Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Zbtb16
Z-value: 1.10
Transcription factors associated with Zbtb16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb16
|
ENSRNOG00000029980 | zinc finger and BTB domain containing 16 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb16 | rn6_v1_chr8_-_53146953_53146953 | -0.31 | 2.1e-08 | Click! |
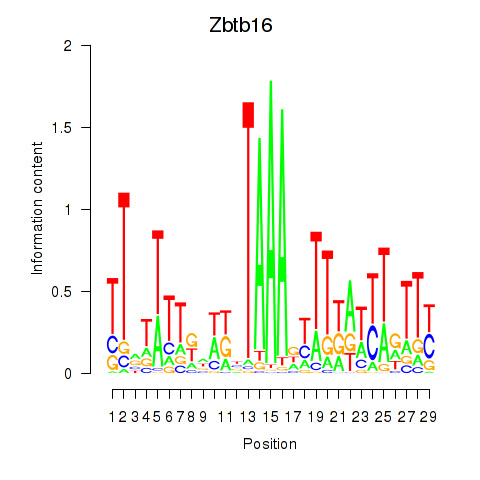
Activity profile of Zbtb16 motif
Sorted Z-values of Zbtb16 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_163762434 | 56.70 |
ENSRNOT00000081854
|
Ly49si1
|
immunoreceptor Ly49si1 |
| chr4_-_164051812 | 56.01 |
ENSRNOT00000085719
|
AABR07062183.1
|
|
| chr10_-_85947938 | 35.57 |
ENSRNOT00000037318
ENSRNOT00000082427 |
Arl5c
|
ADP-ribosylation factor like GTPase 5C |
| chr4_-_163954817 | 35.41 |
ENSRNOT00000079951
|
Ly49si3
|
immunoreceptor Ly49si3 |
| chr4_-_164015365 | 33.84 |
ENSRNOT00000078121
|
Ly49si2
|
immunoreceptor Ly49si2 |
| chr20_+_3875706 | 33.81 |
ENSRNOT00000036900
|
RT1-Ha
|
RT1 class II, locus Ha |
| chr4_-_163849618 | 33.38 |
ENSRNOT00000086363
ENSRNOT00000077637 |
Ly49si1
|
immunoreceptor Ly49si1 |
| chr17_+_43633675 | 32.11 |
ENSRNOT00000072119
|
LOC102549173
|
histone H3.2-like |
| chr4_-_163810403 | 31.84 |
ENSRNOT00000079704
|
AABR07062170.1
|
|
| chr20_-_1878126 | 31.83 |
ENSRNOT00000000995
|
Ubd
|
ubiquitin D |
| chr6_+_122254841 | 29.36 |
ENSRNOT00000005055
|
Gpr65
|
G-protein coupled receptor 65 |
| chr4_-_164123974 | 26.50 |
ENSRNOT00000082447
|
LOC502907
|
similar to immunoreceptor Ly49si1 |
| chr11_-_60547201 | 22.26 |
ENSRNOT00000093151
|
Btla
|
B and T lymphocyte associated |
| chr16_-_75481115 | 22.21 |
ENSRNOT00000035128
|
Defa7
|
defensin alpha 7 |
| chr7_-_118792625 | 21.97 |
ENSRNOT00000007398
|
Myh9
|
myosin, heavy chain 9 |
| chr4_-_164174725 | 20.94 |
ENSRNOT00000080754
|
AABR07062185.1
|
|
| chr4_+_14109864 | 20.94 |
ENSRNOT00000076349
|
RGD1565355
|
similar to fatty acid translocase/CD36 |
| chr13_-_100450209 | 20.65 |
ENSRNOT00000090712
|
Lbr
|
lamin B receptor |
| chr10_+_30038709 | 20.51 |
ENSRNOT00000005898
|
Il12b
|
interleukin 12B |
| chr13_-_61591139 | 19.92 |
ENSRNOT00000005324
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr1_+_81763614 | 18.87 |
ENSRNOT00000027254
|
Cd79a
|
CD79a molecule |
| chrX_+_28486869 | 18.82 |
ENSRNOT00000005620
|
Tlr7
|
toll-like receptor 7 |
| chr11_-_60546997 | 18.82 |
ENSRNOT00000083124
ENSRNOT00000050092 |
Btla
|
B and T lymphocyte associated |
| chr3_-_2853272 | 18.27 |
ENSRNOT00000023022
|
Fcna
|
ficolin A |
| chr9_-_16795887 | 17.90 |
ENSRNOT00000071609
|
Cd79al
|
Cd79a molecule-like |
| chr3_+_100373439 | 17.79 |
ENSRNOT00000085176
|
Kif18a
|
kinesin family member 18A |
| chr1_+_223214132 | 17.55 |
ENSRNOT00000066559
|
AABR07006142.1
|
|
| chr6_-_140572023 | 17.37 |
ENSRNOT00000072338
|
AABR07065772.1
|
|
| chr9_+_92681078 | 16.65 |
ENSRNOT00000034152
|
Sp100
|
SP100 nuclear antigen |
| chr6_-_140407307 | 16.64 |
ENSRNOT00000079087
|
AABR07065768.3
|
|
| chr14_-_82347679 | 16.39 |
ENSRNOT00000032972
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr4_-_163463718 | 15.63 |
ENSRNOT00000085671
|
Klrc1
|
killer cell lectin like receptor C1 |
| chr20_-_14025067 | 15.62 |
ENSRNOT00000093430
ENSRNOT00000074533 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr19_+_26016382 | 15.60 |
ENSRNOT00000004601
|
Klf1
|
Kruppel like factor 1 |
| chr3_+_20375699 | 15.43 |
ENSRNOT00000088492
|
AABR07051731.1
|
|
| chr9_+_91001828 | 15.08 |
ENSRNOT00000080956
|
AABR07068214.1
|
|
| chr7_-_2961873 | 14.53 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chr15_+_31642169 | 14.43 |
ENSRNOT00000072362
|
AABR07017833.1
|
|
| chr18_-_411849 | 14.38 |
ENSRNOT00000033663
|
Mtcp1
|
mature T-cell proliferation 1 |
| chr2_-_54777729 | 14.16 |
ENSRNOT00000082548
|
C7
|
complement C7 |
| chr2_-_202350929 | 14.03 |
ENSRNOT00000026306
|
LOC108348086
|
3 beta-hydroxysteroid dehydrogenase/Delta 5-->4-isomerase type 4 |
| chr5_+_154260062 | 13.98 |
ENSRNOT00000074998
|
Cnr2
|
cannabinoid receptor 2 |
| chr12_-_2438817 | 13.89 |
ENSRNOT00000037059
|
Ccl25
|
C-C motif chemokine ligand 25 |
| chr4_-_165118062 | 13.69 |
ENSRNOT00000087219
|
LOC502908
|
Ly49s8 |
| chr13_+_53351717 | 12.83 |
ENSRNOT00000012038
|
Kif14
|
kinesin family member 14 |
| chr7_+_71065197 | 12.66 |
ENSRNOT00000051075
|
Rdh16
|
retinol dehydrogenase 16 (all-trans) |
| chr3_-_10153554 | 12.53 |
ENSRNOT00000093246
|
Exosc2
|
exosome component 2 |
| chr14_-_86078954 | 12.47 |
ENSRNOT00000018319
|
Polm
|
DNA polymerase mu |
| chr13_-_92210052 | 12.45 |
ENSRNOT00000004670
|
Olr1592
|
olfactory receptor 1592 |
| chr2_-_31753528 | 11.79 |
ENSRNOT00000075057
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr18_+_44716226 | 11.71 |
ENSRNOT00000086431
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr8_-_84320714 | 11.63 |
ENSRNOT00000079356
ENSRNOT00000088487 |
Tinag
|
tubulointerstitial nephritis antigen |
| chr1_+_227640680 | 11.38 |
ENSRNOT00000033613
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C |
| chr18_+_56414488 | 11.36 |
ENSRNOT00000088988
|
Csf1r
|
colony stimulating factor 1 receptor |
| chr2_-_56151531 | 11.24 |
ENSRNOT00000040847
ENSRNOT00000080677 |
Osmr
|
oncostatin M receptor |
| chr11_+_64472072 | 11.17 |
ENSRNOT00000042756
|
RGD1563835
|
similar to ribosomal protein L27 |
| chr3_-_118635268 | 10.99 |
ENSRNOT00000086357
|
Atp8b4
|
ATPase phospholipid transporting 8B4 (putative) |
| chr14_-_88886198 | 10.94 |
ENSRNOT00000012661
|
Tns3
|
tensin 3 |
| chr4_+_171748273 | 10.93 |
ENSRNOT00000009998
|
Dera
|
deoxyribose-phosphate aldolase |
| chr2_+_153830586 | 10.91 |
ENSRNOT00000042576
|
Mme
|
membrane metallo-endopeptidase |
| chr4_+_162937774 | 10.81 |
ENSRNOT00000083188
|
Clec2g
|
C-type lectin domain family 2, member G |
| chr4_-_162680757 | 10.79 |
ENSRNOT00000085918
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr1_+_229416489 | 10.73 |
ENSRNOT00000028617
|
Olr336
|
olfactory receptor 336 |
| chr13_-_99531959 | 10.67 |
ENSRNOT00000005059
|
Wdr26
|
WD repeat domain 26 |
| chr7_-_60860990 | 10.59 |
ENSRNOT00000009511
|
Rap1b
|
RAP1B, member of RAS oncogene family |
| chr10_-_103590607 | 10.51 |
ENSRNOT00000034741
|
Cd300le
|
Cd300 molecule-like family member E |
| chr6_-_99576196 | 10.20 |
ENSRNOT00000040565
|
RGD1562381
|
similar to ribosomal protein S17 |
| chr14_+_46001849 | 10.05 |
ENSRNOT00000076611
|
Rell1
|
RELT-like 1 |
| chr10_+_89484468 | 9.98 |
ENSRNOT00000035523
|
Tmem106a
|
transmembrane protein 106A |
| chr4_-_164304532 | 9.91 |
ENSRNOT00000089484
|
Ly49i5
|
Ly49 inhibitory receptor 5 |
| chr2_+_240396152 | 9.76 |
ENSRNOT00000034565
|
Cenpe
|
centromere protein E |
| chr9_+_65620658 | 9.74 |
ENSRNOT00000084498
|
Casp8
|
caspase 8 |
| chr1_-_22269831 | 9.65 |
ENSRNOT00000081804
|
Stx7
|
syntaxin 7 |
| chr8_-_119326938 | 9.64 |
ENSRNOT00000044467
|
Ccrl2
|
C-C motif chemokine receptor like 2 |
| chr19_-_55389256 | 9.63 |
ENSRNOT00000064180
|
Aprt
|
adenine phosphoribosyl transferase |
| chr2_+_178679395 | 9.23 |
ENSRNOT00000088686
|
Fam198b
|
family with sequence similarity 198, member B |
| chr9_-_20621051 | 9.04 |
ENSRNOT00000015918
|
Tnfrsf21
|
TNF receptor superfamily member 21 |
| chr7_+_12535527 | 9.01 |
ENSRNOT00000093135
|
Polr2e
|
RNA polymerase II subunit E |
| chr10_+_15484795 | 9.00 |
ENSRNOT00000079826
|
Tmem8a
|
transmembrane protein 8A |
| chr6_-_146195819 | 8.97 |
ENSRNOT00000007625
|
Sp4
|
Sp4 transcription factor |
| chr1_+_68436917 | 8.73 |
ENSRNOT00000088586
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr6_-_145777767 | 8.52 |
ENSRNOT00000046799
|
Rpl32
|
ribosomal protein L32 |
| chr9_-_17212628 | 8.48 |
ENSRNOT00000002656
ENSRNOT00000007876 |
RGD1562399
|
similar to 40S ribosomal protein S2 |
| chr2_-_33920837 | 8.44 |
ENSRNOT00000075196
|
Erbin
|
erbb2 interacting protein |
| chr4_-_103569159 | 8.39 |
ENSRNOT00000084036
|
AABR07061072.1
|
|
| chr1_+_247562202 | 8.34 |
ENSRNOT00000021614
|
Pdcd1lg2
|
programmed cell death 1 ligand 2 |
| chr15_-_36798814 | 8.33 |
ENSRNOT00000065764
|
Cenpj
|
centromere protein J |
| chr1_-_134870255 | 8.27 |
ENSRNOT00000055829
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr1_+_68436593 | 8.25 |
ENSRNOT00000080325
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr20_-_5805627 | 8.17 |
ENSRNOT00000085996
|
Clps
|
colipase |
| chr19_-_38484611 | 8.16 |
ENSRNOT00000077960
|
Nfat5
|
nuclear factor of activated T-cells 5 |
| chr19_+_24849356 | 8.14 |
ENSRNOT00000090312
|
Ddx39a
|
DExD-box helicase 39A |
| chr9_+_61738471 | 8.13 |
ENSRNOT00000090305
|
AABR07067762.1
|
|
| chr4_+_163112301 | 8.10 |
ENSRNOT00000087113
|
Clec12a
|
C-type lectin domain family 12, member A |
| chr3_-_72071895 | 7.84 |
ENSRNOT00000082857
|
Selenoh
|
selenoprotein H |
| chr9_+_65172194 | 7.66 |
ENSRNOT00000040493
|
AC128084.1
|
|
| chr12_-_40686249 | 7.56 |
ENSRNOT00000042264
|
LOC100909911
|
40S ribosomal protein S20-like |
| chr6_+_135680615 | 7.56 |
ENSRNOT00000077770
|
Traf3
|
Tnf receptor-associated factor 3 |
| chr8_-_21453190 | 7.52 |
ENSRNOT00000078192
|
Zfp26
|
zinc finger protein 26 |
| chr12_-_46414434 | 7.49 |
ENSRNOT00000041281
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr8_+_116754178 | 7.43 |
ENSRNOT00000068295
ENSRNOT00000084429 |
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr17_-_89163113 | 7.39 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr15_-_27726646 | 7.37 |
ENSRNOT00000082139
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr14_+_104452917 | 7.29 |
ENSRNOT00000050489
|
LOC682793
|
similar to 60S ribosomal protein L38 |
| chr4_-_163649637 | 7.19 |
ENSRNOT00000080873
|
LOC690020
|
similar to killer cell lectin-like receptor, subfamily A, member 17 |
| chr1_+_91716383 | 7.08 |
ENSRNOT00000016919
|
Slc7a9
|
solute carrier family 7 member 9 |
| chr1_-_73732118 | 6.90 |
ENSRNOT00000077964
|
Leng8
|
leukocyte receptor cluster member 8 |
| chr6_+_95826546 | 6.81 |
ENSRNOT00000043587
|
AABR07064810.1
|
|
| chr10_-_76039964 | 6.77 |
ENSRNOT00000003164
|
Msi2
|
musashi RNA-binding protein 2 |
| chr4_-_165522418 | 6.70 |
ENSRNOT00000064420
|
Magohb
|
mago homolog B, exon junction complex core component |
| chr1_-_91588609 | 6.68 |
ENSRNOT00000050931
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr1_-_89258935 | 6.66 |
ENSRNOT00000003180
|
LOC100361079
|
ribosomal protein L36-like |
| chr2_+_26240385 | 6.64 |
ENSRNOT00000024292
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr8_+_23193181 | 6.59 |
ENSRNOT00000071703
|
Zfp872
|
zinc finger protein 872 |
| chr19_-_9843673 | 6.56 |
ENSRNOT00000015795
|
Gins3
|
GINS complex subunit 3 |
| chr3_+_112346627 | 6.43 |
ENSRNOT00000074392
|
Snap23
|
synaptosomal-associated protein 23 |
| chr10_+_104036367 | 6.40 |
ENSRNOT00000004843
|
Armc7
|
armadillo repeat containing 7 |
| chrX_-_1076850 | 6.31 |
ENSRNOT00000090671
|
LOC100363372
|
zinc finger protein 81 (HFZ20)-like |
| chr9_-_88534710 | 6.31 |
ENSRNOT00000020880
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr16_-_80850340 | 6.22 |
ENSRNOT00000000124
|
Champ1
|
chromosome alignment maintaining phosphoprotein 1 |
| chrX_+_77119911 | 6.16 |
ENSRNOT00000080141
|
Atp7a
|
ATPase copper transporting alpha |
| chr1_+_198960542 | 6.09 |
ENSRNOT00000072389
|
Srcap
|
Snf2-related CREBBP activator protein |
| chr4_-_165361623 | 6.01 |
ENSRNOT00000088908
|
Ly49i7
|
immunoreceptor Ly49i7 |
| chr12_-_11808977 | 5.96 |
ENSRNOT00000071795
|
AABR07035375.1
|
|
| chrX_+_40286592 | 5.95 |
ENSRNOT00000087898
|
Mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr4_+_31704747 | 5.94 |
ENSRNOT00000047850
|
AABR07059675.1
|
|
| chr15_-_34263224 | 5.92 |
ENSRNOT00000026001
|
Emc9
|
ER membrane protein complex subunit 9 |
| chr5_+_57657909 | 5.88 |
ENSRNOT00000073017
|
LOC683961
|
similar to ribosomal protein S13 |
| chr2_+_251805392 | 5.88 |
ENSRNOT00000019911
|
Bcl10
|
B-cell CLL/lymphoma 10 |
| chr12_+_11179329 | 5.87 |
ENSRNOT00000001302
|
Zfp394
|
zinc finger protein 394 |
| chr1_-_72329856 | 5.87 |
ENSRNOT00000021391
|
U2af2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr12_-_6879154 | 5.87 |
ENSRNOT00000001207
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr8_+_82038967 | 5.83 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr5_-_60871705 | 5.78 |
ENSRNOT00000016582
|
Exosc3
|
exosome component 3 |
| chr10_-_94652658 | 5.72 |
ENSRNOT00000017371
|
Ern1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr1_-_221233905 | 5.71 |
ENSRNOT00000092740
ENSRNOT00000028331 |
Frmd8
|
FERM domain containing 8 |
| chr2_+_2694480 | 5.60 |
ENSRNOT00000083565
|
AABR07072671.1
|
|
| chr3_-_4341771 | 5.55 |
ENSRNOT00000034694
|
LOC684988
|
similar to ribosomal protein S13 |
| chr11_-_72814850 | 5.55 |
ENSRNOT00000091575
|
LOC100911374
|
UBX domain-containing protein 7-like |
| chr12_-_43576754 | 5.53 |
ENSRNOT00000042331
ENSRNOT00000081780 |
Med13l
|
mediator complex subunit 13-like |
| chr4_-_162726628 | 5.42 |
ENSRNOT00000073877
|
LOC100911272
|
killer cell lectin-like receptor 5-like |
| chrX_+_119390013 | 5.40 |
ENSRNOT00000074269
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr12_-_21973474 | 5.36 |
ENSRNOT00000071110
|
RGD1566138
|
similar to Zinc finger, CW type with PWWP domain 1 |
| chr15_+_37171052 | 5.33 |
ENSRNOT00000011684
|
Zmym2
|
zinc finger MYM-type containing 2 |
| chr16_+_21518873 | 5.33 |
ENSRNOT00000015635
|
Zfp868
|
zinc finger protein 868 |
| chr3_+_156886454 | 5.24 |
ENSRNOT00000022403
|
Lpin3
|
lipin 3 |
| chr17_-_90522091 | 5.21 |
ENSRNOT00000077767
|
Lyst
|
lysosomal trafficking regulator |
| chr5_+_135354700 | 5.20 |
ENSRNOT00000021850
|
Ipp
|
intracisternal A particle-promoted polypeptide |
| chr8_+_119214541 | 5.12 |
ENSRNOT00000050117
|
RGD1564138
|
similar to 60S ribosomal protein L29 (P23) |
| chr12_-_13462038 | 5.06 |
ENSRNOT00000043110
|
LOC100360439
|
ribosomal protein L36-like |
| chr3_+_80670140 | 5.06 |
ENSRNOT00000085614
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr10_+_65552897 | 5.01 |
ENSRNOT00000056217
|
Spag5
|
sperm associated antigen 5 |
| chr4_-_120408232 | 5.00 |
ENSRNOT00000073269
|
LOC100911337
|
40S ribosomal protein S25-like |
| chr1_+_11963836 | 4.93 |
ENSRNOT00000074325
|
AABR07000398.1
|
|
| chr10_-_5344121 | 4.91 |
ENSRNOT00000003479
|
Nubp1
|
nucleotide binding protein 1 |
| chr8_-_13906355 | 4.87 |
ENSRNOT00000029634
|
Cep295
|
centrosomal protein 295 |
| chr14_-_21299068 | 4.83 |
ENSRNOT00000065778
|
Amtn
|
amelotin |
| chr5_-_135994848 | 4.82 |
ENSRNOT00000067675
|
Btbd19
|
BTB domain containing 19 |
| chr3_+_21983841 | 4.81 |
ENSRNOT00000043621
|
RGD1561137
|
similar to 40S ribosomal protein S16 |
| chr5_-_12199283 | 4.81 |
ENSRNOT00000007769
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr6_-_91676344 | 4.80 |
ENSRNOT00000083872
|
Nemf
|
nuclear export mediator factor |
| chr7_+_121361415 | 4.79 |
ENSRNOT00000067904
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr20_-_26852199 | 4.65 |
ENSRNOT00000078739
|
Sirt1
|
sirtuin 1 |
| chr1_-_78110786 | 4.55 |
ENSRNOT00000089718
|
Dhx34
|
DExH-box helicase 34 |
| chr18_-_72649915 | 4.52 |
ENSRNOT00000072205
|
AABR07032582.1
|
|
| chr16_+_19767264 | 4.51 |
ENSRNOT00000051802
ENSRNOT00000092073 |
Ocel1
|
occludin/ELL domain containing 1 |
| chr2_-_235161263 | 4.51 |
ENSRNOT00000080235
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr5_-_50068706 | 4.51 |
ENSRNOT00000084643
|
Orc3
|
origin recognition complex, subunit 3 |
| chr2_+_243425007 | 4.48 |
ENSRNOT00000082894
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr14_+_17195014 | 4.44 |
ENSRNOT00000031667
|
Cxcl11
|
C-X-C motif chemokine ligand 11 |
| chr20_-_3440769 | 4.44 |
ENSRNOT00000084981
|
Ier3
|
immediate early response 3 |
| chr17_-_66288202 | 4.43 |
ENSRNOT00000083172
|
Mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr12_+_47590154 | 4.36 |
ENSRNOT00000045946
|
Git2
|
GIT ArfGAP 2 |
| chr2_-_106703401 | 4.31 |
ENSRNOT00000043933
|
AABR07009638.1
|
|
| chr10_+_66509107 | 4.28 |
ENSRNOT00000017230
|
RGD1565317
|
similar to ubiquitin-like/S30 ribosomal fusion protein |
| chr17_+_1305016 | 4.22 |
ENSRNOT00000025965
|
Ercc6l2
|
ERCC excision repair 6 like 2 |
| chr5_-_138361702 | 4.20 |
ENSRNOT00000067943
ENSRNOT00000064712 |
Ppih
|
peptidylprolyl isomerase H |
| chr13_-_91737053 | 4.19 |
ENSRNOT00000083561
|
Fcer1a
|
Fc fragment of IgE receptor Ia |
| chr17_-_6892497 | 4.06 |
ENSRNOT00000026407
|
Idnk
|
Idnk, gluconokinase |
| chr1_-_238376841 | 4.03 |
ENSRNOT00000076393
|
Tmc1
|
transmembrane channel-like 1 |
| chr18_+_76317134 | 4.01 |
ENSRNOT00000074465
|
Mrps21l
|
mitochondrial ribosomal protein S21-like |
| chr7_-_123476238 | 4.01 |
ENSRNOT00000036917
|
Cenpm
|
centromere protein M |
| chr14_-_45376127 | 3.96 |
ENSRNOT00000059247
|
AABR07015015.2
|
|
| chr10_-_97750679 | 3.95 |
ENSRNOT00000078059
|
Slc16a6
|
solute carrier family 16, member 6 |
| chr2_-_66608324 | 3.93 |
ENSRNOT00000077597
|
Cln5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr12_-_40822159 | 3.89 |
ENSRNOT00000001826
|
Hectd4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr15_+_49010492 | 3.89 |
ENSRNOT00000024527
|
Nuggc
|
nuclear GTPase, germinal center associated |
| chr17_+_53965562 | 3.86 |
ENSRNOT00000090387
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr2_+_236625357 | 3.85 |
ENSRNOT00000081248
|
Papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr13_-_81214494 | 3.79 |
ENSRNOT00000004950
ENSRNOT00000082385 |
Prrx1
|
paired related homeobox 1 |
| chr1_-_84732242 | 3.76 |
ENSRNOT00000025586
|
LOC102553760
|
zinc finger protein 59-like |
| chr6_+_107001763 | 3.75 |
ENSRNOT00000011378
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr16_-_19583386 | 3.73 |
ENSRNOT00000090131
|
Zfp617
|
zinc finger protein 617 |
| chr3_+_111298713 | 3.65 |
ENSRNOT00000043677
|
Dppa3
|
developmental pluripotency-associated 3 |
| chr6_+_10642700 | 3.64 |
ENSRNOT00000040329
|
LOC100359600
|
karyopherin alpha 2-like |
| chr16_+_2278701 | 3.58 |
ENSRNOT00000016233
|
Dennd6a
|
DENN domain containing 6A |
| chr5_-_141363524 | 3.56 |
ENSRNOT00000084109
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr17_+_13519130 | 3.53 |
ENSRNOT00000076794
|
Sema4d
|
semaphorin 4D |
| chr4_+_155531906 | 3.51 |
ENSRNOT00000060937
|
Nanog
|
Nanog homeobox |
| chr13_-_25652473 | 3.50 |
ENSRNOT00000020752
|
Pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb16
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 22.0 | GO:1903919 | protein processing in phagocytic vesicle(GO:1900756) regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 6.1 | 18.3 | GO:0071038 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 5.5 | 16.6 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 5.1 | 20.5 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 4.6 | 18.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 4.3 | 12.8 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 4.0 | 31.8 | GO:0070842 | aggresome assembly(GO:0070842) |
| 3.9 | 15.6 | GO:0031179 | peptide modification(GO:0031179) |
| 3.8 | 18.8 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 3.7 | 41.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 3.2 | 9.6 | GO:0044209 | AMP salvage(GO:0044209) |
| 2.9 | 5.9 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 2.8 | 8.4 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 2.8 | 11.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 2.8 | 13.9 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 2.7 | 10.9 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 2.6 | 5.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 2.4 | 9.7 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 2.4 | 7.2 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 2.3 | 11.4 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 2.3 | 9.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 2.2 | 6.7 | GO:1904638 | response to resveratrol(GO:1904638) |
| 2.2 | 10.9 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 2.1 | 10.6 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 2.1 | 8.3 | GO:0061511 | centriole elongation(GO:0061511) |
| 2.0 | 5.9 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 2.0 | 5.9 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 1.9 | 33.8 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 1.8 | 5.4 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) regulation of metanephros size(GO:0035566) |
| 1.8 | 7.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 1.6 | 4.9 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 1.6 | 9.8 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 1.6 | 4.8 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 1.6 | 12.7 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.6 | 14.0 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 1.5 | 6.2 | GO:1904959 | negative regulation of iron ion transmembrane transport(GO:0034760) elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 1.5 | 4.5 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 1.5 | 7.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 1.5 | 5.8 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.4 | 4.2 | GO:0045401 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 1.3 | 15.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 1.3 | 16.4 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 1.2 | 3.7 | GO:0044726 | protection of DNA demethylation of female pronucleus(GO:0044726) |
| 1.2 | 11.8 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 1.2 | 3.5 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 1.1 | 5.7 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 1.1 | 36.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 1.1 | 4.4 | GO:1904640 | response to methionine(GO:1904640) |
| 1.1 | 16.4 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 1.1 | 3.2 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 1.1 | 17.0 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 1.0 | 3.0 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 1.0 | 5.7 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.9 | 12.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.9 | 8.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.9 | 6.4 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.9 | 3.6 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.9 | 2.6 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.8 | 7.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.8 | 4.1 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.8 | 4.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.8 | 14.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.8 | 6.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.8 | 17.8 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.8 | 4.6 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.7 | 1.5 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.7 | 3.9 | GO:1904426 | positive regulation of GTP binding(GO:1904426) |
| 0.7 | 2.6 | GO:0019042 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.6 | 2.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.6 | 2.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.6 | 1.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.6 | 1.7 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.6 | 2.2 | GO:1900170 | glucocorticoid mediated signaling pathway(GO:0043402) regulation of glucocorticoid mediated signaling pathway(GO:1900169) negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.5 | 3.8 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.5 | 3.2 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.5 | 5.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.5 | 8.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.5 | 1.9 | GO:2000851 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) positive regulation of cortisol secretion(GO:0051464) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.5 | 3.2 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.4 | 3.6 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.4 | 1.8 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.4 | 3.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.4 | 13.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.4 | 1.3 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.4 | 2.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.4 | 8.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.4 | 3.8 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.4 | 12.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.4 | 4.4 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 0.4 | 18.9 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.3 | 2.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 | 11.4 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.3 | 32.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.3 | 3.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.3 | 2.3 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) regulation of hepatic stellate cell activation(GO:2000489) |
| 0.3 | 0.9 | GO:1901090 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.3 | 7.5 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.3 | 4.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 4.4 | GO:0010818 | T cell chemotaxis(GO:0010818) positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.2 | 2.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 1.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 1.0 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.2 | 1.7 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 1.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 | 4.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 0.6 | GO:0030327 | prenylated protein catabolic process(GO:0030327) CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 5.1 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.2 | 0.4 | GO:0045590 | positive regulation of memory T cell differentiation(GO:0043382) negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.2 | 3.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 2.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 1.8 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 6.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.2 | 1.9 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 20.9 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.2 | 10.5 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.2 | 2.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.2 | 9.0 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.2 | 8.5 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.2 | 0.5 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.2 | 3.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 4.4 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 10.9 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 11.7 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 3.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.5 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 2.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.4 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 0.1 | 1.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 2.1 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 6.6 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 3.1 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.1 | 0.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.6 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 4.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 3.2 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 3.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.7 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.1 | 1.7 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 1.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 1.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 2.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 2.9 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.1 | 4.0 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 3.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 27.5 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.1 | 4.6 | GO:0051168 | nuclear export(GO:0051168) |
| 0.1 | 5.2 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.1 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 1.1 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.1 | 1.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 3.0 | GO:0006664 | glycolipid metabolic process(GO:0006664) |
| 0.1 | 2.0 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.1 | 3.0 | GO:1902117 | positive regulation of organelle assembly(GO:1902117) |
| 0.0 | 1.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 4.8 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 6.8 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 8.0 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.4 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 3.4 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 1.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 8.9 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 7.3 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 2.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.6 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 22.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 4.7 | 18.9 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 3.9 | 11.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 3.8 | 11.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 3.2 | 9.7 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 2.4 | 16.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 2.1 | 33.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 2.0 | 14.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 2.0 | 17.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 1.9 | 5.8 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 1.9 | 18.8 | GO:0032009 | early phagosome(GO:0032009) |
| 1.7 | 18.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 1.5 | 18.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.5 | 4.5 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 1.4 | 5.7 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 1.4 | 4.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 1.2 | 9.7 | GO:0070820 | tertiary granule(GO:0070820) |
| 1.2 | 5.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.1 | 20.7 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 1.1 | 3.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 1.0 | 5.9 | GO:0089701 | U2AF(GO:0089701) |
| 1.0 | 6.7 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.9 | 4.7 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.8 | 2.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.8 | 9.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.8 | 9.8 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) mitotic spindle midzone(GO:1990023) |
| 0.7 | 12.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.6 | 31.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.6 | 7.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.5 | 5.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.5 | 3.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.5 | 6.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.5 | 6.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.5 | 3.7 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.4 | 8.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 2.8 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.4 | 9.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.3 | 32.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.3 | 7.1 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.3 | 2.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 4.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 1.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 2.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 0.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 3.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.2 | 3.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 15.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 6.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 1.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 4.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.2 | 5.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 19.0 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 0.6 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 0.8 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 10.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 20.5 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 1.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 6.8 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 44.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 10.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 11.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 2.2 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.1 | 1.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 5.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 2.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 5.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 9.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 7.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 3.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.7 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 6.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 5.3 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 8.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 5.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 3.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 17.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 2.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 1.0 | GO:0044439 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 1.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 3.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 2.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 3.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.0 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 8.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 4.1 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 4.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.9 | GO:0098794 | postsynapse(GO:0098794) |
| 0.0 | 4.5 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.6 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 1.6 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 18.4 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 70.9 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 22.0 | GO:0031762 | follicle-stimulating hormone receptor binding(GO:0031762) |
| 4.7 | 37.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 4.6 | 13.9 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 3.7 | 11.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 3.5 | 14.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 3.2 | 9.6 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 2.6 | 15.6 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 2.4 | 9.7 | GO:0035877 | death effector domain binding(GO:0035877) |
| 2.4 | 18.8 | GO:0035197 | siRNA binding(GO:0035197) |
| 2.0 | 11.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 2.0 | 5.9 | GO:0004464 | arachidonate 5-lipoxygenase activity(GO:0004051) leukotriene-C4 synthase activity(GO:0004464) |
| 1.9 | 30.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 1.8 | 7.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 1.7 | 8.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 1.7 | 31.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 1.7 | 6.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.6 | 4.7 | GO:0043398 | HLH domain binding(GO:0043398) |
| 1.5 | 3.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.4 | 7.2 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 1.3 | 3.8 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.2 | 6.2 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 1.2 | 15.6 | GO:1990405 | protein antigen binding(GO:1990405) |
| 1.2 | 4.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 1.1 | 9.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 1.1 | 4.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.1 | 9.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.1 | 5.4 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 1.1 | 3.2 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 1.0 | 18.3 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.9 | 4.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.9 | 3.5 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.9 | 33.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.8 | 9.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.8 | 5.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.8 | 2.3 | GO:0004096 | catalase activity(GO:0004096) |
| 0.7 | 6.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.7 | 17.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.7 | 10.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.7 | 4.2 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.7 | 7.4 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.6 | 3.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.6 | 4.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.6 | 14.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.6 | 22.2 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.6 | 13.8 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.6 | 4.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.5 | 3.2 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.5 | 7.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 2.6 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.5 | 6.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.5 | 3.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.5 | 136.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.4 | 3.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 4.0 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.4 | 9.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.4 | 2.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.4 | 11.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.4 | 12.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.4 | 1.2 | GO:0052724 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.4 | 3.0 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.4 | 1.8 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.4 | 5.8 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.4 | 9.6 | GO:0004950 | chemokine receptor activity(GO:0004950) |
| 0.3 | 3.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.3 | 5.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 2.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.3 | 5.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 20.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.3 | 1.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.3 | 2.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.3 | 3.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 4.0 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.3 | 1.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.3 | 5.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.3 | 1.8 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 1.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 2.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 1.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 3.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 8.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 8.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 2.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 40.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.2 | 1.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 15.4 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.2 | 1.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 1.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 2.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 11.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.2 | 2.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 10.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 4.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 1.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 3.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 6.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.4 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.1 | 2.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 3.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 27.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 4.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 7.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 2.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 28.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 2.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 2.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 20.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 3.7 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 5.0 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 5.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 3.1 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.6 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 6.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 2.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.8 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 0.9 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 5.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 2.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 9.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.8 | 32.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.7 | 12.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.4 | 5.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 6.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.4 | 16.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 8.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 10.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.3 | 11.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.3 | 7.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.3 | 17.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 4.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 10.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 9.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 7.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 4.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 4.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 4.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 7.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 5.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 18.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 1.7 | 21.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.9 | 9.7 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.8 | 7.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.8 | 11.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.8 | 17.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.7 | 25.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.7 | 49.4 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.7 | 4.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.7 | 10.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.6 | 6.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.6 | 7.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.6 | 9.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.6 | 9.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.6 | 17.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 15.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.5 | 9.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.5 | 5.9 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.4 | 5.9 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.4 | 5.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.4 | 5.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.4 | 18.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 7.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 17.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.4 | 15.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 16.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.3 | 7.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.3 | 3.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 7.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 49.0 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.2 | 2.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 9.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 7.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 3.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 7.6 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.2 | 4.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 8.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 8.7 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.1 | 2.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 4.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 13.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 12.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 5.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 4.4 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 2.5 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 0.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 4.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |