Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
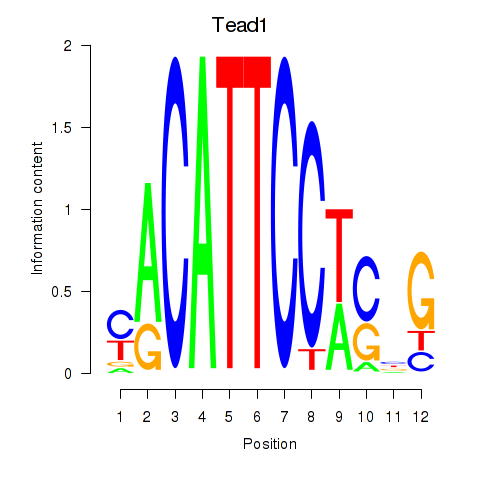
Results for Tead1
Z-value: 1.28
Transcription factors associated with Tead1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tead1
|
ENSRNOG00000015488 | TEA domain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tead1 | rn6_v1_chr1_+_177569618_177569618 | 0.68 | 1.3e-44 | Click! |
Activity profile of Tead1 motif
Sorted Z-values of Tead1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_21854763 | 45.18 |
ENSRNOT00000089196
ENSRNOT00000020528 |
Ctgf
|
connective tissue growth factor |
| chr1_+_214203861 | 39.80 |
ENSRNOT00000023219
|
Rassf7
|
Ras association domain family member 7 |
| chr11_+_64522130 | 39.41 |
ENSRNOT00000004315
|
Upk1b
|
uroplakin 1B |
| chr18_+_57286322 | 39.15 |
ENSRNOT00000026174
|
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr16_+_47368768 | 38.24 |
ENSRNOT00000017966
|
Wwc2
|
WW and C2 domain containing 2 |
| chr13_-_108841482 | 36.78 |
ENSRNOT00000004632
ENSRNOT00000086775 |
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr10_-_88874528 | 32.42 |
ENSRNOT00000026783
|
Ptrf
|
polymerase I and transcript release factor |
| chr20_-_3605638 | 32.11 |
ENSRNOT00000074460
|
Sfta2
|
surfactant associated 2 |
| chr6_+_109617596 | 30.97 |
ENSRNOT00000085031
ENSRNOT00000089972 ENSRNOT00000082402 |
Flvcr2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr10_+_84182118 | 30.24 |
ENSRNOT00000011027
|
Hoxb3
|
homeo box B3 |
| chr1_-_252550394 | 29.88 |
ENSRNOT00000083468
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr6_+_109617355 | 27.28 |
ENSRNOT00000011599
|
Flvcr2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr9_-_55256340 | 26.69 |
ENSRNOT00000028907
|
Sdpr
|
serum deprivation response |
| chr11_-_60679555 | 26.25 |
ENSRNOT00000059735
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr11_-_77703255 | 25.65 |
ENSRNOT00000083319
|
Cldn16
|
claudin 16 |
| chr4_-_64981384 | 24.60 |
ENSRNOT00000017338
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr10_+_15672382 | 24.15 |
ENSRNOT00000027965
|
Rhbdf1
|
rhomboid 5 homolog 1 |
| chr13_+_52662996 | 23.91 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr17_-_10004321 | 23.63 |
ENSRNOT00000042394
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr3_+_79860179 | 22.46 |
ENSRNOT00000081160
ENSRNOT00000068444 |
Rapsn
|
receptor-associated protein of the synapse |
| chr15_-_33218456 | 22.43 |
ENSRNOT00000017753
|
Ajuba
|
ajuba LIM protein |
| chr7_-_143016040 | 22.22 |
ENSRNOT00000029697
|
Krt80
|
keratin 80 |
| chr3_+_80362858 | 21.85 |
ENSRNOT00000021353
ENSRNOT00000086242 |
Lrp4
|
LDL receptor related protein 4 |
| chr4_+_44597123 | 20.26 |
ENSRNOT00000078250
|
Cav1
|
caveolin 1 |
| chr2_-_251532312 | 19.90 |
ENSRNOT00000019501
|
Cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr5_-_158439078 | 19.81 |
ENSRNOT00000025517
|
Klhdc7a
|
kelch domain containing 7A |
| chr8_+_111210811 | 19.46 |
ENSRNOT00000011347
|
Amotl2
|
angiomotin like 2 |
| chr9_-_52238564 | 19.01 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr4_-_71763679 | 18.95 |
ENSRNOT00000024037
|
Epha1
|
Eph receptor A1 |
| chr18_-_28454756 | 18.45 |
ENSRNOT00000040091
|
Spata24
|
spermatogenesis associated 24 |
| chr13_-_86671515 | 18.34 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr19_-_55367353 | 17.41 |
ENSRNOT00000091139
|
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr3_+_152857592 | 16.84 |
ENSRNOT00000027445
|
Myl9
|
myosin light chain 9 |
| chr10_+_57322331 | 16.69 |
ENSRNOT00000041493
|
Kif1c
|
kinesin family member 1C |
| chr1_-_18058055 | 16.56 |
ENSRNOT00000020988
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr9_-_119332967 | 15.83 |
ENSRNOT00000021048
|
Myl12a
|
myosin light chain 12A |
| chr4_+_181286455 | 15.82 |
ENSRNOT00000083633
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr13_+_112031594 | 15.79 |
ENSRNOT00000008440
|
Lamb3
|
laminin subunit beta 3 |
| chr1_-_219682287 | 15.79 |
ENSRNOT00000026092
|
Rhod
|
ras homolog family member D |
| chr1_-_131460473 | 15.45 |
ENSRNOT00000084336
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_-_122721496 | 14.65 |
ENSRNOT00000079697
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr8_-_61595032 | 13.61 |
ENSRNOT00000023428
|
Snx33
|
sorting nexin 33 |
| chr4_+_157125998 | 13.44 |
ENSRNOT00000078349
|
C1r
|
complement C1r |
| chr4_+_120672152 | 13.26 |
ENSRNOT00000077231
|
Mgll
|
monoglyceride lipase |
| chr1_+_101517714 | 13.22 |
ENSRNOT00000028423
|
Plekha4
|
pleckstrin homology domain containing A4 |
| chr1_-_261371508 | 12.94 |
ENSRNOT00000019978
|
Avpi1
|
arginine vasopressin-induced 1 |
| chrX_+_77119911 | 12.49 |
ENSRNOT00000080141
|
Atp7a
|
ATPase copper transporting alpha |
| chr9_+_15166118 | 11.43 |
ENSRNOT00000020432
|
Mdfi
|
MyoD family inhibitor |
| chr10_-_85636902 | 10.98 |
ENSRNOT00000017241
|
Pcgf2
|
polycomb group ring finger 2 |
| chr1_-_216703534 | 10.52 |
ENSRNOT00000027972
|
Phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr19_-_37938857 | 10.11 |
ENSRNOT00000026730
|
Slc12a4
|
solute carrier family 12 member 4 |
| chr19_+_43163562 | 10.03 |
ENSRNOT00000048900
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr19_-_11341863 | 9.49 |
ENSRNOT00000025694
|
Mt4
|
metallothionein 4 |
| chr2_-_116002561 | 9.43 |
ENSRNOT00000012924
|
Prkci
|
protein kinase C, iota |
| chr13_+_73929136 | 9.39 |
ENSRNOT00000005425
|
Nphs2
|
nephrosis 2, idiopathic, steroid-resistant |
| chr7_-_67345295 | 8.78 |
ENSRNOT00000087045
ENSRNOT00000005829 |
Avpr1a
|
arginine vasopressin receptor 1A |
| chr20_-_27757149 | 8.55 |
ENSRNOT00000088287
|
Dse
|
dermatan sulfate epimerase |
| chr19_+_43163129 | 8.11 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr5_+_75392790 | 8.07 |
ENSRNOT00000048457
|
Musk
|
muscle associated receptor tyrosine kinase |
| chr7_+_116745061 | 7.46 |
ENSRNOT00000076174
|
Rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr10_+_74413989 | 7.29 |
ENSRNOT00000036098
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr5_+_164796185 | 6.96 |
ENSRNOT00000010779
|
Nppb
|
natriuretic peptide B |
| chr10_-_56000225 | 6.80 |
ENSRNOT00000013439
|
Tmem88
|
transmembrane protein 88 |
| chr10_-_70646495 | 6.51 |
ENSRNOT00000071218
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr5_+_28850950 | 6.41 |
ENSRNOT00000009759
|
Tmem64
|
transmembrane protein 64 |
| chr18_-_55771730 | 6.31 |
ENSRNOT00000026426
|
Smim3
|
small integral membrane protein 3 |
| chr5_+_138300107 | 5.43 |
ENSRNOT00000047151
|
Cldn19
|
claudin 19 |
| chr13_+_68709243 | 5.13 |
ENSRNOT00000066419
|
Ivns1abp
|
influenza virus NS1A binding protein |
| chr2_+_198231291 | 5.08 |
ENSRNOT00000078288
ENSRNOT00000046739 |
Otud7b
|
OTU deubiquitinase 7B |
| chr10_+_104523996 | 4.61 |
ENSRNOT00000065339
ENSRNOT00000086747 |
Itgb4
|
integrin subunit beta 4 |
| chr7_-_117070936 | 4.19 |
ENSRNOT00000048446
|
Fam83h
|
family with sequence similarity 83, member H |
| chr4_-_40036563 | 3.69 |
ENSRNOT00000084531
|
Tmem168
|
transmembrane protein 168 |
| chr12_-_16126953 | 3.68 |
ENSRNOT00000001682
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr8_-_55681930 | 3.49 |
ENSRNOT00000065744
|
Colca2
|
colorectal cancer associated 2 |
| chrX_-_14783792 | 2.92 |
ENSRNOT00000087609
|
LOC691895
|
similar to ferritin, heavy polypeptide-like 17 |
| chr20_+_46428124 | 2.86 |
ENSRNOT00000000327
|
Foxo3
|
forkhead box O3 |
| chr10_-_62699723 | 2.85 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr3_+_93648343 | 2.08 |
ENSRNOT00000091164
|
AABR07053136.1
|
|
| chr5_+_169475270 | 1.97 |
ENSRNOT00000014625
|
Icmt
|
isoprenylcysteine carboxyl methyltransferase |
| chr8_-_66893083 | 1.81 |
ENSRNOT00000037028
ENSRNOT00000091755 |
Kif23
|
kinesin family member 23 |
| chr15_+_52265557 | 1.75 |
ENSRNOT00000015969
|
Nudt18
|
nudix hydrolase 18 |
| chr8_-_73194837 | 1.46 |
ENSRNOT00000024885
ENSRNOT00000081450 ENSRNOT00000064613 |
Tln2
|
talin 2 |
| chr13_+_26172243 | 1.25 |
ENSRNOT00000003840
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr1_-_217720414 | 0.91 |
ENSRNOT00000078810
|
Ppfia1
|
PTPRF interacting protein alpha 1 |
| chr1_+_99729704 | 0.45 |
ENSRNOT00000037867
|
Klk9
|
kallikrein related-peptidase 9 |
| chr5_+_59080765 | 0.36 |
ENSRNOT00000021888
ENSRNOT00000064419 |
Rgp1
|
RGP1 homolog, RAB6A GEF complex partner 1 |
| chr17_+_86408151 | 0.35 |
ENSRNOT00000022734
|
Otud1
|
OTU deubiquitinase 1 |
| chr1_+_46835971 | 0.22 |
ENSRNOT00000066798
|
Synj2
|
synaptojanin 2 |
| chr19_+_54766589 | 0.10 |
ENSRNOT00000025894
|
Banp
|
Btg3 associated nuclear protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tead1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.1 | 60.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 9.0 | 45.2 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 8.0 | 23.9 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 7.6 | 30.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 7.5 | 29.9 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 6.8 | 20.3 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) regulation of inward rectifier potassium channel activity(GO:1901979) positive regulation of gap junction assembly(GO:1903598) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 6.0 | 29.9 | GO:0090131 | glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 5.9 | 23.6 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 5.8 | 58.3 | GO:0015886 | heme transport(GO:0015886) |
| 5.1 | 15.4 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 4.8 | 19.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 3.6 | 39.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 3.3 | 19.9 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 3.3 | 13.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 3.1 | 12.5 | GO:0051542 | negative regulation of iron ion transmembrane transport(GO:0034760) elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 2.9 | 8.8 | GO:0045819 | cellular response to water deprivation(GO:0042631) positive regulation of glycogen catabolic process(GO:0045819) |
| 2.8 | 8.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 2.5 | 17.4 | GO:0033634 | positive regulation of integrin activation(GO:0033625) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 2.3 | 9.4 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 2.2 | 22.5 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 1.9 | 36.8 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 1.8 | 25.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.8 | 10.5 | GO:0060721 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 1.7 | 5.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 1.6 | 6.4 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 1.5 | 13.6 | GO:0036089 | cleavage furrow formation(GO:0036089) macropinocytosis(GO:0044351) |
| 1.3 | 9.4 | GO:0034351 | negative regulation of glial cell apoptotic process(GO:0034351) |
| 1.2 | 3.7 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.2 | 16.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 1.0 | 7.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.6 | 1.8 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.6 | 11.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.6 | 1.7 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.5 | 4.6 | GO:0061450 | nail development(GO:0035878) trophoblast cell migration(GO:0061450) |
| 0.5 | 32.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.5 | 2.9 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.5 | 16.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.4 | 5.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.4 | 1.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.4 | 14.4 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.4 | 15.8 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.3 | 24.1 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.3 | 44.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.3 | 24.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.3 | 49.9 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.2 | 2.9 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 1.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 2.0 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 12.9 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 4.2 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.1 | 11.0 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.1 | 13.4 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 8.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 6.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 17.5 | GO:0016055 | Wnt signaling pathway(GO:0016055) |
| 0.1 | 5.1 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.1 | 39.4 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.0 | 0.7 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 9.5 | GO:0006875 | cellular metal ion homeostasis(GO:0006875) |
| 0.0 | 9.2 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 10.2 | GO:0016567 | protein ubiquitination(GO:0016567) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 29.9 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 6.0 | 23.9 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 4.8 | 19.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.3 | 20.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.8 | 21.9 | GO:0016600 | flotillin complex(GO:0016600) |
| 1.8 | 15.8 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 1.6 | 11.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 1.5 | 22.5 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 1.5 | 29.6 | GO:0016460 | myosin II complex(GO:0016460) |
| 1.3 | 26.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.0 | 22.2 | GO:0030057 | desmosome(GO:0030057) |
| 1.0 | 17.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.9 | 9.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.8 | 45.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.7 | 13.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.5 | 9.4 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.5 | 7.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.5 | 1.8 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.4 | 12.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.4 | 48.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.3 | 4.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 26.7 | GO:0005901 | caveola(GO:0005901) |
| 0.3 | 39.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.3 | 16.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 19.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 16.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 37.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 46.4 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.1 | 8.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 6.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 3.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 46.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 19.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 4.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 10.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 79.7 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.1 | 13.2 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.1 | 16.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 28.0 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 58.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 6.0 | 23.9 | GO:0030172 | troponin C binding(GO:0030172) |
| 3.2 | 19.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 3.0 | 23.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 2.9 | 8.8 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 2.8 | 8.5 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 2.5 | 12.5 | GO:0032767 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 2.5 | 22.5 | GO:0043495 | protein anchor(GO:0043495) |
| 2.4 | 69.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 1.9 | 17.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.8 | 20.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 1.7 | 22.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 1.5 | 24.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 1.5 | 16.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.3 | 6.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.3 | 15.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 1.3 | 21.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 1.0 | 13.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 1.0 | 15.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.9 | 26.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.9 | 11.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.8 | 4.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.8 | 18.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.6 | 1.7 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.5 | 5.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.5 | 38.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.5 | 26.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.5 | 9.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.4 | 30.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.3 | 19.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 7.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.3 | 23.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.3 | 2.0 | GO:0051998 | cAMP response element binding protein binding(GO:0008140) carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.3 | 15.8 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.2 | 18.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 38.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.9 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 14.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 15.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 49.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 3.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 2.9 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 15.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 15.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 10.2 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 16.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 7.5 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 2.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 20.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.9 | 19.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.9 | 45.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.9 | 20.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.8 | 19.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.6 | 18.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.4 | 19.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.4 | 16.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.4 | 14.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.3 | 17.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 50.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 28.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.3 | 16.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 15.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 2.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 33.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 1.7 | 46.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 1.7 | 23.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 1.6 | 45.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 1.5 | 32.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 1.2 | 40.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.9 | 19.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.7 | 20.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 19.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 20.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.3 | 3.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 12.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 15.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 8.5 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.2 | 1.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 7.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 8.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 4.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |