Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
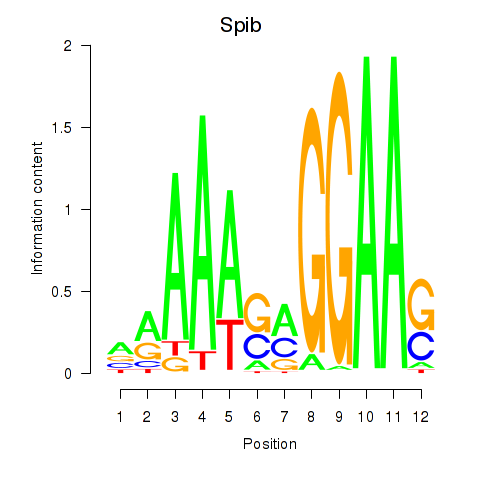
Results for Spib
Z-value: 2.50
Transcription factors associated with Spib
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Spib
|
ENSRNOG00000019660 | Spi-B transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Spib | rn6_v1_chr1_-_100537377_100537377 | 0.92 | 2.0e-131 | Click! |
Activity profile of Spib motif
Sorted Z-values of Spib motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_78196300 | 114.72 |
ENSRNOT00000048695
|
P2ry10
|
purinergic receptor P2Y10 |
| chr7_-_107616038 | 105.69 |
ENSRNOT00000088752
|
Sla
|
src-like adaptor |
| chr9_-_9675110 | 92.12 |
ENSRNOT00000073294
|
Vav1
|
vav guanine nucleotide exchange factor 1 |
| chr7_+_145068286 | 91.25 |
ENSRNOT00000088956
ENSRNOT00000065753 |
Nckap1l
|
NCK associated protein 1 like |
| chr3_+_28627084 | 90.54 |
ENSRNOT00000049884
|
Arhgap15
|
Rho GTPase activating protein 15 |
| chr14_-_23604834 | 89.49 |
ENSRNOT00000002760
|
Stap1
|
signal transducing adaptor family member 1 |
| chr5_+_149047681 | 89.02 |
ENSRNOT00000015198
|
Laptm5
|
lysosomal protein transmembrane 5 |
| chr1_-_101236065 | 88.44 |
ENSRNOT00000066834
|
Cd37
|
CD37 molecule |
| chr8_+_133029625 | 86.08 |
ENSRNOT00000008809
|
Ccr3
|
C-C motif chemokine receptor 3 |
| chr10_-_5260608 | 86.04 |
ENSRNOT00000003572
|
Ciita
|
class II, major histocompatibility complex, transactivator |
| chr2_+_186776644 | 84.13 |
ENSRNOT00000046778
|
Fcrl3
|
Fc receptor-like 3 |
| chr12_-_2826378 | 79.26 |
ENSRNOT00000061749
|
Clec4m
|
C-type lectin domain family 4 member M |
| chr14_-_100184192 | 78.80 |
ENSRNOT00000007044
|
Plek
|
pleckstrin |
| chr1_+_242959488 | 71.89 |
ENSRNOT00000015668
|
Dock8
|
dedicator of cytokinesis 8 |
| chr7_-_11330278 | 71.78 |
ENSRNOT00000027730
|
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr11_-_66759402 | 70.67 |
ENSRNOT00000003326
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr4_-_155923079 | 69.85 |
ENSRNOT00000013308
|
Clec4a3
|
C-type lectin domain family 4, member A3 |
| chr10_+_47930633 | 68.58 |
ENSRNOT00000003515
|
Grap
|
GRB2-related adaptor protein |
| chr14_+_73674753 | 67.94 |
ENSRNOT00000040676
|
RGD1561694
|
similar to High mobility group protein 2 (HMG-2) |
| chr17_-_46115004 | 66.11 |
ENSRNOT00000087838
|
Aoah
|
acyloxyacyl hydrolase |
| chr11_+_67082193 | 65.37 |
ENSRNOT00000003129
|
Cd86
|
CD86 molecule |
| chr8_+_60760078 | 65.16 |
ENSRNOT00000063930
|
Pstpip1
|
proline-serine-threonine phosphatase-interacting protein 1 |
| chr9_+_40972089 | 64.13 |
ENSRNOT00000067928
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chrX_+_54390733 | 63.81 |
ENSRNOT00000004977
|
RGD1565785
|
similar to chromosome X open reading frame 21 |
| chr14_-_82347679 | 63.30 |
ENSRNOT00000032972
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr16_-_19349080 | 61.40 |
ENSRNOT00000038494
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr2_-_203413124 | 60.70 |
ENSRNOT00000077898
ENSRNOT00000056140 |
Cd101
|
CD101 molecule |
| chr5_+_157222636 | 60.20 |
ENSRNOT00000022579
|
Pla2g2d
|
phospholipase A2, group IID |
| chr13_-_89306219 | 60.10 |
ENSRNOT00000004183
ENSRNOT00000079247 |
Fcrla
|
Fc receptor-like A |
| chr1_-_47502952 | 59.90 |
ENSRNOT00000025580
|
Tagap
|
T-cell activation RhoGTPase activating protein |
| chr10_+_55013703 | 59.64 |
ENSRNOT00000032785
|
Pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr1_-_227506822 | 58.66 |
ENSRNOT00000091506
|
Ms4a7
|
membrane spanning 4-domains A7 |
| chr13_-_89343868 | 56.49 |
ENSRNOT00000058497
ENSRNOT00000035400 |
Fcgr2b
|
Fc fragment of IgG receptor IIb |
| chr3_-_153001309 | 55.87 |
ENSRNOT00000027581
|
Sla2
|
Src-like-adaptor 2 |
| chr7_-_75422268 | 55.12 |
ENSRNOT00000080218
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr4_+_66670618 | 55.04 |
ENSRNOT00000010796
|
Tbxas1
|
thromboxane A synthase 1 |
| chr10_-_57436368 | 54.74 |
ENSRNOT00000056608
|
Scimp
|
SLP adaptor and CSK interacting membrane protein |
| chr1_+_226634009 | 53.38 |
ENSRNOT00000028094
|
Cyb561a3
|
cytochrome b561 family, member A3 |
| chr1_-_221015929 | 53.30 |
ENSRNOT00000028137
|
Sipa1
|
signal-induced proliferation-associated 1 |
| chr2_+_55835151 | 52.84 |
ENSRNOT00000018634
|
Fyb
|
FYN binding protein |
| chr20_-_5163247 | 52.76 |
ENSRNOT00000001135
|
Aif1
|
allograft inflammatory factor 1 |
| chr2_-_198439454 | 52.67 |
ENSRNOT00000028780
|
Fcgr1a
|
Fc fragment of IgG receptor Ia |
| chr1_-_20417637 | 51.96 |
ENSRNOT00000036049
|
RGD1559962
|
similar to High mobility group protein 2 (HMG-2) |
| chr19_+_19395655 | 51.51 |
ENSRNOT00000019130
|
Snx20
|
sorting nexin 20 |
| chr7_-_75421874 | 49.88 |
ENSRNOT00000012775
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr8_+_102304095 | 49.62 |
ENSRNOT00000011358
|
Slc9a9
|
solute carrier family 9 member A9 |
| chr5_-_155258392 | 48.57 |
ENSRNOT00000017065
|
C1qc
|
complement C1q C chain |
| chr9_+_94745217 | 48.34 |
ENSRNOT00000051338
|
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr3_+_79918969 | 47.97 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr5_-_155252003 | 47.59 |
ENSRNOT00000017060
|
C1qb
|
complement C1q B chain |
| chr4_-_157252565 | 47.54 |
ENSRNOT00000079947
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr1_-_260254600 | 47.47 |
ENSRNOT00000019014
|
Blnk
|
B-cell linker |
| chr9_+_47134034 | 45.69 |
ENSRNOT00000020108
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr4_-_157252104 | 45.31 |
ENSRNOT00000082739
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr1_-_43638161 | 44.90 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr8_+_49378644 | 43.32 |
ENSRNOT00000007588
|
Jaml
|
junction adhesion molecule like |
| chr10_+_83655460 | 42.87 |
ENSRNOT00000008011
|
Gngt2
|
G protein subunit gamma transducin 2 |
| chrX_+_15155230 | 42.52 |
ENSRNOT00000073289
ENSRNOT00000051439 |
Was
|
Wiskott-Aldrich syndrome |
| chr17_-_34905117 | 42.18 |
ENSRNOT00000088931
|
Irf4
|
interferon regulatory factor 4 |
| chr10_+_104932616 | 41.98 |
ENSRNOT00000075706
|
RGD1562667
|
similar to leukocyte mono-Ig-like receptor2 |
| chr7_+_70614617 | 40.94 |
ENSRNOT00000035382
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr13_-_91198036 | 39.31 |
ENSRNOT00000004204
|
LOC108348047
|
low affinity immunoglobulin gamma Fc region receptor III |
| chr4_+_163112301 | 38.29 |
ENSRNOT00000087113
|
Clec12a
|
C-type lectin domain family 12, member A |
| chr9_-_61528882 | 37.83 |
ENSRNOT00000015432
|
Ankrd44
|
ankyrin repeat domain 44 |
| chr10_-_103685844 | 37.47 |
ENSRNOT00000064284
|
Cd300lf
|
Cd300 molecule-like family member F |
| chr3_+_11756384 | 36.89 |
ENSRNOT00000087762
|
Sh2d3c
|
SH2 domain containing 3C |
| chr12_-_6879154 | 36.85 |
ENSRNOT00000001207
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr4_+_22898527 | 36.06 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr3_+_2642531 | 34.98 |
ENSRNOT00000081798
|
Fut7
|
fucosyltransferase 7 |
| chr10_-_70506963 | 31.19 |
ENSRNOT00000076883
ENSRNOT00000012874 |
Slfn14
|
schlafen family member 14 |
| chr6_-_102196138 | 30.86 |
ENSRNOT00000014132
|
Tmem229b
|
transmembrane protein 229B |
| chr8_-_48634797 | 30.44 |
ENSRNOT00000012868
|
Hinfp
|
histone H4 transcription factor |
| chr13_-_91228901 | 30.35 |
ENSRNOT00000071728
ENSRNOT00000073643 ENSRNOT00000071897 |
LOC100911825
|
low affinity immunoglobulin gamma Fc region receptor III-like |
| chr4_+_115024927 | 29.83 |
ENSRNOT00000090987
|
Mob1a
|
MOB kinase activator 1A |
| chr13_-_91197744 | 27.59 |
ENSRNOT00000047349
|
LOC108348047
|
low affinity immunoglobulin gamma Fc region receptor III |
| chr5_-_160423811 | 27.27 |
ENSRNOT00000018864
|
Efhd2
|
EF-hand domain family, member D2 |
| chr4_-_57625147 | 27.15 |
ENSRNOT00000074649
ENSRNOT00000078961 |
Ube2h
|
ubiquitin-conjugating enzyme E2H |
| chr10_-_56289882 | 27.03 |
ENSRNOT00000090762
ENSRNOT00000056903 |
Tnfsf13
|
tumor necrosis factor superfamily member 13 |
| chr14_+_76657311 | 26.83 |
ENSRNOT00000076730
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr1_+_221420271 | 25.79 |
ENSRNOT00000028481
|
LOC687780
|
similar to Finkel-Biskis-Reilly murine sarcoma virusubiquitously expressed |
| chr10_+_56187679 | 25.60 |
ENSRNOT00000085115
|
Tp53
|
tumor protein p53 |
| chr3_-_11102515 | 25.15 |
ENSRNOT00000035580
|
Fam78a
|
family with sequence similarity 78, member A |
| chrX_-_77700269 | 24.77 |
ENSRNOT00000092418
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr3_+_103753238 | 24.53 |
ENSRNOT00000007144
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr13_+_48287873 | 24.47 |
ENSRNOT00000068223
|
Fam72a
|
family with sequence similarity 72, member A |
| chr1_-_53520788 | 24.41 |
ENSRNOT00000060121
|
Gpr31
|
G protein-coupled receptor 31 |
| chr6_-_103313074 | 24.12 |
ENSRNOT00000083677
|
Zfp36l1
|
zinc finger protein 36, C3H type-like 1 |
| chr20_-_13706205 | 23.70 |
ENSRNOT00000038623
|
Derl3
|
derlin 3 |
| chr19_+_25815207 | 22.86 |
ENSRNOT00000003980
|
Lyl1
|
LYL1, basic helix-loop-helix family member |
| chr1_-_242440885 | 22.77 |
ENSRNOT00000076537
|
Pip5k1b
|
phosphatidylinositol-4-phosphate 5-kinase type 1 beta |
| chr16_-_85306366 | 22.25 |
ENSRNOT00000089650
|
Tnfsf13b
|
tumor necrosis factor superfamily member 13b |
| chr5_+_25168295 | 22.08 |
ENSRNOT00000020245
|
Fsbp
|
fibrinogen silencer binding protein |
| chr6_+_137323713 | 21.72 |
ENSRNOT00000029017
|
Pld4
|
phospholipase D family, member 4 |
| chr1_-_64446818 | 21.51 |
ENSRNOT00000081980
|
Myadm
|
myeloid-associated differentiation marker |
| chr3_-_23297774 | 21.24 |
ENSRNOT00000019162
|
Rpl35
|
ribosomal protein L35 |
| chr3_+_103773459 | 21.15 |
ENSRNOT00000079727
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr1_-_142183884 | 21.13 |
ENSRNOT00000016032
|
Fes
|
FES proto-oncogene, tyrosine kinase |
| chr2_+_34312766 | 20.80 |
ENSRNOT00000060962
|
Cenpk
|
centromere protein K |
| chr10_+_74959285 | 20.72 |
ENSRNOT00000010296
|
Rnf43
|
ring finger protein 43 |
| chr1_-_242441247 | 20.63 |
ENSRNOT00000068645
|
Pip5k1b
|
phosphatidylinositol-4-phosphate 5-kinase type 1 beta |
| chrX_+_113948654 | 20.56 |
ENSRNOT00000068431
|
Tmem164
|
transmembrane protein 164 |
| chr9_+_10471742 | 20.38 |
ENSRNOT00000072276
|
Safb2
|
scaffold attachment factor B2 |
| chr5_-_141430659 | 19.37 |
ENSRNOT00000034944
|
Akirin1
|
akirin 1 |
| chr2_+_38230757 | 18.75 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chr6_-_47904163 | 18.58 |
ENSRNOT00000011333
|
Rps7
|
ribosomal protein S7 |
| chr9_-_46951847 | 18.09 |
ENSRNOT00000051231
|
RGD1564447
|
similar to Zgc:56193 |
| chr6_+_134804141 | 17.81 |
ENSRNOT00000086143
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr14_+_58877806 | 17.64 |
ENSRNOT00000051559
|
RGD1562755
|
similar to 60S ribosomal protein L23a |
| chr17_+_81352700 | 17.58 |
ENSRNOT00000024736
|
Mrc1
|
mannose receptor, C type 1 |
| chr10_+_23661013 | 17.38 |
ENSRNOT00000076664
|
Ebf1
|
early B-cell factor 1 |
| chr3_+_160467552 | 17.32 |
ENSRNOT00000066657
|
Stk4
|
serine/threonine kinase 4 |
| chr13_-_95250235 | 17.27 |
ENSRNOT00000085648
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr17_-_46794845 | 17.01 |
ENSRNOT00000077910
ENSRNOT00000090663 ENSRNOT00000078331 |
Elmo1
|
engulfment and cell motility 1 |
| chr11_-_33003021 | 17.01 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr12_+_1889331 | 16.94 |
ENSRNOT00000066281
|
Arhgef18
|
Rho/Rac guanine nucleotide exchange factor 18 |
| chrX_+_128493614 | 16.69 |
ENSRNOT00000044240
|
Stag2
|
stromal antigen 2 |
| chr4_+_90990088 | 16.52 |
ENSRNOT00000030320
|
Mmrn1
|
multimerin 1 |
| chr13_-_89433815 | 16.18 |
ENSRNOT00000091541
|
Fcgr2b
|
Fc fragment of IgG receptor IIb |
| chr8_-_45375435 | 16.16 |
ENSRNOT00000010873
|
Ubash3b
|
ubiquitin associated and SH3 domain containing, B |
| chr1_+_197999037 | 16.10 |
ENSRNOT00000091065
|
Apobr
|
apolipoprotein B receptor |
| chr3_+_118317761 | 15.37 |
ENSRNOT00000012700
|
Fgf7
|
fibroblast growth factor 7 |
| chr1_-_224974203 | 14.92 |
ENSRNOT00000065694
|
Tmem179b
|
transmembrane protein 179B |
| chr15_-_3650819 | 14.84 |
ENSRNOT00000014273
|
Plau
|
plasminogen activator, urokinase |
| chr5_+_61425746 | 14.22 |
ENSRNOT00000064113
|
RGD1305807
|
hypothetical LOC298077 |
| chr4_-_15505362 | 13.99 |
ENSRNOT00000009763
|
Hgf
|
hepatocyte growth factor |
| chr4_+_95884743 | 13.84 |
ENSRNOT00000008585
|
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1` |
| chr10_+_31880918 | 13.60 |
ENSRNOT00000059448
|
Timd4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr5_-_137238354 | 13.10 |
ENSRNOT00000039235
|
Szt2
|
seizure threshold 2 homolog (mouse) |
| chr2_+_157854667 | 12.92 |
ENSRNOT00000039294
|
AC119603.1
|
|
| chr2_-_34313094 | 12.84 |
ENSRNOT00000016863
|
Ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr13_+_49250301 | 12.82 |
ENSRNOT00000036110
|
Rbbp5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr6_-_47904437 | 12.23 |
ENSRNOT00000092867
|
Rps7
|
ribosomal protein S7 |
| chr1_+_199217016 | 12.23 |
ENSRNOT00000025732
|
Orai3
|
ORAI calcium release-activated calcium modulator 3 |
| chr7_+_109411475 | 12.22 |
ENSRNOT00000051728
|
RGD1564548
|
similar to H3 histone, family 3B |
| chr12_+_25497104 | 12.08 |
ENSRNOT00000002028
|
Ncf1
|
neutrophil cytosolic factor 1 |
| chr8_-_76940094 | 11.93 |
ENSRNOT00000082709
ENSRNOT00000084313 |
Rnf111
|
ring finger protein 111 |
| chrX_-_63204530 | 11.72 |
ENSRNOT00000076175
|
Zfx
|
zinc finger protein X-linked |
| chr13_-_69172643 | 11.54 |
ENSRNOT00000049703
|
AABR07021384.1
|
|
| chr3_-_130114770 | 11.39 |
ENSRNOT00000010638
|
Jag1
|
jagged 1 |
| chr15_+_44441856 | 11.29 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr4_-_120041238 | 11.20 |
ENSRNOT00000073799
|
LOC100911337
|
40S ribosomal protein S25-like |
| chr1_-_164441167 | 11.16 |
ENSRNOT00000023935
|
Rps3
|
ribosomal protein S3 |
| chr3_-_140141679 | 10.94 |
ENSRNOT00000014632
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr1_+_197999336 | 10.84 |
ENSRNOT00000023555
|
Apobr
|
apolipoprotein B receptor |
| chr8_+_114916122 | 10.35 |
ENSRNOT00000074194
|
Tlr9
|
toll-like receptor 9 |
| chr2_+_189454340 | 9.68 |
ENSRNOT00000087063
|
Nup210l
|
nucleoporin 210-like |
| chr1_+_221710670 | 9.61 |
ENSRNOT00000064798
|
Map4k2
|
mitogen activated protein kinase kinase kinase kinase 2 |
| chr4_+_145330457 | 9.35 |
ENSRNOT00000011937
|
Arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr3_-_80875817 | 9.34 |
ENSRNOT00000091265
|
Dgkz
|
diacylglycerol kinase zeta |
| chr1_-_143169657 | 9.13 |
ENSRNOT00000025888
|
Rps17
|
ribosomal protein S17 |
| chr2_-_118882562 | 9.09 |
ENSRNOT00000058860
|
Kcnmb3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr2_+_187740531 | 8.89 |
ENSRNOT00000092653
|
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr17_+_90670872 | 8.39 |
ENSRNOT00000081530
|
Gpr137b
|
G protein-coupled receptor 137B |
| chr5_+_64892802 | 8.33 |
ENSRNOT00000059869
|
Rnf20
|
ring finger protein 20 |
| chr3_+_47439076 | 8.00 |
ENSRNOT00000011794
|
Tank
|
TRAF family member-associated NFKB activator |
| chr2_-_189765415 | 7.98 |
ENSRNOT00000020815
|
Slc27a3
|
solute carrier family 27 member 3 |
| chr19_+_19672856 | 7.72 |
ENSRNOT00000064275
ENSRNOT00000086560 |
Brd7
|
bromodomain containing 7 |
| chr13_-_67206688 | 7.55 |
ENSRNOT00000003630
ENSRNOT00000090693 |
Pla2g4a
|
phospholipase A2 group IVA |
| chr20_-_5466265 | 7.53 |
ENSRNOT00000000554
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr8_-_108261021 | 7.48 |
ENSRNOT00000040710
|
LOC100912210
|
40S ribosomal protein S25-like |
| chr3_+_28416954 | 7.28 |
ENSRNOT00000043533
|
Kynu
|
kynureninase |
| chr7_+_122818975 | 7.15 |
ENSRNOT00000000206
|
Ep300
|
E1A binding protein p300 |
| chrX_+_156655960 | 7.03 |
ENSRNOT00000085723
|
Mecp2
|
methyl CpG binding protein 2 |
| chr16_+_40050734 | 6.74 |
ENSRNOT00000067375
|
Spcs3
|
signal peptidase complex subunit 3 |
| chr4_-_120414118 | 6.72 |
ENSRNOT00000072795
|
LOC100911337
|
40S ribosomal protein S25-like |
| chr7_+_122160171 | 6.66 |
ENSRNOT00000074499
|
LOC100362980
|
CG3918-like |
| chr13_-_90977734 | 6.63 |
ENSRNOT00000011869
|
Slamf8
|
SLAM family member 8 |
| chr1_-_280233755 | 6.46 |
ENSRNOT00000064463
|
Shtn1
|
shootin 1 |
| chr3_-_103203299 | 6.30 |
ENSRNOT00000047208
|
Olr783
|
olfactory receptor 783 |
| chr15_+_34270648 | 5.87 |
ENSRNOT00000026333
|
Rnf31
|
ring finger protein 31 |
| chr1_+_234749568 | 5.74 |
ENSRNOT00000016871
|
Ostf1
|
osteoclast stimulating factor 1 |
| chr8_+_114897011 | 5.73 |
ENSRNOT00000074683
|
Twf2
|
twinfilin actin-binding protein 2 |
| chr7_-_70407177 | 5.23 |
ENSRNOT00000049895
|
Os9
|
OS9, endoplasmic reticulum lectin |
| chr13_-_79744939 | 4.99 |
ENSRNOT00000076706
|
Suco
|
SUN domain containing ossification factor |
| chr17_-_76281660 | 4.78 |
ENSRNOT00000058070
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr4_-_100099517 | 4.71 |
ENSRNOT00000014277
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr16_-_85305782 | 4.69 |
ENSRNOT00000067511
ENSRNOT00000076737 |
Tnfsf13b
|
tumor necrosis factor superfamily member 13b |
| chr10_-_64642292 | 4.52 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chr3_-_59688692 | 4.28 |
ENSRNOT00000078752
|
Sp3
|
Sp3 transcription factor |
| chr3_-_151224123 | 4.15 |
ENSRNOT00000026091
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr19_-_41029206 | 4.15 |
ENSRNOT00000023383
|
Vac14
|
Vac14, PIKFYVE complex component |
| chr7_-_116936674 | 4.02 |
ENSRNOT00000029456
|
Eef1d
|
eukaryotic translation elongation factor 1 delta |
| chr16_+_83358116 | 3.94 |
ENSRNOT00000031109
|
Rab20
|
RAB20, member RAS oncogene family |
| chr1_-_254671778 | 3.93 |
ENSRNOT00000025493
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr11_+_81972219 | 3.89 |
ENSRNOT00000002452
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr8_-_39243882 | 3.80 |
ENSRNOT00000082086
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr15_-_52317219 | 3.65 |
ENSRNOT00000016555
|
Dmtn
|
dematin actin binding protein |
| chr9_-_42805673 | 3.35 |
ENSRNOT00000020558
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr6_+_108831108 | 3.28 |
ENSRNOT00000038594
|
LOC103692678
|
U6 snRNA-associated Sm-like protein LSm5 pseudogene |
| chr3_-_125470413 | 3.01 |
ENSRNOT00000028893
|
Trmt6
|
tRNA methyltransferase 6 |
| chr6_+_129609375 | 3.00 |
ENSRNOT00000083732
|
Papola
|
poly (A) polymerase alpha |
| chr17_-_1648973 | 2.61 |
ENSRNOT00000036212
|
Slc35d2
|
solute carrier family 35 member D2 |
| chr4_-_63039422 | 2.47 |
ENSRNOT00000015808
|
Mtpn
|
myotrophin |
| chr2_-_196270826 | 2.38 |
ENSRNOT00000028609
|
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1, alpha |
| chr1_-_103128743 | 2.00 |
ENSRNOT00000075006
|
Spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr12_-_19582185 | 1.94 |
ENSRNOT00000060020
ENSRNOT00000085853 |
RGD1305455
|
similar to hypothetical protein FLJ10925 |
| chr2_-_120316357 | 1.86 |
ENSRNOT00000065469
|
Ccdc39
|
coiled-coil domain containing 39 |
| chr1_-_84812486 | 1.72 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chr4_+_72656125 | 1.39 |
ENSRNOT00000047359
|
Olr820
|
olfactory receptor 820 |
| chr1_-_198008893 | 1.37 |
ENSRNOT00000025950
|
Il27
|
interleukin 27 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Spib
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 30.4 | 91.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 29.8 | 89.5 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 23.2 | 92.8 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 22.1 | 88.4 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 21.8 | 65.4 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 20.6 | 82.5 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 18.7 | 37.5 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 18.2 | 72.7 | GO:0002434 | immune complex clearance(GO:0002434) negative regulation of hypersensitivity(GO:0002884) |
| 17.8 | 53.3 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 17.7 | 70.7 | GO:0042531 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 17.6 | 52.8 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 17.6 | 52.7 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 17.5 | 105.0 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 16.8 | 84.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 15.2 | 45.7 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 15.2 | 45.7 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 14.4 | 43.3 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 14.3 | 86.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 14.1 | 42.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 12.3 | 61.4 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 12.1 | 48.3 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 12.0 | 48.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 10.8 | 54.0 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 9.2 | 36.9 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 8.8 | 97.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 8.8 | 79.3 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 7.4 | 14.8 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 7.2 | 21.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 7.1 | 42.5 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 6.8 | 95.2 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 6.7 | 46.9 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 6.4 | 25.6 | GO:1901003 | negative regulation of fermentation(GO:1901003) negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 6.1 | 86.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 6.1 | 55.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 6.0 | 24.1 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) mesendoderm development(GO:0048382) regulation of intracellular mRNA localization(GO:1904580) |
| 5.7 | 17.0 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 5.2 | 10.3 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 4.9 | 29.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 4.9 | 63.3 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 4.8 | 71.9 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 4.7 | 14.2 | GO:0046865 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 4.5 | 323.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 4.3 | 17.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 4.3 | 52.0 | GO:0006265 | DNA topological change(GO:0006265) |
| 4.1 | 24.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 4.0 | 12.1 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 3.8 | 49.6 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 3.8 | 11.4 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 3.7 | 11.2 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 3.5 | 24.5 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 3.2 | 92.1 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 2.8 | 30.8 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 2.7 | 10.9 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 2.7 | 8.0 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 2.6 | 28.9 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 2.6 | 13.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 2.4 | 7.2 | GO:0018076 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 2.3 | 7.0 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) negative regulation of dendrite extension(GO:1903860) negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 2.3 | 22.9 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 2.2 | 6.6 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 2.1 | 21.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 2.1 | 8.3 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 2.1 | 31.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 2.0 | 30.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 1.9 | 7.5 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 1.9 | 11.3 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 1.9 | 16.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 1.8 | 16.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 1.7 | 11.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 1.6 | 6.5 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.5 | 47.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 1.5 | 4.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of neutrophil activation(GO:1902564) |
| 1.5 | 16.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 1.5 | 7.3 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 1.3 | 3.9 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 1.3 | 7.5 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 1.1 | 45.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 1.1 | 48.5 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 1.0 | 60.7 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 1.0 | 20.7 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 1.0 | 27.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 1.0 | 13.8 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 1.0 | 5.9 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.9 | 13.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.8 | 3.3 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.8 | 30.1 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.8 | 18.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.8 | 52.8 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.8 | 3.9 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.8 | 17.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.7 | 9.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.7 | 5.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.7 | 4.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.7 | 6.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.7 | 2.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.7 | 17.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.6 | 66.1 | GO:0044264 | cellular polysaccharide metabolic process(GO:0044264) |
| 0.6 | 9.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.6 | 12.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.6 | 64.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.6 | 3.9 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.6 | 11.7 | GO:0060746 | parental behavior(GO:0060746) |
| 0.5 | 17.0 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.5 | 4.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.4 | 54.7 | GO:0007127 | meiosis I(GO:0007127) |
| 0.4 | 55.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.4 | 61.3 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.3 | 9.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.3 | 4.7 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.3 | 7.7 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.3 | 2.5 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.3 | 47.5 | GO:0042113 | B cell activation(GO:0042113) |
| 0.3 | 12.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 12.8 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.2 | 21.7 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.2 | 34.7 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.2 | 5.0 | GO:0010714 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.2 | 4.0 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.1 | 4.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 2.9 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.1 | 3.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 16.5 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 24.5 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.1 | 1.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 8.9 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 1.3 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 12.7 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.9 | 59.6 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 11.6 | 92.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 10.1 | 91.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 7.2 | 65.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 7.0 | 98.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 4.8 | 105.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 3.9 | 23.7 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 2.8 | 11.3 | GO:1990005 | granular vesicle(GO:1990005) |
| 2.5 | 365.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 2.5 | 88.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 2.3 | 52.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 2.1 | 8.3 | GO:0033503 | HULC complex(GO:0033503) |
| 1.5 | 42.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 1.3 | 4.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.3 | 9.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 1.3 | 96.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 1.2 | 3.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 1.2 | 12.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 1.2 | 5.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 1.1 | 10.3 | GO:0032009 | early phagosome(GO:0032009) |
| 1.1 | 92.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 1.1 | 6.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 1.1 | 17.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.0 | 5.2 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 1.0 | 111.6 | GO:0016605 | PML body(GO:0016605) |
| 1.0 | 3.0 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 1.0 | 12.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.9 | 21.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.8 | 51.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.8 | 75.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.7 | 233.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.6 | 4.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.6 | 30.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.6 | 35.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.6 | 39.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.5 | 31.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.5 | 13.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.5 | 30.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.4 | 40.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 4.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.4 | 7.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 7.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.3 | 13.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.3 | 99.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.3 | 2.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 5.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.3 | 7.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 55.4 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.3 | 28.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.2 | 23.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.2 | 28.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.2 | 3.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 62.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 6.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 186.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.2 | 17.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 39.9 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.2 | 23.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.2 | 3.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.2 | 41.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 36.0 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.1 | 49.9 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 16.3 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 28.1 | GO:0005768 | endosome(GO:0005768) |
| 0.1 | 14.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 4.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 3.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 16.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.9 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 27.7 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 3.9 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 33.6 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 29.8 | 89.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 18.5 | 222.6 | GO:0019864 | IgG binding(GO:0019864) |
| 15.2 | 45.7 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 13.0 | 52.0 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 12.3 | 36.9 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) leukotriene-C4 synthase activity(GO:0004464) |
| 12.1 | 48.3 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) PTB domain binding(GO:0051425) |
| 11.4 | 45.7 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 10.7 | 64.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 10.3 | 92.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 9.9 | 59.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 9.4 | 37.5 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 8.6 | 86.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 8.1 | 97.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 8.1 | 201.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 7.0 | 105.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 6.0 | 48.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 5.8 | 35.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 5.7 | 45.8 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 5.4 | 26.9 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 5.0 | 49.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 5.0 | 24.8 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 4.8 | 96.8 | GO:0005537 | mannose binding(GO:0005537) |
| 4.5 | 267.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 4.2 | 92.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 3.6 | 90.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 3.5 | 17.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 3.4 | 30.8 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 3.2 | 25.6 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 3.1 | 15.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 2.7 | 21.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 2.7 | 8.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 2.6 | 60.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 2.4 | 7.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 2.2 | 11.2 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 2.0 | 24.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 1.9 | 42.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 1.6 | 9.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 1.6 | 40.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 1.5 | 11.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 1.3 | 42.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 1.3 | 54.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 1.2 | 101.9 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 1.1 | 88.1 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 1.0 | 55.0 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 1.0 | 13.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.9 | 55.0 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.9 | 17.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.9 | 8.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.8 | 7.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.7 | 70.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.7 | 22.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.7 | 12.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.7 | 17.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.6 | 14.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.6 | 31.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.5 | 20.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.5 | 3.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.5 | 11.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 23.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.4 | 101.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.4 | 65.2 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.4 | 2.9 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 43.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.4 | 87.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.4 | 60.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.3 | 9.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 7.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 13.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.3 | 50.9 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.3 | 3.9 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.3 | 12.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.3 | 3.9 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 75.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.3 | 40.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.2 | 48.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 4.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 16.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 17.3 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 8.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 4.1 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 20.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 76.0 | GO:0005102 | receptor binding(GO:0005102) |
| 0.0 | 21.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 14.7 | GO:0003682 | chromatin binding(GO:0003682) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 449.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 4.8 | 62.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 4.0 | 150.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 3.2 | 161.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 2.8 | 47.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 2.5 | 59.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 2.2 | 81.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 2.0 | 43.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 1.8 | 111.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 1.5 | 42.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 1.3 | 17.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 1.3 | 132.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 1.3 | 47.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 1.0 | 157.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.7 | 14.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.7 | 24.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.7 | 11.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.6 | 12.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.4 | 7.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.4 | 16.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.4 | 16.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 5.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.3 | 16.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.3 | 21.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.3 | 11.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.3 | 11.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 9.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.2 | 56.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 1.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 1.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 16.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.1 | 145.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 8.7 | 96.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 8.1 | 97.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 7.9 | 157.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 5.9 | 17.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 5.4 | 65.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 4.4 | 105.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 4.2 | 67.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 4.1 | 119.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 3.2 | 180.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 3.1 | 53.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 3.0 | 47.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 2.1 | 21.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 2.0 | 42.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 1.9 | 24.8 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 1.9 | 72.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 1.7 | 86.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 1.7 | 55.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 1.6 | 43.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 1.4 | 14.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 1.3 | 24.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 1.3 | 10.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 1.3 | 11.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 1.3 | 71.8 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 1.2 | 25.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 1.2 | 68.6 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 1.1 | 16.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 1.0 | 11.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.9 | 15.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.9 | 83.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.9 | 15.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.8 | 95.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.8 | 14.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.8 | 42.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.8 | 71.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.7 | 101.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.7 | 24.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.5 | 4.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.5 | 20.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.5 | 6.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.4 | 16.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.4 | 27.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.4 | 9.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 21.2 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.3 | 3.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.3 | 3.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 4.8 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.2 | 17.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 3.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 22.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 3.9 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 1.0 | REACTOME TRANSLATION | Genes involved in Translation |