Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
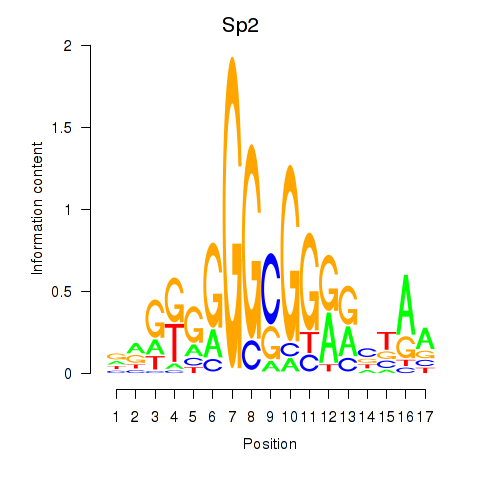
Results for Sp2
Z-value: 0.81
Transcription factors associated with Sp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sp2
|
ENSRNOG00000010492 | Sp2 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp2 | rn6_v1_chr10_-_84920886_84920886 | -0.61 | 2.4e-34 | Click! |
Activity profile of Sp2 motif
Sorted Z-values of Sp2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_80195532 | 53.95 |
ENSRNOT00000022528
|
Rtn2
|
reticulon 2 |
| chr6_+_137184820 | 31.03 |
ENSRNOT00000073796
|
Adssl1
|
adenylosuccinate synthase like 1 |
| chr13_-_90602365 | 30.65 |
ENSRNOT00000009344
|
Casq1
|
calsequestrin 1 |
| chr10_-_8498422 | 27.83 |
ENSRNOT00000082332
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr1_-_80609836 | 20.58 |
ENSRNOT00000091647
ENSRNOT00000046169 |
Apoc1
|
apolipoprotein C1 |
| chr13_+_104284660 | 20.39 |
ENSRNOT00000005400
|
Dusp10
|
dual specificity phosphatase 10 |
| chr12_+_45905371 | 20.10 |
ENSRNOT00000039275
|
Hspb8
|
heat shock protein family B (small) member 8 |
| chr15_+_34251606 | 19.87 |
ENSRNOT00000025725
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr2_-_53413638 | 18.81 |
ENSRNOT00000021081
|
Ghr
|
growth hormone receptor |
| chr1_-_198232344 | 17.52 |
ENSRNOT00000080988
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr8_-_53146953 | 16.63 |
ENSRNOT00000045356
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr8_-_25904564 | 16.60 |
ENSRNOT00000082744
ENSRNOT00000064783 |
Tbx20
|
T-box 20 |
| chr9_+_74124016 | 16.42 |
ENSRNOT00000019023
|
Cps1
|
carbamoyl-phosphate synthase 1 |
| chr1_-_215836641 | 16.19 |
ENSRNOT00000080246
|
Igf2
|
insulin-like growth factor 2 |
| chr3_+_111160205 | 15.79 |
ENSRNOT00000019392
|
Chac1
|
ChaC glutathione-specific gamma-glutamylcyclotransferase 1 |
| chr7_+_117759083 | 14.48 |
ENSRNOT00000050556
|
Gpt
|
glutamic--pyruvic transaminase |
| chr7_+_26375866 | 14.45 |
ENSRNOT00000059639
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr10_+_56445647 | 13.70 |
ENSRNOT00000056870
|
Tmem256
|
transmembrane protein 256 |
| chr18_-_28444880 | 11.93 |
ENSRNOT00000060696
|
Prob1
|
proline rich basic protein 1 |
| chrX_-_134866210 | 11.09 |
ENSRNOT00000005331
|
Apln
|
apelin |
| chr19_+_31524671 | 10.69 |
ENSRNOT00000024616
|
Hhip
|
Hedgehog-interacting protein |
| chr8_+_91464229 | 10.47 |
ENSRNOT00000013249
|
Bckdhb
|
branched chain keto acid dehydrogenase E1 subunit beta |
| chr11_-_83867203 | 10.42 |
ENSRNOT00000002394
|
Chrd
|
chordin |
| chr20_-_7397448 | 10.22 |
ENSRNOT00000059426
|
LOC294154
|
similar to chromosome 6 open reading frame 106 isoform a |
| chr20_-_29199224 | 9.72 |
ENSRNOT00000071477
|
Mcu
|
mitochondrial calcium uniporter |
| chr17_-_78735324 | 9.22 |
ENSRNOT00000036299
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr9_-_9985630 | 9.22 |
ENSRNOT00000071780
|
Crb3
|
crumbs 3, cell polarity complex component |
| chr19_+_37226186 | 8.90 |
ENSRNOT00000075933
ENSRNOT00000065013 |
Hsf4
|
heat shock transcription factor 4 |
| chr9_-_17698569 | 8.89 |
ENSRNOT00000087528
|
Mrpl14
|
mitochondrial ribosomal protein L14 |
| chr9_+_65478496 | 8.77 |
ENSRNOT00000016060
|
Ndufb3
|
NADH:ubiquinone oxidoreductase subunit B3 |
| chr3_-_163935617 | 8.76 |
ENSRNOT00000074023
|
Kcnb1
|
potassium voltage-gated channel subfamily B member 1 |
| chr10_-_97896525 | 8.28 |
ENSRNOT00000005172
|
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr2_+_95045034 | 8.17 |
ENSRNOT00000081305
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr9_-_99651813 | 7.66 |
ENSRNOT00000022089
|
Ndufa10
|
NADH:ubiquinone oxidoreductase subunit A10 |
| chr19_-_38321528 | 7.62 |
ENSRNOT00000031977
|
Smpd3
|
sphingomyelin phosphodiesterase 3 |
| chrX_-_142131545 | 7.51 |
ENSRNOT00000077402
|
Fgf13
|
fibroblast growth factor 13 |
| chr1_-_64099277 | 7.24 |
ENSRNOT00000084846
|
Tsen34
|
tRNA splicing endonuclease subunit 34 |
| chr3_+_80362858 | 7.02 |
ENSRNOT00000021353
ENSRNOT00000086242 |
Lrp4
|
LDL receptor related protein 4 |
| chr13_+_52588917 | 6.82 |
ENSRNOT00000011999
|
Phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr15_-_3435888 | 6.74 |
ENSRNOT00000016709
|
Adk
|
adenosine kinase |
| chr1_+_141832774 | 6.65 |
ENSRNOT00000073302
|
Fuom
|
fucose mutarotase |
| chr10_-_16689321 | 6.64 |
ENSRNOT00000028173
|
Bnip1
|
BCL2/adenovirus E1B interacting protein 1 |
| chr10_-_89084885 | 6.55 |
ENSRNOT00000027452
|
Plekhh3
|
pleckstrin homology, MyTH4 and FERM domain containing H3 |
| chrX_-_34794589 | 6.34 |
ENSRNOT00000008703
|
Rai2
|
retinoic acid induced 2 |
| chr20_-_5533448 | 6.33 |
ENSRNOT00000000568
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr8_-_85720790 | 6.32 |
ENSRNOT00000071049
|
Ooep
|
oocyte expressed protein |
| chr8_-_22937909 | 6.21 |
ENSRNOT00000015684
|
Tmem205
|
transmembrane protein 205 |
| chr13_-_91872954 | 6.16 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr6_-_55647665 | 5.91 |
ENSRNOT00000007414
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr1_+_101405314 | 5.85 |
ENSRNOT00000028194
|
Ntf4
|
neurotrophin 4 |
| chr1_-_64030175 | 5.82 |
ENSRNOT00000089950
|
Tsen34l1
|
tRNA splicing endonuclease subunit 34-like 1 |
| chr15_+_344360 | 5.70 |
ENSRNOT00000077671
|
Kcnma1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr15_+_344685 | 5.69 |
ENSRNOT00000065542
ENSRNOT00000066928 |
Kcnma1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr1_+_126523169 | 5.67 |
ENSRNOT00000051000
|
Tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr1_+_238843338 | 5.56 |
ENSRNOT00000024583
|
Zfand5
|
zinc finger AN1-type containing 5 |
| chr17_-_55709740 | 5.52 |
ENSRNOT00000033359
|
RGD1562037
|
similar to OTTHUMP00000046255 |
| chr8_+_36625733 | 5.47 |
ENSRNOT00000016248
|
Cdon
|
cell adhesion associated, oncogene regulated |
| chr1_-_241460868 | 5.30 |
ENSRNOT00000054776
|
RGD1560242
|
similar to RIKEN cDNA 1700028P14 |
| chr2_+_205592182 | 5.29 |
ENSRNOT00000081050
|
Dennd2c
|
DENN domain containing 2C |
| chr4_-_113610243 | 5.21 |
ENSRNOT00000008813
|
Hk2
|
hexokinase 2 |
| chrX_+_157150655 | 5.15 |
ENSRNOT00000090795
|
Pnck
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr1_-_54748763 | 5.10 |
ENSRNOT00000074549
|
LOC100911027
|
protein MAL2-like |
| chr3_+_172385672 | 5.09 |
ENSRNOT00000090989
|
Gnas
|
GNAS complex locus |
| chr10_+_12929471 | 5.06 |
ENSRNOT00000036563
|
Zscan10
|
zinc finger and SCAN domain containing 10 |
| chr2_+_188844073 | 5.06 |
ENSRNOT00000028117
|
Kcnn3
|
potassium calcium-activated channel subfamily N member 3 |
| chr7_+_94130852 | 5.00 |
ENSRNOT00000011485
|
Mal2
|
mal, T-cell differentiation protein 2 |
| chr1_-_192025350 | 4.65 |
ENSRNOT00000071946
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr7_-_77162148 | 4.58 |
ENSRNOT00000008350
|
Klf10
|
Kruppel-like factor 10 |
| chr13_+_103300932 | 4.57 |
ENSRNOT00000085214
ENSRNOT00000038264 |
Eprs
|
glutamyl-prolyl-tRNA synthetase |
| chr8_-_111850393 | 4.56 |
ENSRNOT00000044956
|
Cdv3
|
CDV3 homolog |
| chr1_-_264756546 | 4.53 |
ENSRNOT00000020020
|
Mrpl43
|
mitochondrial ribosomal protein L43 |
| chr4_-_152380184 | 4.52 |
ENSRNOT00000091473
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr7_-_98709649 | 4.48 |
ENSRNOT00000077440
|
Tmem65
|
transmembrane protein 65 |
| chr8_+_115155470 | 4.46 |
ENSRNOT00000017139
|
Pcbp4
|
poly(rC) binding protein 4 |
| chr6_-_55001464 | 4.43 |
ENSRNOT00000006618
|
Ahr
|
aryl hydrocarbon receptor |
| chr7_+_2752680 | 4.32 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr8_-_127782070 | 4.20 |
ENSRNOT00000045493
|
Plcd1
|
phospholipase C, delta 1 |
| chr12_-_25638797 | 4.18 |
ENSRNOT00000002033
|
Gatsl2
|
GATS protein-like 2 |
| chr3_+_151032952 | 4.17 |
ENSRNOT00000064013
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr1_-_101351879 | 4.16 |
ENSRNOT00000028142
|
Ppfia3
|
PTPRF interacting protein alpha 3 |
| chr16_+_19051965 | 4.04 |
ENSRNOT00000016399
|
Slc35e1
|
solute carrier family 35, member E1 |
| chr9_-_9702306 | 3.93 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr13_+_48607308 | 3.89 |
ENSRNOT00000063882
|
Slc41a1
|
solute carrier family 41 member 1 |
| chrX_-_10630281 | 3.89 |
ENSRNOT00000080651
ENSRNOT00000048980 |
Usp9x
|
ubiquitin specific peptidase 9, X-linked |
| chrX_+_20484517 | 3.88 |
ENSRNOT00000093701
|
FAM120C
|
family with sequence similarity 120C |
| chr2_+_187322416 | 3.88 |
ENSRNOT00000025183
|
Crabp2
|
cellular retinoic acid binding protein 2 |
| chr9_+_43889473 | 3.82 |
ENSRNOT00000024330
|
Inpp4a
|
inositol polyphosphate-4-phosphatase type I A |
| chr10_+_46783979 | 3.74 |
ENSRNOT00000005237
|
Gid4
|
GID complex subunit 4 |
| chr16_-_71809925 | 3.70 |
ENSRNOT00000022425
|
Tm2d2
|
TM2 domain containing 2 |
| chr6_-_72462657 | 3.61 |
ENSRNOT00000088720
ENSRNOT00000077578 |
Strn3
|
striatin 3 |
| chr1_-_221420115 | 3.60 |
ENSRNOT00000028475
|
Mrpl49
|
mitochondrial ribosomal protein L49 |
| chr2_+_233615739 | 3.59 |
ENSRNOT00000051009
|
Pitx2
|
paired-like homeodomain 2 |
| chr1_+_219759183 | 3.42 |
ENSRNOT00000026316
|
Pc
|
pyruvate carboxylase |
| chr16_+_71810377 | 3.42 |
ENSRNOT00000041330
|
Adam9
|
ADAM metallopeptidase domain 9 |
| chr10_-_59743315 | 3.41 |
ENSRNOT00000093646
|
Emc6
|
ER membrane protein complex subunit 6 |
| chr5_-_151768123 | 3.38 |
ENSRNOT00000079380
|
Nudc
|
nuclear distribution C, dynein complex regulator |
| chr5_+_136406308 | 3.35 |
ENSRNOT00000072492
|
Eri3
|
ERI1 exoribonuclease family member 3 |
| chr10_-_86004096 | 3.32 |
ENSRNOT00000091978
ENSRNOT00000066855 |
Stac2
|
SH3 and cysteine rich domain 2 |
| chr1_+_264756499 | 3.32 |
ENSRNOT00000031018
|
Peo1
|
progressive external ophthalmoplegia 1 |
| chr8_+_64364741 | 3.27 |
ENSRNOT00000082840
|
Celf6
|
CUGBP, Elav-like family member 6 |
| chr17_+_82065937 | 3.21 |
ENSRNOT00000051349
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chr7_+_144531814 | 3.20 |
ENSRNOT00000033300
|
Hoxc13
|
homeobox C13 |
| chr4_+_77211692 | 3.10 |
ENSRNOT00000007620
|
Cul1
|
cullin 1 |
| chr5_+_136406130 | 3.10 |
ENSRNOT00000093099
|
Eri3
|
ERI1 exoribonuclease family member 3 |
| chr9_+_45672157 | 3.05 |
ENSRNOT00000017882
|
Pdcl3
|
phosducin-like 3 |
| chr17_+_49610548 | 3.01 |
ENSRNOT00000017955
|
Yae1d1
|
Yae1 domain containing 1 |
| chr1_+_192025357 | 2.97 |
ENSRNOT00000025072
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr1_+_249574954 | 2.94 |
ENSRNOT00000074100
|
Cstf2t
|
cleavage stimulation factor subunit 2, tau variant |
| chr8_-_1450138 | 2.91 |
ENSRNOT00000008062
|
Aasdhppt
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr8_-_111850075 | 2.87 |
ENSRNOT00000082097
|
Cdv3
|
CDV3 homolog |
| chr1_-_198120061 | 2.87 |
ENSRNOT00000026231
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr6_+_137243185 | 2.87 |
ENSRNOT00000030879
|
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr5_-_151397603 | 2.83 |
ENSRNOT00000076866
|
Gpr3
|
G protein-coupled receptor 3 |
| chr5_+_171472273 | 2.81 |
ENSRNOT00000055386
|
LOC100911486
|
multiple epidermal growth factor-like domains protein 6-like |
| chr4_+_342302 | 2.81 |
ENSRNOT00000009233
|
Insig1
|
insulin induced gene 1 |
| chr12_+_38274297 | 2.80 |
ENSRNOT00000087905
ENSRNOT00000057788 |
Rsrc2
|
arginine and serine rich coiled-coil 2 |
| chr2_+_211050360 | 2.79 |
ENSRNOT00000026928
|
Psma5
|
proteasome subunit alpha 5 |
| chr1_-_218810118 | 2.78 |
ENSRNOT00000065950
ENSRNOT00000020886 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr6_+_92568975 | 2.78 |
ENSRNOT00000036933
|
Abhd12b
|
abhydrolase domain containing 12B |
| chr1_+_165625058 | 2.76 |
ENSRNOT00000025104
|
Rab6a
|
RAB6A, member RAS oncogene family |
| chr15_-_33775109 | 2.70 |
ENSRNOT00000033722
|
Jph4
|
junctophilin 4 |
| chr16_-_7758189 | 2.70 |
ENSRNOT00000026588
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr1_-_229639187 | 2.65 |
ENSRNOT00000016804
|
Zfp91
|
zinc finger protein 91 |
| chr8_+_132441285 | 2.65 |
ENSRNOT00000087488
ENSRNOT00000068233 |
Lars2
|
leucyl-tRNA synthetase 2 |
| chrX_-_154722220 | 2.62 |
ENSRNOT00000089743
ENSRNOT00000087893 |
Fmr1
|
fragile X mental retardation 1 |
| chr5_-_165918445 | 2.61 |
ENSRNOT00000018167
|
Pex14
|
peroxisomal biogenesis factor 14 |
| chr19_-_43149210 | 2.58 |
ENSRNOT00000038452
|
Pdpr
|
pyruvate dehydrogenase phosphatase regulatory subunit |
| chr17_+_9830332 | 2.57 |
ENSRNOT00000021946
|
Rab24
|
RAB24, member RAS oncogene family |
| chr20_-_10680283 | 2.55 |
ENSRNOT00000001579
|
Sik1
|
salt-inducible kinase 1 |
| chr2_+_218951451 | 2.51 |
ENSRNOT00000019190
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr6_+_104718512 | 2.31 |
ENSRNOT00000007947
|
Smoc1
|
SPARC related modular calcium binding 1 |
| chr13_+_100817359 | 2.27 |
ENSRNOT00000004330
|
Tp53bp2
|
tumor protein p53 binding protein, 2 |
| chrX_-_20070537 | 2.25 |
ENSRNOT00000093602
ENSRNOT00000003397 |
Gnl3l
|
G protein nucleolar 3 like |
| chr8_+_12994155 | 2.25 |
ENSRNOT00000011855
|
Cwc15
|
CWC15 spliceosome-associated protein |
| chr3_+_134413170 | 2.25 |
ENSRNOT00000074338
|
AABR07054000.1
|
|
| chr2_-_187786700 | 2.20 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr8_+_116054465 | 2.19 |
ENSRNOT00000040056
|
Cish
|
cytokine inducible SH2-containing protein |
| chr5_-_172623899 | 2.17 |
ENSRNOT00000080591
|
Ski
|
SKI proto-oncogene |
| chr13_+_76942928 | 2.12 |
ENSRNOT00000040505
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr10_-_88307132 | 2.11 |
ENSRNOT00000040845
|
Jup
|
junction plakoglobin |
| chr19_+_55197704 | 2.05 |
ENSRNOT00000045657
|
Zc3h18
|
zinc finger CCCH-type containing 18 |
| chr12_+_22259713 | 2.00 |
ENSRNOT00000001913
|
Pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr1_+_221710670 | 1.89 |
ENSRNOT00000064798
|
Map4k2
|
mitogen activated protein kinase kinase kinase kinase 2 |
| chr10_-_88307412 | 1.86 |
ENSRNOT00000089933
|
Jup
|
junction plakoglobin |
| chr5_+_106409456 | 1.83 |
ENSRNOT00000058387
|
Focad
|
focadhesin |
| chrX_+_156863754 | 1.81 |
ENSRNOT00000083611
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr13_-_35048444 | 1.80 |
ENSRNOT00000009963
|
Gli2
|
GLI family zinc finger 2 |
| chr8_-_49075892 | 1.79 |
ENSRNOT00000082687
|
Arcn1
|
archain 1 |
| chr1_-_57489727 | 1.75 |
ENSRNOT00000002037
|
Psmb1
|
proteasome subunit beta 1 |
| chr10_-_59743037 | 1.75 |
ENSRNOT00000026168
|
Emc6
|
ER membrane protein complex subunit 6 |
| chr17_-_67904674 | 1.74 |
ENSRNOT00000078532
|
Klf6
|
Kruppel-like factor 6 |
| chr15_+_106257650 | 1.73 |
ENSRNOT00000014859
ENSRNOT00000072538 |
Ipo5
|
importin 5 |
| chr1_-_100559942 | 1.73 |
ENSRNOT00000026862
|
Nr1h2
|
nuclear receptor subfamily 1, group H, member 2 |
| chr13_+_76943633 | 1.68 |
ENSRNOT00000076908
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr20_+_11972381 | 1.66 |
ENSRNOT00000001642
|
Adarb1
|
adenosine deaminase, RNA-specific, B1 |
| chr2_+_218951141 | 1.62 |
ENSRNOT00000091001
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr3_+_2676296 | 1.61 |
ENSRNOT00000020497
|
Clic3
|
chloride intracellular channel 3 |
| chr16_+_36116258 | 1.57 |
ENSRNOT00000017652
|
Sap30
|
Sin3A associated protein 30 |
| chr11_+_46009322 | 1.57 |
ENSRNOT00000002233
|
Tmem45a
|
transmembrane protein 45A |
| chr14_-_10749120 | 1.55 |
ENSRNOT00000003046
|
Cops4
|
COP9 signalosome subunit 4 |
| chr3_+_176989672 | 1.51 |
ENSRNOT00000020373
|
Tpd52l2
|
tumor protein D52-like 2 |
| chr20_-_3822754 | 1.51 |
ENSRNOT00000000541
ENSRNOT00000077357 |
Slc39a7
|
solute carrier family 39 member 7 |
| chr9_-_82514399 | 1.49 |
ENSRNOT00000026799
|
Dnpep
|
aspartyl aminopeptidase |
| chrX_-_157139291 | 1.46 |
ENSRNOT00000092123
|
Slc6a8
|
solute carrier family 6 member 8 |
| chr10_+_56591292 | 1.44 |
ENSRNOT00000023379
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1 |
| chr15_-_93667355 | 1.44 |
ENSRNOT00000093571
ENSRNOT00000089876 |
Fbxl3
|
F-box and leucine-rich repeat protein 3 |
| chr10_-_14324170 | 1.36 |
ENSRNOT00000035513
ENSRNOT00000090587 |
Hn1l
|
hematological and neurological expressed 1-like |
| chr3_-_60795951 | 1.32 |
ENSRNOT00000002174
|
Atf2
|
activating transcription factor 2 |
| chr5_+_58995249 | 1.31 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr7_-_143863186 | 1.30 |
ENSRNOT00000017096
|
Rarg
|
retinoic acid receptor, gamma |
| chr2_+_22427474 | 1.27 |
ENSRNOT00000061761
|
Mtx3
|
metaxin 3 |
| chr7_+_41114697 | 1.27 |
ENSRNOT00000041354
|
Atp2b1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr4_-_82141385 | 1.24 |
ENSRNOT00000008447
|
Hoxa3
|
homeobox A3 |
| chr1_+_193076430 | 1.24 |
ENSRNOT00000086492
ENSRNOT00000017885 |
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr17_+_82066152 | 1.22 |
ENSRNOT00000083034
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chr1_+_226742343 | 1.21 |
ENSRNOT00000074079
|
NEWGENE_1309258
|
VPS37C, ESCRT-I subunit |
| chr18_-_6587080 | 1.20 |
ENSRNOT00000040815
|
LOC103694404
|
60S ribosomal protein L39 |
| chr10_-_54467956 | 1.19 |
ENSRNOT00000065383
|
Usp43
|
ubiquitin specific peptidase 43 |
| chr4_+_180887182 | 1.17 |
ENSRNOT00000042270
ENSRNOT00000002476 |
Fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr3_+_56861396 | 1.13 |
ENSRNOT00000000008
ENSRNOT00000084375 |
Gad1
|
glutamate decarboxylase 1 |
| chr17_+_86199623 | 1.12 |
ENSRNOT00000022727
|
Ptf1a
|
pancreas specific transcription factor, 1a |
| chr17_-_18421861 | 1.11 |
ENSRNOT00000060500
ENSRNOT00000036560 |
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr1_+_225163391 | 1.10 |
ENSRNOT00000027305
|
Eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chrX_-_152641679 | 1.10 |
ENSRNOT00000080277
|
Gabra3
|
gamma-aminobutyric acid type A receptor alpha3 subunit |
| chr5_-_33182147 | 1.05 |
ENSRNOT00000080358
|
Maged2
|
MAGE family member D2 |
| chr18_+_65155685 | 1.01 |
ENSRNOT00000081797
|
Tcf4
|
transcription factor 4 |
| chr3_+_33641616 | 0.99 |
ENSRNOT00000051953
|
Epc2
|
enhancer of polycomb homolog 2 |
| chr1_+_198120099 | 0.97 |
ENSRNOT00000073652
|
Bola2
|
bolA family member 2 |
| chr14_+_44479614 | 0.94 |
ENSRNOT00000003691
|
Ugdh
|
UDP-glucose 6-dehydrogenase |
| chr4_+_153217782 | 0.83 |
ENSRNOT00000015499
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr12_+_37923528 | 0.82 |
ENSRNOT00000045223
|
Abcb9
|
ATP binding cassette subfamily B member 9 |
| chr1_-_214511529 | 0.78 |
ENSRNOT00000026390
|
Chid1
|
chitinase domain containing 1 |
| chr19_-_25955371 | 0.74 |
ENSRNOT00000004042
ENSRNOT00000084123 |
Rad23a
|
RAD23 homolog A, nucleotide excision repair protein |
| chr10_-_46783889 | 0.70 |
ENSRNOT00000004948
|
Atpaf2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr16_-_14210449 | 0.67 |
ENSRNOT00000016494
|
Ccser2
|
coiled-coil serine-rich protein 2 |
| chr5_+_156668712 | 0.66 |
ENSRNOT00000067228
|
Ddost
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase non-catalytic subunit |
| chr10_-_82326771 | 0.66 |
ENSRNOT00000004673
|
Acsf2
|
acyl-CoA synthetase family member 2 |
| chr5_+_136112417 | 0.61 |
ENSRNOT00000025990
|
Tmem53
|
transmembrane protein 53 |
| chr5_+_47853818 | 0.59 |
ENSRNOT00000009228
ENSRNOT00000079656 |
Casp8ap2
|
caspase 8 associated protein 2 |
| chr1_-_92119951 | 0.51 |
ENSRNOT00000018153
ENSRNOT00000092121 |
Zfp507
|
zinc finger protein 507 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sp2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.3 | 31.0 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 7.7 | 30.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 6.7 | 20.2 | GO:0060577 | pulmonary vein morphogenesis(GO:0060577) |
| 5.1 | 20.6 | GO:0010916 | regulation of phosphatidylcholine catabolic process(GO:0010899) regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 5.1 | 20.4 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 4.8 | 14.4 | GO:0042560 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 3.8 | 11.4 | GO:0060082 | eye blink reflex(GO:0060082) |
| 3.6 | 14.5 | GO:0042853 | L-alanine catabolic process(GO:0042853) |
| 3.4 | 10.2 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 3.3 | 16.6 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 3.3 | 16.4 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 3.3 | 13.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 2.8 | 19.9 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 2.8 | 11.1 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 2.2 | 6.7 | GO:0044209 | purine deoxyribonucleotide biosynthetic process(GO:0009153) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) AMP salvage(GO:0044209) adenosine biosynthetic process(GO:0046086) |
| 2.2 | 13.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 2.1 | 18.8 | GO:0006600 | allantoin metabolic process(GO:0000255) isoleucine metabolic process(GO:0006549) creatine metabolic process(GO:0006600) creatinine metabolic process(GO:0046449) |
| 2.1 | 10.4 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 2.0 | 15.8 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 2.0 | 13.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.8 | 7.0 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 1.7 | 5.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 1.7 | 6.7 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 1.5 | 5.9 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 1.4 | 4.2 | GO:0019541 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 1.4 | 4.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 1.3 | 5.2 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 1.3 | 3.9 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 1.3 | 7.6 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 1.3 | 17.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.2 | 27.8 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 1.1 | 3.4 | GO:0051088 | positive regulation of macrophage fusion(GO:0034241) PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 1.1 | 10.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 1.1 | 4.4 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 1.1 | 5.5 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 1.1 | 7.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.0 | 9.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.9 | 4.6 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.9 | 56.6 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) |
| 0.9 | 6.5 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.9 | 4.5 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.9 | 2.6 | GO:1905244 | negative regulation of long term synaptic depression(GO:1900453) regulation of intracellular transport of viral material(GO:1901252) regulation of modification of synaptic structure(GO:1905244) |
| 0.9 | 10.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.9 | 3.5 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.9 | 3.4 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.8 | 4.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.7 | 2.9 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.7 | 2.8 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.6 | 3.9 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.6 | 1.8 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.6 | 2.9 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.5 | 2.7 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.5 | 7.5 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.5 | 3.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.5 | 2.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.4 | 1.3 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.4 | 2.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.4 | 1.3 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.4 | 5.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 1.2 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.4 | 3.2 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.4 | 4.3 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.4 | 3.0 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.4 | 10.7 | GO:0045879 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.4 | 2.9 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.4 | 2.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.3 | 1.7 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.3 | 7.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.3 | 0.9 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.3 | 2.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 3.9 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.3 | 5.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 1.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 3.4 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.2 | 1.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 4.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 3.9 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.2 | 1.7 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.2 | 3.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 1.3 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.2 | 1.0 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.2 | 5.6 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.2 | 2.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 4.5 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.2 | 1.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.2 | 1.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 2.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 0.7 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.2 | 4.6 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.2 | 1.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 6.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 8.9 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 6.8 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 2.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 1.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 6.3 | GO:0035088 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.1 | 2.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.6 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 5.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.4 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 6.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 3.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.3 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.1 | 0.3 | GO:0032258 | CVT pathway(GO:0032258) |
| 0.1 | 0.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.8 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 2.8 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.1 | 0.8 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.1 | 1.0 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 | 0.4 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 1.9 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 2.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.9 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.1 | 5.1 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 3.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 1.3 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.5 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 2.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.5 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 1.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 2.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 3.9 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.7 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 2.2 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.2 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 1.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 2.9 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 30.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 3.8 | 18.8 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 2.6 | 13.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 2.1 | 10.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 1.6 | 9.7 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.6 | 45.2 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 1.3 | 7.9 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 1.1 | 20.6 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 1.0 | 5.2 | GO:1903349 | omegasome membrane(GO:1903349) omegasome(GO:1990462) |
| 1.0 | 17.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 1.0 | 4.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 1.0 | 2.9 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.9 | 2.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.9 | 9.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.9 | 4.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.9 | 2.6 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.9 | 2.6 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.8 | 8.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.7 | 10.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.6 | 7.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.6 | 8.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.6 | 3.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.4 | 19.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.4 | 4.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.3 | 2.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 16.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 2.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.3 | 17.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.3 | 8.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.3 | 0.8 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.2 | 1.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 10.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 2.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 19.4 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 39.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 3.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 3.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 6.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 17.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.2 | 1.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.9 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 3.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 5.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 5.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 5.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 4.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 7.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 21.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 10.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 15.5 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 2.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 45.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 6.3 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.7 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 12.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0014704 | intercalated disc(GO:0014704) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.5 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 4.7 | 18.8 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 3.9 | 15.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 3.4 | 10.2 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 3.2 | 9.7 | GO:0015292 | uniporter activity(GO:0015292) |
| 2.8 | 11.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 2.7 | 10.7 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 2.6 | 13.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 2.5 | 17.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 2.3 | 20.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 1.8 | 14.4 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 1.7 | 6.7 | GO:0042806 | fucose binding(GO:0042806) |
| 1.5 | 4.4 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 1.4 | 4.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 1.4 | 16.4 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 1.3 | 10.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.3 | 20.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 1.3 | 3.8 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 1.2 | 16.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 1.2 | 10.5 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 1.1 | 3.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.1 | 7.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 1.0 | 4.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.0 | 21.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.9 | 2.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.9 | 5.2 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.9 | 4.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.8 | 10.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.7 | 5.9 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.7 | 5.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.7 | 31.0 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.6 | 2.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.6 | 15.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.5 | 21.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.4 | 6.7 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.4 | 8.8 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.4 | 1.7 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.4 | 3.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.4 | 7.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 2.9 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.4 | 1.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 3.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.4 | 2.6 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.4 | 3.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 1.7 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.3 | 1.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 4.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 1.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 0.9 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.3 | 2.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.3 | 11.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.3 | 4.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 3.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 1.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 7.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 1.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 3.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 3.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 2.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 8.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 2.6 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 4.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 0.8 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.2 | 6.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 5.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 2.9 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 1.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 2.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 11.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 3.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 22.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 2.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 2.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 2.8 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.6 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 4.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 9.2 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 1.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 15.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 3.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 6.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 22.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 7.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 2.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 1.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.9 | GO:0051287 | NAD binding(GO:0051287) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 18.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.5 | 20.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 19.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.3 | 21.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 12.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 2.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.2 | 10.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 4.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 3.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 7.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 4.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 6.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 3.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 6.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 6.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 3.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 31.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.0 | 21.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.8 | 14.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.8 | 16.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.6 | 10.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.6 | 20.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 4.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 6.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 7.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.3 | 16.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 4.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 2.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 10.6 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.2 | 5.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 7.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 2.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 4.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 6.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 8.2 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 1.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 17.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 5.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 4.0 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 2.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 5.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 14.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 0.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 5.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 3.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.4 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.7 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |