Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Tbp
Z-value: 2.98
Transcription factors associated with Tbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbp
|
ENSRNOG00000001489 | TATA box binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbp | rn6_v1_chr1_+_57491643_57491643 | -0.34 | 6.7e-10 | Click! |
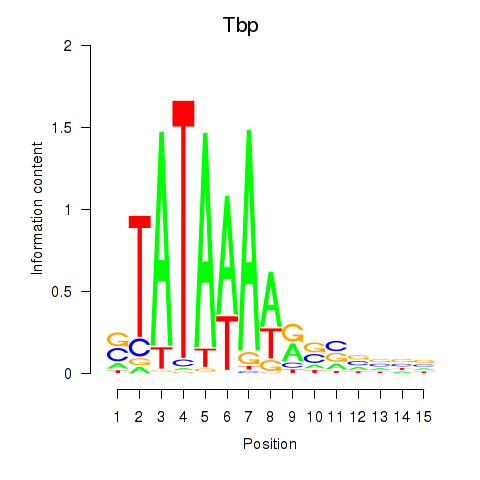
Activity profile of Tbp motif
Sorted Z-values of Tbp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_109729019 | 111.44 |
ENSRNOT00000054959
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chr10_+_53818818 | 98.00 |
ENSRNOT00000057260
|
Myh8
|
myosin heavy chain 8 |
| chr2_-_105089659 | 96.03 |
ENSRNOT00000043381
|
Cpb1
|
carboxypeptidase B1 |
| chr1_+_80321585 | 86.77 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr17_+_44794130 | 83.31 |
ENSRNOT00000077571
|
Hist1h2ac
|
histone cluster 1, H2ac |
| chr10_+_53621375 | 82.54 |
ENSRNOT00000004147
|
Myh3
|
myosin heavy chain 3 |
| chr4_+_70776046 | 79.32 |
ENSRNOT00000040403
|
Prss1
|
protease, serine 1 |
| chr2_-_190100276 | 77.74 |
ENSRNOT00000015351
|
S100a9
|
S100 calcium binding protein A9 |
| chr3_-_105512939 | 76.52 |
ENSRNOT00000011773
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr1_-_213650247 | 76.36 |
ENSRNOT00000019679
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb |
| chr12_-_2438817 | 72.92 |
ENSRNOT00000037059
|
Ccl25
|
C-C motif chemokine ligand 25 |
| chr4_-_69268336 | 71.37 |
ENSRNOT00000018042
|
Prss3b
|
protease, serine, 3B |
| chr17_+_44766313 | 71.17 |
ENSRNOT00000091688
|
Hist1h1b
|
histone cluster 1 H1 family member b |
| chr3_+_72385666 | 70.02 |
ENSRNOT00000011168
|
Prg2
|
proteoglycan 2 |
| chr15_-_34612432 | 68.19 |
ENSRNOT00000090206
|
Mcpt1l1
|
mast cell protease 1-like 1 |
| chr5_-_58198782 | 67.55 |
ENSRNOT00000023951
|
Ccl21
|
C-C motif chemokine ligand 21 |
| chr17_-_44793927 | 65.49 |
ENSRNOT00000086309
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr7_-_118108864 | 64.36 |
ENSRNOT00000006184
|
Mb
|
myoglobin |
| chr4_+_70755795 | 64.27 |
ENSRNOT00000043527
|
LOC683849
|
similar to Anionic trypsin II precursor (Pretrypsinogen II) |
| chr2_+_198390166 | 64.11 |
ENSRNOT00000081042
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr9_-_73948583 | 63.33 |
ENSRNOT00000018097
|
Myl1
|
myosin, light chain 1 |
| chr17_-_43798383 | 63.09 |
ENSRNOT00000075069
|
LOC684828
|
similar to Histone H1.2 (H1 VAR.1) (H1c) |
| chr8_+_119030875 | 62.87 |
ENSRNOT00000028458
|
Myl3
|
myosin light chain 3 |
| chr17_+_44758460 | 62.02 |
ENSRNOT00000089436
|
Hist1h2an
|
histone cluster 1, H2an |
| chr4_+_14070553 | 60.40 |
ENSRNOT00000077505
|
Cd36
|
CD36 molecule |
| chr10_-_88163712 | 58.77 |
ENSRNOT00000005382
ENSRNOT00000084493 |
Krt17
|
keratin 17 |
| chr8_+_5606592 | 58.13 |
ENSRNOT00000011727
|
Mmp12
|
matrix metallopeptidase 12 |
| chr10_+_31880918 | 57.98 |
ENSRNOT00000059448
|
Timd4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr7_-_29171783 | 57.38 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr13_-_50916982 | 57.37 |
ENSRNOT00000004408
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr17_-_43807540 | 57.33 |
ENSRNOT00000074763
|
LOC684762
|
similar to CG31613-PA |
| chr4_+_14151343 | 56.98 |
ENSRNOT00000061687
ENSRNOT00000076573 ENSRNOT00000077219 ENSRNOT00000008319 |
Cd36
|
CD36 molecule |
| chr4_+_14212925 | 56.33 |
ENSRNOT00000076946
|
LOC103690020
|
platelet glycoprotein 4-like |
| chr3_-_7141522 | 55.76 |
ENSRNOT00000014572
|
Cel
|
carboxyl ester lipase |
| chr8_+_50537009 | 55.10 |
ENSRNOT00000080658
|
Apoa4
|
apolipoprotein A4 |
| chr2_-_105047984 | 54.36 |
ENSRNOT00000014970
|
Cpa3
|
carboxypeptidase A3 |
| chr1_+_168945449 | 54.08 |
ENSRNOT00000087661
ENSRNOT00000019913 |
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr10_-_88036040 | 53.11 |
ENSRNOT00000018851
|
Krt13
|
keratin 13 |
| chr1_-_100537377 | 52.97 |
ENSRNOT00000026599
|
Spib
|
Spi-B transcription factor |
| chr1_-_85220237 | 52.66 |
ENSRNOT00000026907
|
Sycn
|
syncollin |
| chr10_-_88050622 | 52.38 |
ENSRNOT00000019037
|
Krt15
|
keratin 15 |
| chr2_-_198380836 | 52.17 |
ENSRNOT00000040906
|
LOC102548682
|
histone H4-like |
| chr4_+_70689737 | 52.16 |
ENSRNOT00000018852
|
Prss2
|
protease, serine, 2 |
| chr10_+_11240138 | 50.60 |
ENSRNOT00000048687
|
Srl
|
sarcalumenin |
| chr1_+_168957460 | 49.94 |
ENSRNOT00000090745
|
LOC103694857
|
hemoglobin subunit beta-2 |
| chr17_+_43627930 | 49.93 |
ENSRNOT00000081719
|
LOC102549061
|
histone H2B type 1-N-like |
| chr2_+_181987217 | 49.71 |
ENSRNOT00000034521
|
Fgg
|
fibrinogen gamma chain |
| chr4_-_70628470 | 49.67 |
ENSRNOT00000029319
|
Try5
|
trypsin 5 |
| chr5_+_164808323 | 49.59 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr2_-_88135410 | 48.36 |
ENSRNOT00000014180
|
Car3
|
carbonic anhydrase 3 |
| chr20_+_30791422 | 48.17 |
ENSRNOT00000047394
ENSRNOT00000000683 |
Tbata
|
thymus, brain and testes associated |
| chr17_-_43614844 | 47.33 |
ENSRNOT00000023054
|
Hist1h1a
|
histone cluster 1 H1 family member a |
| chr17_-_44758170 | 47.04 |
ENSRNOT00000091176
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr15_+_35032049 | 45.95 |
ENSRNOT00000091757
|
Mcpt2
|
mast cell protease 2 |
| chr10_-_87067456 | 44.77 |
ENSRNOT00000014163
|
Ccr7
|
C-C motif chemokine receptor 7 |
| chr6_-_128003418 | 44.32 |
ENSRNOT00000013896
|
Serpina3c
|
serine (or cysteine) proteinase inhibitor, clade A, member 3C |
| chr5_+_79179417 | 44.12 |
ENSRNOT00000010454
|
Orm1
|
orosomucoid 1 |
| chr10_-_38774449 | 44.06 |
ENSRNOT00000049820
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr1_+_83653234 | 43.95 |
ENSRNOT00000085008
ENSRNOT00000084230 ENSRNOT00000090071 |
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr17_+_43734461 | 43.82 |
ENSRNOT00000072564
|
Hist1h1d
|
histone cluster 1, H1d |
| chr7_+_11908107 | 43.67 |
ENSRNOT00000041106
|
Mknk2
|
MAP kinase-interacting serine/threonine kinase 2 |
| chr19_-_11341863 | 43.64 |
ENSRNOT00000025694
|
Mt4
|
metallothionein 4 |
| chr9_+_9721105 | 43.63 |
ENSRNOT00000073042
ENSRNOT00000075494 |
C3
|
complement C3 |
| chr2_+_93792601 | 43.33 |
ENSRNOT00000014701
ENSRNOT00000077311 |
Fabp4
|
fatty acid binding protein 4 |
| chr10_+_82292110 | 42.40 |
ENSRNOT00000004435
|
Chad
|
chondroadherin |
| chr13_+_90260783 | 41.18 |
ENSRNOT00000050547
|
Cd84
|
CD84 molecule |
| chr1_+_168964202 | 40.66 |
ENSRNOT00000089102
|
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr5_-_19368431 | 40.65 |
ENSRNOT00000012819
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr2_-_123281856 | 40.48 |
ENSRNOT00000079745
|
Ccna2
|
cyclin A2 |
| chr17_+_43673294 | 40.37 |
ENSRNOT00000074109
|
Hist2h4a
|
histone cluster 2, H4 |
| chr6_+_33176778 | 39.61 |
ENSRNOT00000046811
ENSRNOT00000007371 |
Apob
|
apolipoprotein B |
| chrX_-_157013443 | 39.28 |
ENSRNOT00000082711
|
Srpk3
|
SRSF protein kinase 3 |
| chr15_-_23969011 | 39.27 |
ENSRNOT00000014821
|
Gch1
|
GTP cyclohydrolase 1 |
| chr10_-_88172910 | 39.19 |
ENSRNOT00000046956
|
Krt42
|
keratin 42 |
| chr3_-_48451650 | 38.34 |
ENSRNOT00000007356
|
Gcg
|
glucagon |
| chr2_+_189997129 | 38.16 |
ENSRNOT00000015958
|
S100a4
|
S100 calcium-binding protein A4 |
| chr1_+_91363492 | 37.91 |
ENSRNOT00000014517
|
Cebpa
|
CCAAT/enhancer binding protein alpha |
| chr1_-_25839198 | 37.20 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr4_-_69196430 | 37.03 |
ENSRNOT00000017673
|
LOC312273
|
Trypsin V-A |
| chr7_-_54778848 | 36.66 |
ENSRNOT00000005399
|
Glipr1
|
GLI pathogenesis-related 1 |
| chr7_+_121841855 | 36.60 |
ENSRNOT00000024673
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr2_+_198388809 | 36.23 |
ENSRNOT00000083087
|
Hist2h2aa3
|
histone cluster 2, H2aa3 |
| chr14_-_18853315 | 36.14 |
ENSRNOT00000003794
|
Ppbp
|
pro-platelet basic protein |
| chr6_-_139142218 | 35.60 |
ENSRNOT00000006975
|
Ighg
|
Immunoglobulin heavy chain (gamma polypeptide) |
| chr9_-_42839837 | 35.38 |
ENSRNOT00000038610
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr7_+_37812831 | 35.36 |
ENSRNOT00000005910
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr17_+_44522140 | 34.93 |
ENSRNOT00000080490
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr1_-_168972725 | 34.93 |
ENSRNOT00000090422
|
Hbb
|
hemoglobin subunit beta |
| chr10_+_70262361 | 34.89 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr3_+_111422267 | 34.48 |
ENSRNOT00000052312
|
Nusap1
|
nucleolar and spindle associated protein 1 |
| chr13_-_61070599 | 34.46 |
ENSRNOT00000005251
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr2_-_198382190 | 34.39 |
ENSRNOT00000044268
|
Hist2h2aa2
|
histone cluster 2, H2aa2 |
| chr17_+_43632397 | 34.24 |
ENSRNOT00000013790
|
Hist1h2ah
|
histone cluster 1, H2ah |
| chr2_+_198417619 | 33.86 |
ENSRNOT00000085945
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr4_+_109467272 | 33.50 |
ENSRNOT00000008212
|
Reg3b
|
regenerating family member 3 beta |
| chr3_-_156340913 | 33.49 |
ENSRNOT00000021452
|
Mafb
|
MAF bZIP transcription factor B |
| chr1_-_89369960 | 33.44 |
ENSRNOT00000028545
|
Hamp
|
hepcidin antimicrobial peptide |
| chr3_-_111422203 | 33.39 |
ENSRNOT00000084290
|
Oip5
|
Opa interacting protein 5 |
| chr10_-_87541851 | 33.24 |
ENSRNOT00000089610
|
RGD1561684
|
similar to keratin associated protein 2-4 |
| chr17_+_43617247 | 32.96 |
ENSRNOT00000075065
|
Hist1h2ao
|
histone cluster 1, H2ao |
| chr7_+_141249044 | 32.50 |
ENSRNOT00000084911
|
Aqp5
|
aquaporin 5 |
| chr19_+_54314865 | 32.08 |
ENSRNOT00000024069
|
Irf8
|
interferon regulatory factor 8 |
| chr7_-_143141589 | 32.07 |
ENSRNOT00000068505
ENSRNOT00000077207 |
Kb23
|
type II keratin 23 |
| chr2_-_251532312 | 32.05 |
ENSRNOT00000019501
|
Cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr3_-_121882726 | 31.83 |
ENSRNOT00000006308
|
Il1b
|
interleukin 1 beta |
| chr17_+_81352700 | 30.85 |
ENSRNOT00000024736
|
Mrc1
|
mannose receptor, C type 1 |
| chr17_-_43821536 | 30.71 |
ENSRNOT00000072286
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr17_-_44815995 | 30.66 |
ENSRNOT00000091201
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr2_-_45518502 | 30.18 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr1_+_201620642 | 30.12 |
ENSRNOT00000093674
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr16_-_75637789 | 30.08 |
ENSRNOT00000058029
|
Defb4
|
defensin beta 4 |
| chr11_-_32550539 | 30.06 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chr8_+_50559126 | 29.91 |
ENSRNOT00000024918
|
Apoa5
|
apolipoprotein A5 |
| chr1_+_261281543 | 29.91 |
ENSRNOT00000018890
|
Ankrd2
|
ankyrin repeat domain 2 |
| chr5_+_164796185 | 29.86 |
ENSRNOT00000010779
|
Nppb
|
natriuretic peptide B |
| chr9_+_46840992 | 29.83 |
ENSRNOT00000019415
|
Il1r2
|
interleukin 1 receptor type 2 |
| chr7_-_143497108 | 29.62 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr1_+_101324652 | 29.49 |
ENSRNOT00000028116
|
Hrc
|
histidine rich calcium binding protein |
| chr12_+_23473270 | 29.23 |
ENSRNOT00000001935
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr1_-_81627710 | 28.86 |
ENSRNOT00000071704
|
LOC100909700
|
CD177 antigen-like |
| chr8_+_22035256 | 28.62 |
ENSRNOT00000028066
|
Icam1
|
intercellular adhesion molecule 1 |
| chr6_-_128149220 | 28.49 |
ENSRNOT00000014204
|
Gsc
|
goosecoid homeobox |
| chr1_-_80594136 | 28.48 |
ENSRNOT00000024800
|
Apoc2
|
apolipoprotein C2 |
| chr10_-_87928251 | 28.30 |
ENSRNOT00000064120
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr6_+_33885495 | 28.10 |
ENSRNOT00000086633
|
Sdc1
|
syndecan 1 |
| chr8_+_5768811 | 27.67 |
ENSRNOT00000013936
|
Mmp8
|
matrix metallopeptidase 8 |
| chr8_+_50525091 | 26.95 |
ENSRNOT00000074357
|
Apoa1
|
apolipoprotein A1 |
| chr2_-_53313884 | 26.89 |
ENSRNOT00000046951
|
Ghr
|
growth hormone receptor |
| chr15_+_30270740 | 26.84 |
ENSRNOT00000070991
|
AABR07017707.1
|
|
| chr1_+_279798187 | 26.76 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr17_-_44744902 | 26.61 |
ENSRNOT00000085381
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr7_+_61236037 | 26.49 |
ENSRNOT00000009776
|
Il22
|
interleukin 22 |
| chr7_+_143122269 | 26.28 |
ENSRNOT00000082542
ENSRNOT00000045495 ENSRNOT00000081386 ENSRNOT00000067422 |
Krt86
|
keratin 86 |
| chr4_-_109532234 | 26.19 |
ENSRNOT00000008665
|
Reg3g
|
regenerating family member 3 gamma |
| chr10_+_69476775 | 26.15 |
ENSRNOT00000031626
|
Ccl12
|
chemokine (C-C motif) ligand 12 |
| chr10_-_70744315 | 25.92 |
ENSRNOT00000014865
|
Ccl5
|
C-C motif chemokine ligand 5 |
| chr7_-_143228060 | 25.84 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr17_-_43627629 | 25.75 |
ENSRNOT00000022965
|
Hist1h2af
|
histone cluster 1, H2af |
| chr10_-_87954055 | 25.70 |
ENSRNOT00000018048
|
Krt34
|
keratin 34 |
| chr5_+_149047681 | 25.37 |
ENSRNOT00000015198
|
Laptm5
|
lysosomal protein transmembrane 5 |
| chr3_+_149643481 | 25.31 |
ENSRNOT00000020713
|
Bpifa5
|
BPI fold containing family A, member 5 |
| chr10_-_87510629 | 25.07 |
ENSRNOT00000084197
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr7_-_143112960 | 25.00 |
ENSRNOT00000012845
ENSRNOT00000068748 ENSRNOT00000093507 |
Krt81
|
keratin 81 |
| chr20_+_10438444 | 24.97 |
ENSRNOT00000071248
ENSRNOT00000075545 |
Cryaa
|
crystallin, alpha A |
| chr16_+_81616604 | 24.94 |
ENSRNOT00000026392
ENSRNOT00000057740 |
Adprhl1
Grtp1
|
ADP-ribosylhydrolase like 1 growth hormone regulated TBC protein 1 |
| chr3_-_121836086 | 24.90 |
ENSRNOT00000006113
|
Il1a
|
interleukin 1 alpha |
| chr4_+_71675383 | 24.60 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr3_+_19045214 | 24.60 |
ENSRNOT00000070878
|
AABR07051670.1
|
|
| chr1_+_89008117 | 24.48 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chr9_-_23493081 | 24.28 |
ENSRNOT00000072144
|
Rhag
|
Rh-associated glycoprotein |
| chr10_+_14088319 | 24.24 |
ENSRNOT00000019508
|
Rps2
|
ribosomal protein S2 |
| chr16_-_10706073 | 24.15 |
ENSRNOT00000089114
|
Fam25a
|
family with sequence similarity 25, member A |
| chr4_-_164406146 | 24.15 |
ENSRNOT00000090110
|
Klra22
|
killer cell lectin-like receptor subfamily A, member 22 |
| chr4_+_179481263 | 23.91 |
ENSRNOT00000021284
|
Etfrf1
|
electron transfer flavoprotein regulatory factor 1 |
| chr17_-_43770561 | 23.88 |
ENSRNOT00000088408
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr20_+_33077106 | 23.78 |
ENSRNOT00000000456
|
Vgll2
|
vestigial-like family member 2 |
| chr10_-_87521514 | 23.57 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr2_-_198360678 | 23.30 |
ENSRNOT00000051917
|
Hist2h2ac
|
histone cluster 2 H2A family member c |
| chr10_+_110138586 | 23.12 |
ENSRNOT00000086096
|
Slc16a3
|
solute carrier family 16 member 3 |
| chr4_-_163570803 | 23.09 |
ENSRNOT00000082002
ENSRNOT00000078642 |
Klri2
|
killer cell lectin-like receptor family I member 2 |
| chr1_+_272799784 | 22.99 |
ENSRNOT00000016052
|
Ins1
|
insulin 1 |
| chrX_-_23187341 | 22.73 |
ENSRNOT00000000180
|
Alas2
|
5'-aminolevulinate synthase 2 |
| chr20_+_5441876 | 22.68 |
ENSRNOT00000092476
|
Rps18
|
ribosomal protein S18 |
| chr8_-_66863476 | 22.55 |
ENSRNOT00000018820
|
Rplp1
|
ribosomal protein lateral stalk subunit P1 |
| chr6_+_26602144 | 22.40 |
ENSRNOT00000008037
|
Ucn
|
urocortin |
| chr11_+_47243342 | 22.34 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr1_-_82987253 | 21.91 |
ENSRNOT00000051619
|
Cd177
|
CD177 molecule |
| chr10_-_87195075 | 21.43 |
ENSRNOT00000014851
|
Krt24
|
keratin 24 |
| chrX_-_156440461 | 21.36 |
ENSRNOT00000083951
|
Rpl10
|
ribosomal protein L10 |
| chr15_-_37325178 | 21.29 |
ENSRNOT00000011699
|
Gja3
|
gap junction protein, alpha 3 |
| chr5_+_152533349 | 21.12 |
ENSRNOT00000067524
|
Trim63
|
tripartite motif containing 63 |
| chr18_+_27424328 | 20.89 |
ENSRNOT00000033784
|
Kif20a
|
kinesin family member 20A |
| chr10_-_87517199 | 20.87 |
ENSRNOT00000080171
|
LOC100365588
|
keratin associated protein 1-3-like |
| chr15_-_29532988 | 20.75 |
ENSRNOT00000074782
|
AABR07017639.1
|
|
| chr10_-_88000423 | 20.74 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr4_-_69211671 | 20.05 |
ENSRNOT00000017852
|
LOC286960
|
preprotrypsinogen IV |
| chr7_-_143392777 | 19.92 |
ENSRNOT00000086504
ENSRNOT00000038105 |
Krt72
|
keratin 72 |
| chr20_+_4855829 | 19.78 |
ENSRNOT00000001110
|
LOC103694380
|
tumor necrosis factor-like |
| chr14_-_18839420 | 19.72 |
ENSRNOT00000034090
|
Cxcl3
|
chemokine (C-X-C motif) ligand 3 |
| chr10_-_89454681 | 19.59 |
ENSRNOT00000028109
|
Brca1
|
BRCA1, DNA repair associated |
| chr17_-_43689311 | 19.50 |
ENSRNOT00000028779
|
Hist1h2bcl1
|
histone cluster 1, H2bc-like 1 |
| chr14_+_22375955 | 19.46 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr2_+_208749996 | 19.31 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr10_-_87564327 | 19.29 |
ENSRNOT00000064760
ENSRNOT00000068237 |
LOC680160
|
similar to keratin associated protein 4-7 |
| chr5_-_79691258 | 19.11 |
ENSRNOT00000072920
|
Tnfsf8
|
tumor necrosis factor superfamily member 8 |
| chr13_-_47623849 | 18.51 |
ENSRNOT00000006106
|
Il24
|
interleukin 24 |
| chr10_-_87232723 | 18.32 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr10_-_87535438 | 18.06 |
ENSRNOT00000086873
|
Krtap2-4
|
keratin associated protein 2-4 |
| chr10_-_87962846 | 18.06 |
ENSRNOT00000018176
|
Krt31
|
keratin 31 |
| chr12_-_23841049 | 17.97 |
ENSRNOT00000031555
|
Hspb1
|
heat shock protein family B (small) member 1 |
| chr7_-_143453544 | 17.64 |
ENSRNOT00000034450
ENSRNOT00000083956 |
Krt1
Krt5
|
keratin 1 keratin 5 |
| chr1_+_99749936 | 17.54 |
ENSRNOT00000025299
|
Klk7
|
kallikrein-related peptidase 7 |
| chr15_+_44441856 | 17.54 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr3_+_61590376 | 17.46 |
ENSRNOT00000002155
|
Hoxd13
|
homeo box D13 |
| chr15_+_35001838 | 17.39 |
ENSRNOT00000038253
|
Mcpt1l4
|
mast cell protease 1-like 4 |
| chr1_-_169005190 | 17.22 |
ENSRNOT00000043719
|
Hbe1
|
hemoglobin subunit epsilon 1 |
| chr3_+_114236718 | 17.12 |
ENSRNOT00000024201
|
Duoxa2
|
dual oxidase maturation factor 2 |
| chr2_+_40000313 | 17.08 |
ENSRNOT00000014270
|
Depdc1b
|
DEP domain containing 1B |
| chr3_+_79918969 | 16.93 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr9_-_97151832 | 16.90 |
ENSRNOT00000040169
|
Asb18
|
ankyrin repeat and SOCS box-containing 18 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 28.1 | 140.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 24.8 | 74.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 23.3 | 70.0 | GO:0002215 | defense response to nematode(GO:0002215) |
| 21.5 | 64.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 19.6 | 117.4 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 19.1 | 76.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 18.4 | 55.1 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) multicellular organism lipid catabolic process(GO:0044240) |
| 18.0 | 71.8 | GO:1903576 | response to L-arginine(GO:1903576) |
| 16.5 | 49.6 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 16.0 | 272.3 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 15.9 | 31.8 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 15.8 | 79.2 | GO:0001794 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 14.9 | 44.8 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 14.5 | 58.1 | GO:0072641 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 14.2 | 56.9 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 14.1 | 14.1 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 14.0 | 195.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 13.4 | 106.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 13.1 | 39.3 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 12.4 | 37.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 10.3 | 41.2 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 10.2 | 71.2 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 9.9 | 29.8 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 9.8 | 29.5 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 9.8 | 98.0 | GO:0033275 | actin-myosin filament sliding(GO:0033275) |
| 9.5 | 28.5 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) positive regulation of phospholipid catabolic process(GO:0060697) |
| 9.1 | 36.5 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 8.7 | 43.4 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 8.4 | 33.5 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 8.4 | 33.4 | GO:1903413 | negative regulation of iron ion transmembrane transport(GO:0034760) cellular response to bile acid(GO:1903413) |
| 8.3 | 24.9 | GO:1904444 | regulation of establishment of Sertoli cell barrier(GO:1904444) |
| 8.3 | 24.8 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 8.1 | 56.8 | GO:0015669 | gas transport(GO:0015669) carbon dioxide transport(GO:0015670) |
| 8.0 | 32.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 7.9 | 39.6 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 7.7 | 69.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 7.5 | 15.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 7.5 | 22.4 | GO:0035483 | gastric emptying(GO:0035483) |
| 7.2 | 28.6 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 7.0 | 28.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 6.9 | 34.7 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 6.9 | 96.4 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 6.8 | 54.4 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 6.5 | 19.6 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 6.5 | 25.9 | GO:0042531 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) diapedesis(GO:0050904) cellular response to vitamin K(GO:0071307) |
| 6.4 | 76.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 6.3 | 44.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 6.2 | 87.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 6.2 | 24.9 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 6.0 | 18.0 | GO:0071348 | cellular response to interleukin-11(GO:0071348) response to butyrate(GO:1903544) |
| 5.8 | 17.5 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 5.7 | 34.5 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 5.7 | 22.7 | GO:1901423 | response to benzene(GO:1901423) |
| 5.6 | 16.9 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 5.3 | 21.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 5.3 | 15.8 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 5.0 | 15.0 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 5.0 | 5.0 | GO:2000229 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 5.0 | 25.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 5.0 | 29.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 4.9 | 14.7 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 4.9 | 9.8 | GO:0009644 | response to high light intensity(GO:0009644) |
| 4.8 | 57.4 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 4.7 | 113.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 4.7 | 42.4 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 4.7 | 14.1 | GO:1904383 | response to sodium phosphate(GO:1904383) response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) |
| 4.6 | 13.7 | GO:0006407 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) rRNA export from nucleus(GO:0006407) |
| 4.5 | 13.6 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 4.5 | 22.5 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 4.5 | 26.8 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 4.4 | 53.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 4.4 | 13.1 | GO:0006738 | nicotinamide riboside catabolic process(GO:0006738) urate biosynthetic process(GO:0034418) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 4.2 | 8.5 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 4.2 | 37.9 | GO:0000050 | urea cycle(GO:0000050) |
| 4.2 | 29.2 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 4.1 | 49.7 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) negative regulation of platelet aggregation(GO:0090331) |
| 4.1 | 24.8 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 4.1 | 45.2 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 4.1 | 12.2 | GO:0061470 | astrocyte chemotaxis(GO:0035700) T follicular helper cell differentiation(GO:0061470) regulation of astrocyte chemotaxis(GO:2000458) |
| 4.0 | 12.0 | GO:1904528 | positive regulation of microtubule binding(GO:1904528) |
| 3.9 | 270.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 3.7 | 26.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 3.7 | 14.6 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 3.6 | 309.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 3.6 | 14.3 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 3.5 | 35.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 3.5 | 17.5 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 3.4 | 16.8 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 3.4 | 10.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 3.3 | 23.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 3.3 | 19.8 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 3.3 | 13.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 3.2 | 9.6 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 3.2 | 35.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 3.1 | 9.3 | GO:1902949 | regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 3.0 | 39.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 3.0 | 26.9 | GO:0000255 | allantoin metabolic process(GO:0000255) isoleucine metabolic process(GO:0006549) creatine metabolic process(GO:0006600) creatinine metabolic process(GO:0046449) |
| 3.0 | 11.9 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 2.9 | 17.5 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 2.9 | 17.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 2.8 | 8.3 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 2.7 | 10.7 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 2.7 | 8.0 | GO:1900111 | regulation of histone H3-K9 dimethylation(GO:1900109) positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 2.6 | 89.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 2.6 | 28.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 2.5 | 30.4 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 2.5 | 10.1 | GO:0035519 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) protein K29-linked ubiquitination(GO:0035519) |
| 2.5 | 72.9 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 2.5 | 30.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 2.4 | 7.3 | GO:1900195 | spindle assembly involved in female meiosis(GO:0007056) positive regulation of oocyte maturation(GO:1900195) |
| 2.4 | 43.3 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 2.4 | 16.6 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 2.3 | 43.7 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 2.3 | 6.9 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 2.3 | 27.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 2.2 | 91.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 2.2 | 15.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 2.1 | 6.4 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 2.1 | 29.6 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 2.1 | 4.2 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 2.1 | 4.2 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 2.1 | 6.2 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 2.0 | 56.4 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 2.0 | 53.0 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 1.9 | 3.8 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 1.9 | 24.6 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.9 | 11.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) pteridine-containing compound biosynthetic process(GO:0042559) |
| 1.9 | 5.6 | GO:0070666 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 1.8 | 5.3 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 1.7 | 19.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 1.7 | 6.7 | GO:0044691 | tooth eruption(GO:0044691) |
| 1.7 | 30.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 1.7 | 11.6 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 1.6 | 83.6 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 1.6 | 67.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 1.5 | 15.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 1.5 | 20.9 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 1.5 | 5.9 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 1.5 | 11.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 1.4 | 4.3 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 1.4 | 5.5 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 1.3 | 106.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 1.3 | 14.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 1.3 | 12.7 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 1.3 | 15.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 1.2 | 3.7 | GO:0061056 | deltoid tuberosity development(GO:0035993) sclerotome development(GO:0061056) |
| 1.2 | 8.3 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 1.2 | 10.5 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 1.1 | 12.4 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 1.1 | 3.2 | GO:1902047 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 1.0 | 1.0 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 1.0 | 26.8 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 1.0 | 58.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.9 | 14.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.9 | 3.5 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.9 | 6.0 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.8 | 32.0 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.8 | 18.3 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.8 | 21.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.8 | 37.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.7 | 9.7 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.6 | 25.1 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.6 | 4.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.6 | 2.9 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.6 | 4.0 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.6 | 11.3 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.6 | 15.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.5 | 4.9 | GO:0031424 | keratinization(GO:0031424) |
| 0.5 | 6.4 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.5 | 1.6 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.5 | 10.0 | GO:0072606 | interleukin-8 secretion(GO:0072606) |
| 0.5 | 19.0 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.5 | 10.4 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.5 | 21.3 | GO:0009268 | response to pH(GO:0009268) |
| 0.4 | 2.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.4 | 4.0 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.4 | 16.2 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.4 | 8.7 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.4 | 42.6 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.4 | 3.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.4 | 7.8 | GO:0006415 | translational termination(GO:0006415) |
| 0.4 | 40.7 | GO:0007586 | digestion(GO:0007586) |
| 0.4 | 8.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.3 | 1.0 | GO:1904350 | regulation of protein catabolic process in the vacuole(GO:1904350) |
| 0.3 | 2.4 | GO:0048739 | elastic fiber assembly(GO:0048251) cardiac muscle fiber development(GO:0048739) |
| 0.3 | 1.3 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.3 | 6.5 | GO:0030903 | notochord development(GO:0030903) |
| 0.3 | 13.7 | GO:2000242 | negative regulation of reproductive process(GO:2000242) |
| 0.3 | 3.2 | GO:2000018 | regulation of male gonad development(GO:2000018) |
| 0.3 | 9.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 12.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.3 | 78.5 | GO:0016485 | protein processing(GO:0016485) |
| 0.3 | 14.5 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.3 | 3.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.3 | 14.1 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.2 | 12.1 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) negative regulation of vasculature development(GO:1901343) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.2 | 15.0 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.2 | 7.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.2 | 27.3 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.2 | 24.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.2 | 1.5 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.2 | 9.4 | GO:0042303 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.1 | 16.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 5.5 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 0.7 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.6 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.1 | 22.7 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.1 | 8.6 | GO:0017158 | regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.1 | 1.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.4 | GO:0060043 | regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.1 | 2.1 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 6.0 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 1.6 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 3.9 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 3.9 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 2.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 12.0 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.1 | GO:0046687 | response to chromate(GO:0046687) |
| 0.0 | 2.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 0.7 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.8 | 195.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 16.8 | 252.1 | GO:0032982 | myosin filament(GO:0032982) |
| 13.9 | 180.1 | GO:0042627 | chylomicron(GO:0042627) |
| 13.5 | 40.5 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 12.6 | 37.9 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 12.4 | 37.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 11.7 | 327.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 11.6 | 808.8 | GO:0000786 | nucleosome(GO:0000786) |
| 9.1 | 686.0 | GO:0045095 | keratin filament(GO:0045095) |
| 8.9 | 35.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 6.8 | 33.8 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 6.2 | 49.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 5.0 | 80.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 4.9 | 19.6 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 4.4 | 17.5 | GO:1990005 | granular vesicle(GO:1990005) |
| 3.9 | 11.6 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 3.7 | 33.4 | GO:0045179 | apical cortex(GO:0045179) |
| 3.5 | 109.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 2.9 | 17.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 2.8 | 14.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 2.6 | 7.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 2.5 | 10.0 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 2.5 | 14.7 | GO:0071547 | piP-body(GO:0071547) |
| 2.3 | 112.7 | GO:0016459 | myosin complex(GO:0016459) |
| 2.2 | 18.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 2.1 | 51.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 2.1 | 15.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 2.0 | 20.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 2.0 | 13.7 | GO:0001652 | granular component(GO:0001652) |
| 1.9 | 9.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.8 | 7.3 | GO:0042585 | germinal vesicle(GO:0042585) |
| 1.7 | 22.4 | GO:0043196 | varicosity(GO:0043196) |
| 1.7 | 12.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.6 | 15.8 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 1.6 | 113.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 1.5 | 33.4 | GO:0010369 | chromocenter(GO:0010369) |
| 1.5 | 6.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 1.4 | 120.5 | GO:0042641 | actomyosin(GO:0042641) |
| 1.3 | 5.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.1 | 28.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 1.1 | 8.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 1.0 | 21.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.9 | 14.0 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.8 | 32.2 | GO:0000791 | euchromatin(GO:0000791) |
| 0.8 | 29.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.8 | 13.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.8 | 7.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.8 | 3.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.7 | 29.8 | GO:0031672 | A band(GO:0031672) |
| 0.7 | 76.2 | GO:0005901 | caveola(GO:0005901) |
| 0.7 | 37.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.7 | 12.1 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.7 | 2.7 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.7 | 16.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.6 | 12.4 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.6 | 15.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.6 | 53.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.6 | 40.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.6 | 794.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.5 | 6.0 | GO:0000801 | central element(GO:0000801) |
| 0.5 | 54.7 | GO:0016605 | PML body(GO:0016605) |
| 0.5 | 10.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.5 | 24.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.5 | 10.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.4 | 24.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.4 | 20.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 0.7 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.3 | 4.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 8.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 18.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 2.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 44.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 9.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 21.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.2 | 59.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.2 | 4.2 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.2 | 23.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.2 | 15.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 7.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 26.8 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 10.3 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 5.0 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 30.2 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 14.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.3 | GO:0036126 | sperm flagellum(GO:0036126) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.3 | 72.9 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 23.5 | 117.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 19.9 | 59.7 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 19.5 | 58.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 17.4 | 52.1 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 17.3 | 207.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 14.5 | 43.6 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 13.7 | 82.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 13.0 | 77.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 12.4 | 86.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 9.8 | 58.8 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 9.6 | 38.4 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 9.5 | 47.6 | GO:0032564 | dATP binding(GO:0032564) |
| 8.7 | 26.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 8.2 | 180.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 7.9 | 39.6 | GO:0035473 | lipase binding(GO:0035473) |
| 7.6 | 22.7 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 7.5 | 30.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 7.5 | 29.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 7.5 | 44.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 6.7 | 26.9 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 6.0 | 48.4 | GO:0016151 | nickel cation binding(GO:0016151) |
| 5.6 | 39.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 5.6 | 72.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 5.6 | 22.4 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 5.6 | 150.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 5.5 | 38.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 5.3 | 69.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 5.2 | 25.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 5.0 | 15.1 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 4.9 | 14.7 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 4.5 | 44.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 4.4 | 13.1 | GO:0002060 | purine nucleobase binding(GO:0002060) purine-nucleoside phosphorylase activity(GO:0004731) |
| 4.3 | 30.1 | GO:0035375 | zymogen binding(GO:0035375) |
| 4.2 | 12.7 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 4.2 | 37.9 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 4.1 | 12.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 4.0 | 43.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 3.8 | 79.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 3.7 | 29.8 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 3.7 | 14.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 3.7 | 29.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 3.4 | 10.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 3.3 | 16.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 3.3 | 16.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 3.2 | 9.6 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 3.2 | 56.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 3.2 | 15.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 3.0 | 66.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 3.0 | 24.3 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 3.0 | 35.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 2.9 | 23.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 2.7 | 76.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 2.7 | 80.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 2.5 | 74.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 2.4 | 16.8 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 2.2 | 6.5 | GO:0004655 | porphobilinogen synthase activity(GO:0004655) |
| 2.1 | 481.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 2.1 | 16.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 2.1 | 14.6 | GO:0008061 | chitin binding(GO:0008061) |
| 2.0 | 408.6 | GO:0042393 | histone binding(GO:0042393) |
| 2.0 | 14.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 1.7 | 70.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 1.7 | 6.9 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 1.7 | 15.4 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 1.7 | 184.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 1.6 | 21.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 1.4 | 4.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 1.4 | 7.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 1.4 | 25.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 1.3 | 8.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 1.3 | 23.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 1.3 | 24.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 1.2 | 32.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 1.2 | 14.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 1.2 | 5.9 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.2 | 10.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 1.1 | 25.9 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 1.1 | 14.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 1.1 | 3.2 | GO:0042979 | ornithine decarboxylase activator activity(GO:0042978) ornithine decarboxylase regulator activity(GO:0042979) |
| 1.1 | 8.5 | GO:0038085 | vascular endothelial growth factor-activated receptor activity(GO:0005021) vascular endothelial growth factor binding(GO:0038085) |
| 1.1 | 28.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 1.0 | 45.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 1.0 | 5.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 1.0 | 32.2 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 1.0 | 58.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 1.0 | 13.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 1.0 | 2.9 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.0 | 34.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.9 | 26.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.9 | 24.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.9 | 32.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.8 | 113.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.8 | 5.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.8 | 6.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.8 | 48.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.7 | 49.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.7 | 6.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.7 | 23.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.7 | 9.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.7 | 7.8 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.7 | 57.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.7 | 91.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.7 | 2.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.7 | 16.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.7 | 2.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.7 | 5.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.7 | 3.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.6 | 11.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.6 | 19.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.6 | 33.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.6 | 419.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.6 | 281.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.6 | 2.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.6 | 106.8 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.6 | 14.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.6 | 12.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.6 | 9.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.5 | 2.7 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.5 | 8.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.4 | 10.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.4 | 3.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.4 | 19.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.4 | 11.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.4 | 4.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.4 | 12.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.4 | 7.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 10.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 29.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 62.6 | GO:0000982 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.3 | 6.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.3 | 19.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.2 | 5.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 10.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 18.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 0.4 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 12.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.2 | 10.2 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 27.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 4.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 6.4 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 1.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 23.5 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 2.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 3.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 13.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 3.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 11.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0008527 | taste receptor activity(GO:0008527) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 154.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 3.2 | 77.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 2.9 | 49.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 2.8 | 50.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 2.8 | 112.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 2.6 | 36.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 2.5 | 170.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 2.1 | 68.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 1.9 | 85.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 1.9 | 28.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 1.8 | 53.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 1.3 | 47.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 1.1 | 77.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 1.1 | 49.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 1.0 | 29.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.9 | 19.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.8 | 5.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.8 | 14.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.8 | 39.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.8 | 8.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.7 | 17.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.6 | 143.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.6 | 12.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.6 | 16.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.6 | 165.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.5 | 18.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.5 | 9.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.5 | 13.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.5 | 57.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.5 | 12.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.5 | 34.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.5 | 14.3 | PID ATM PATHWAY | ATM pathway |
| 0.5 | 13.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.5 | 10.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.5 | 17.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 14.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.3 | 2.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 4.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 4.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 5.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 6.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 3.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 3.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 7.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.8 | 162.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 12.4 | 322.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 9.7 | 378.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 9.0 | 153.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 5.8 | 52.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 4.7 | 223.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 4.0 | 32.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 3.4 | 43.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 3.3 | 49.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 3.0 | 32.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 2.8 | 41.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 2.7 | 33.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 2.7 | 40.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 2.6 | 71.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 2.3 | 32.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 2.2 | 35.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 2.0 | 153.7 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 2.0 | 47.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 1.9 | 26.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 1.9 | 15.4 | REACTOME OPSINS | Genes involved in Opsins |
| 1.8 | 12.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 1.8 | 49.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 1.7 | 29.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 1.7 | 23.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 1.7 | 23.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.6 | 67.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 1.6 | 14.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 1.5 | 82.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 1.5 | 34.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 1.5 | 26.8 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 1.5 | 36.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 1.4 | 9.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 1.3 | 13.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.2 | 21.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 1.2 | 15.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 1.1 | 9.1 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 1.0 | 14.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.9 | 58.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.9 | 14.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.9 | 19.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.9 | 12.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.7 | 14.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.7 | 29.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.7 | 20.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 10.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.5 | 5.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.5 | 14.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.5 | 6.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.4 | 7.8 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.4 | 12.7 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.4 | 10.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.3 | 7.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.2 | 4.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 6.4 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.2 | 12.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 6.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 5.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 2.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 24.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 4.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 3.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |