein L3-like |
| chr13_-_104080631 | 6.66 |
ENSRNOT00000032865
|
Lyplal1
|
lysophospholipase-like 1 |
| chr5_-_56536772 | 6.65 |
ENSRNOT00000060765
|
Ddx58
|
DEXD/H-box helicase 58 |
| chr6_+_48866601 | 6.64 |
ENSRNOT00000077321
ENSRNOT00000079891 |
Pxdn
|
peroxidasin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb18
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 26.9 | 134.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 17.2 | 68.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 16.6 | 66.4 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 16.4 | 65.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 14.9 | 74.6 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 14.4 | 57.6 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 13.7 | 41.1 | GO:0046865 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 13.5 | 54.1 | GO:0048252 | lauric acid metabolic process(GO:0048252) |
| 12.5 | 37.5 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 10.0 | 49.8 | GO:0009758 | carbohydrate utilization(GO:0009758) fructose transport(GO:0015755) |
| 9.8 | 137.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 8.7 | 34.7 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 8.3 | 41.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 8.1 | 81.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 8.0 | 55.9 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 7.8 | 31.0 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 7.5 | 15.0 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) |
| 7.1 | 21.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 6.9 | 27.5 | GO:0002434 | immune complex clearance(GO:0002434) |
| 6.8 | 27.3 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 6.3 | 38.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 6.0 | 29.8 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 5.9 | 35.5 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 5.8 | 189.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 5.7 | 17.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 5.2 | 15.7 | GO:0018924 | mandelate metabolic process(GO:0018924) |
| 5.1 | 46.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 4.9 | 19.5 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 4.8 | 14.4 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 4.7 | 18.9 | GO:0043215 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) response to cyclosporin A(GO:1905237) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 4.6 | 18.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 4.5 | 35.9 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 4.4 | 53.4 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 4.4 | 44.2 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 4.4 | 13.1 | GO:0035483 | gastric emptying(GO:0035483) |
| 4.3 | 13.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 4.2 | 12.7 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 4.2 | 16.7 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 4.1 | 16.6 | GO:1901491 | notochord formation(GO:0014028) axial mesoderm morphogenesis(GO:0048319) negative regulation of lymphangiogenesis(GO:1901491) |
| 4.1 | 16.5 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 3.9 | 27.1 | GO:0019374 | galactolipid metabolic process(GO:0019374) |
| 3.7 | 41.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 3.6 | 25.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 3.6 | 14.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 3.6 | 14.2 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 3.5 | 14.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 3.3 | 13.4 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 3.0 | 9.1 | GO:0030091 | protein repair(GO:0030091) |
| 2.9 | 5.7 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 2.8 | 5.7 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 2.7 | 8.0 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 2.6 | 7.9 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 2.6 | 7.9 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 2.6 | 15.6 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 2.6 | 7.8 | GO:0016128 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 2.6 | 12.8 | GO:0044691 | tooth eruption(GO:0044691) |
| 2.5 | 7.6 | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 2.4 | 38.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 2.2 | 8.8 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 2.2 | 4.3 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 2.1 | 8.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 2.0 | 4.1 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 2.0 | 14.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 2.0 | 19.9 | GO:0015677 | copper ion import(GO:0015677) |
| 2.0 | 9.9 | GO:0042737 | drug catabolic process(GO:0042737) |
| 1.9 | 9.7 | GO:0045297 | mating plug formation(GO:0042628) post-mating behavior(GO:0045297) |
| 1.9 | 7.7 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 1.9 | 17.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 1.8 | 5.5 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 1.8 | 9.1 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 1.8 | 5.4 | GO:1902953 | endoplasmic reticulum membrane organization(GO:0090158) positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 1.8 | 80.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 1.8 | 19.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 1.7 | 6.7 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 1.7 | 6.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.6 | 4.9 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 1.6 | 6.3 | GO:1990375 | baculum development(GO:1990375) |
| 1.6 | 6.3 | GO:0009597 | detection of virus(GO:0009597) |
| 1.6 | 4.7 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 1.5 | 100.6 | GO:0032094 | response to food(GO:0032094) |
| 1.5 | 3.0 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) |
| 1.4 | 8.6 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.4 | 7.0 | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste(GO:0001582) |
| 1.4 | 42.1 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 1.4 | 4.2 | GO:0019401 | fumarate metabolic process(GO:0006106) glycerol biosynthetic process(GO:0006114) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) alditol biosynthetic process(GO:0019401) |
| 1.3 | 17.5 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 1.3 | 3.9 | GO:0032242 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 1.3 | 11.5 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 1.3 | 20.5 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 1.3 | 3.8 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 1.2 | 3.7 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 1.2 | 2.4 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 1.2 | 15.6 | GO:0099562 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 1.2 | 7.1 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 1.1 | 3.2 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 1.1 | 8.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.0 | 3.1 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 1.0 | 19.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 1.0 | 3.0 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 1.0 | 9.0 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 1.0 | 15.9 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.9 | 4.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.9 | 3.7 | GO:0060701 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.9 | 2.7 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.9 | 7.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.9 | 2.6 | GO:0033123 | positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.9 | 4.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.8 | 4.2 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.8 | 4.8 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.8 | 4.7 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.8 | 1.5 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.8 | 7.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.8 | 0.8 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.7 | 3.7 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.7 | 3.6 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.7 | 10.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.7 | 12.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.7 | 4.1 | GO:0002001 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) |
| 0.7 | 6.8 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.7 | 6.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.7 | 16.8 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.7 | 4.6 | GO:0010587 | miRNA catabolic process(GO:0010587) positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.7 | 2.0 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.6 | 2.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.6 | 13.2 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.6 | 3.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.6 | 49.1 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.6 | 3.1 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.6 | 2.5 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.6 | 3.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.6 | 9.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.6 | 5.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.6 | 4.6 | GO:0060482 | lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.6 | 2.3 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.6 | 11.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.5 | 2.2 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.5 | 1.6 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.5 | 12.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.5 | 4.3 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.5 | 8.5 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.5 | 1.4 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.5 | 4.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.5 | 4.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.4 | 21.7 | GO:0006956 | complement activation(GO:0006956) |
| 0.4 | 11.7 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.4 | 17.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.4 | 15.1 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.4 | 8.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.4 | 4.9 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.4 | 1.5 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.4 | 13.0 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.4 | 6.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.4 | 1.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.4 | 2.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 3.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 1.7 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 3.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.3 | 1.6 | GO:0035983 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.3 | 1.0 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.3 | 1.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.3 | 3.2 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.3 | 3.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 4.8 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.3 | 413.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.3 | 38.1 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.3 | 5.3 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.3 | 1.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.3 | 1.5 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.3 | 3.6 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.3 | 11.4 | GO:0060479 | lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.3 | 1.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 5.6 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.3 | 2.0 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) |
| 0.3 | 5.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 5.6 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.3 | 3.3 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.3 | 2.4 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.3 | 4.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.3 | 3.4 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.3 | 4.7 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.3 | 1.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 2.9 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.2 | 3.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 1.0 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 | 5.0 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.2 | 0.7 | GO:0032197 | transposition, RNA-mediated(GO:0032197) |
| 0.2 | 1.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 3.3 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.2 | 0.9 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.2 | 12.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.2 | 5.4 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.2 | 15.7 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.2 | 26.1 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.2 | 2.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 3.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 1.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 9.2 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.2 | 38.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.2 | 14.4 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.2 | 2.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 2.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.2 | 1.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 0.6 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 3.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 5.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 5.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 5.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 6.2 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 8.6 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 1.6 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.2 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 2.5 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.1 | 2.3 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 7.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 8.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 3.9 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 0.3 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 9.4 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 0.3 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.1 | 5.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 1.0 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 2.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.3 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 2.0 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 1.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.8 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 1.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0035745 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.2 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 3.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 2.9 | GO:0051282 | sequestering of calcium ion(GO:0051208) release of sequestered calcium ion into cytosol(GO:0051209) regulation of sequestering of calcium ion(GO:0051282) negative regulation of sequestering of calcium ion(GO:0051283) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 2.7 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.5 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.8 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.7 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.8 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 1.2 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.7 | 38.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 5.5 | 27.6 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 4.0 | 79.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 3.9 | 30.9 | GO:0043203 | axon hillock(GO:0043203) |
| 3.8 | 49.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 3.6 | 17.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 2.9 | 389.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 2.5 | 7.6 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 2.5 | 7.6 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 2.3 | 32.0 | GO:0031983 | vesicle lumen(GO:0031983) |
| 2.2 | 58.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 2.1 | 15.0 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 2.0 | 3.9 | GO:0097342 | ripoptosome(GO:0097342) |
| 1.5 | 27.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 1.4 | 7.0 | GO:1903767 | sweet taste receptor complex(GO:1903767) taste receptor complex(GO:1903768) |
| 1.3 | 15.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.1 | 9.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 1.0 | 20.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.0 | 114.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.8 | 50.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.8 | 33.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.8 | 8.6 | GO:0002177 | manchette(GO:0002177) |
| 0.8 | 12.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.8 | 3.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.7 | 36.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.6 | 2.6 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.6 | 6.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.6 | 6.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.6 | 6.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.6 | 38.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.6 | 3.9 | GO:0070449 | elongin complex(GO:0070449) |
| 0.5 | 9.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.5 | 1.8 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.4 | 17.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.4 | 3.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.4 | 4.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.4 | 6.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.4 | 17.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.4 | 1.9 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.4 | 15.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.4 | 8.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.4 | 2.9 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.4 | 2.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 4.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 5.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 2.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.3 | 1.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 8.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.3 | 47.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.3 | 5.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 1.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.3 | 211.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.3 | 3.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 1.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 5.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 1.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 17.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 3.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 1.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 107.7 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.2 | 0.9 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 3.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 257.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.2 | 3.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 40.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 2.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 2.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 4.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 1.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 9.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.7 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 4.0 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 2.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 10.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 4.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 9.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 5.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 37.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 7.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 11.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 4.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 2.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 4.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 5.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 95.1 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 0.5 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 2.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 1.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 3.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.9 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.1 | 2.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 87.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.6 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.0 | 12.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 142.0 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 1.8 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 44.8 | 134.5 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 30.7 | 92.0 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 21.8 | 65.4 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 18.0 | 54.1 | GO:0008405 | arachidonic acid 11,12-epoxygenase activity(GO:0008405) |
| 14.7 | 44.2 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 11.7 | 81.8 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 10.3 | 41.3 | GO:0034618 | arginine binding(GO:0034618) |
| 9.3 | 27.8 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 7.4 | 66.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 6.5 | 19.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 6.4 | 19.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 6.2 | 37.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 5.7 | 17.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 5.5 | 49.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 5.5 | 27.6 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 5.3 | 21.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 5.2 | 15.7 | GO:0052853 | very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 5.1 | 35.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 4.7 | 37.9 | GO:0008430 | selenium binding(GO:0008430) |
| 4.5 | 13.4 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 4.3 | 13.0 | GO:0004335 | galactokinase activity(GO:0004335) |
| 4.3 | 34.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 4.2 | 12.7 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 4.1 | 109.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 4.0 | 28.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 3.8 | 38.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 3.8 | 18.9 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 3.6 | 17.8 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 3.4 | 27.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 3.3 | 93.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 3.3 | 19.9 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 3.0 | 15.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 2.9 | 35.2 | GO:0019864 | IgG binding(GO:0019864) |
| 2.9 | 8.8 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 2.9 | 34.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 2.9 | 17.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 2.8 | 55.5 | GO:0005537 | mannose binding(GO:0005537) |
| 2.7 | 8.0 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 2.6 | 25.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 2.4 | 2.4 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 2.4 | 7.2 | GO:0005534 | galactose binding(GO:0005534) |
| 2.3 | 9.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 2.3 | 41.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 2.2 | 6.5 | GO:0038100 | nodal binding(GO:0038100) |
| 2.1 | 64.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 2.1 | 6.2 | GO:0031765 | galanin receptor binding(GO:0031763) type 2 galanin receptor binding(GO:0031765) type 3 galanin receptor binding(GO:0031766) |
| 2.0 | 231.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 2.0 | 14.2 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 2.0 | 41.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 2.0 | 15.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.9 | 19.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.9 | 5.6 | GO:0047708 | biotinidase activity(GO:0047708) |
| 1.8 | 5.4 | GO:0047522 | 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 1.8 | 23.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 1.8 | 7.0 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 1.7 | 39.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 1.7 | 5.0 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 1.6 | 27.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 1.6 | 7.9 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 1.6 | 6.3 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 1.6 | 4.7 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.5 | 9.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 1.5 | 3.1 | GO:0031762 | follicle-stimulating hormone receptor binding(GO:0031762) |
| 1.5 | 29.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 1.4 | 5.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.4 | 14.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 1.4 | 7.1 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 1.4 | 4.2 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.3 | 6.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 1.3 | 7.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 1.3 | 5.0 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 1.3 | 11.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 1.2 | 34.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 1.2 | 15.6 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 1.2 | 22.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 1.1 | 5.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.1 | 35.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 1.1 | 12.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.1 | 15.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.1 | 3.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) high-density lipoprotein particle receptor activity(GO:0070506) |
| 1.1 | 15.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 1.0 | 4.2 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 1.0 | 22.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 1.0 | 3.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 1.0 | 3.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.9 | 11.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.9 | 44.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.9 | 3.7 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.9 | 31.5 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.9 | 8.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.9 | 2.6 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.8 | 5.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.8 | 5.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.8 | 2.3 | GO:0071209 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.8 | 6.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.8 | 17.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.7 | 11.6 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.7 | 8.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.7 | 4.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.7 | 11.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.7 | 6.9 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.6 | 142.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.6 | 7.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.6 | 2.9 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.6 | 4.6 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.6 | 9.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.6 | 8.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.6 | 1.7 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.5 | 3.8 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.5 | 31.6 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.5 | 12.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.5 | 9.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.4 | 2.6 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.4 | 1.7 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.4 | 7.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.4 | 1.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.4 | 3.9 | GO:0070513 | death domain binding(GO:0070513) |
| 0.4 | 17.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.3 | 2.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.3 | 11.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 3.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 6.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 39.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.3 | 1.5 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.3 | 3.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.3 | 381.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.3 | 24.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.3 | 4.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 1.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 6.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 1.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 3.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 2.2 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.2 | 2.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 3.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 3.4 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 16.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.2 | 3.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.2 | 23.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 6.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.2 | 3.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 1.1 | GO:0034235 | GPI-anchor transamidase activity(GO:0003923) GPI anchor binding(GO:0034235) |
| 0.2 | 2.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 3.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 3.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.8 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 5.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 23.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 0.5 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.2 | 2.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 4.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 4.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 5.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 5.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 3.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 5.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 6.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 3.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 2.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 3.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 7.6 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 1.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 6.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 1.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.7 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 1.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 2.1 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 5.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 6.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 1.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.6 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 2.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 10.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 7.8 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.1 | 13.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 2.2 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 11.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 2.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 2.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 6.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 377.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 1.5 | 82.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.9 | 9.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.9 | 21.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.7 | 35.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.7 | 61.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.6 | 49.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.6 | 11.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.6 | 8.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.6 | 9.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.5 | 14.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.5 | 76.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.4 | 3.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.4 | 89.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 6.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 15.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 4.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 5.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 15.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 4.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 3.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 5.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 2.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 133.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 7.3 | 65.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 5.3 | 63.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 5.1 | 51.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 5.0 | 85.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 4.9 | 44.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 4.8 | 62.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 4.8 | 138.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 3.0 | 27.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 2.8 | 139.3 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 2.4 | 12.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 2.1 | 39.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 2.0 | 13.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 1.9 | 75.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 1.8 | 27.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 1.8 | 1.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 1.4 | 53.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 1.4 | 11.0 | REACTOME DEFENSINS | Genes involved in Defensins |
| 1.3 | 36.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 1.1 | 19.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 1.1 | 14.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.0 | 34.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.9 | 138.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.9 | 13.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.8 | 5.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.7 | 8.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.7 | 42.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.7 | 36.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.7 | 9.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.7 | 12.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.6 | 6.8 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.6 | 8.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.6 | 17.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.5 | 15.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.4 | 13.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.4 | 6.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.4 | 7.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 25.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.3 | 14.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.3 | 7.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 16.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 6.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 2.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 8.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 3.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 6.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 1.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 0.9 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.2 | 4.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 3.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 6.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 13.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 2.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 2.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.0 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 3.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 5.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.7 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |